Back to Journals » Cancer Management and Research » Volume 12
circ_0136666 Facilitates the Progression of Colorectal Cancer via miR-383/CREB1 Axis
Authors Li Y, Zang H, Zhang X, Huang G
Received 29 February 2020
Accepted for publication 9 July 2020
Published 4 August 2020 Volume 2020:12 Pages 6795—6806
DOI https://doi.org/10.2147/CMAR.S251952
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Dr Ahmet Emre Eşkazan
Yuhui Li, Hongliang Zang, Xue Zhang, Guomin Huang
Department of General Surgery, China-Japan Union Hospital of Jilin University, Changchun, Jilin, People’s Republic of China
Correspondence: Guomin Huang
Department of General Surgery, China-Japan Union Hospital of Jilin University, No. 829 Xinmin Avenue, Changchun, Jilin 130012, People’s Republic of China
Tel +86-13504426968
Email [email protected]
Background: The changes in dietary patterns cause an increased incidence of colorectal cancer (CRC) globally. We aimed to explore the mechanism behind circular RNA circ_0136666 in the progression of CRC.
Materials and Methods: The expression of circ_0136666, miR-383 and cAMP response element binding protein 1 (CREB1) was detected by real-time quantitative polymerase chain reaction (RT-qPCR). Cell proliferation, apoptosis and glycolysis were measured by 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), flow cytometry and glucose or lactate detection kit, respectively. The combination between miR-383 and circ_0136666 or CREB1 in 293T cells was predicted by Circular RNA Interactome or Starbase software and confirmed by dual-luciferase reporter assay. Western blot assay was performed to detect the abundance of CREB1, hexokinase 2 (HK2) and lactate dehydrogenase A (LDHA) in CRC cells. Murine xenograft model was established to verify the function of circ_0136666 in vivo.
Results: circ_0136666 was aberrantly up-regulated in CRC tissues and cells, and it promoted the proliferation and glycolysis and inhibited the apoptosis of CRC cells. circ_0136666 accelerated the progression of CRC through directly targeting and down-regulating miR-383. CREB1 could bind to miR-383 in 293T cells. The overexpression of CREB1 reversed the inhibitory effects of miR-383 accumulation on the proliferation and glycolysis and the promoting impact on the apoptosis of CRC cells. The enrichment of CREB1 was modulated by circ_0136666/miR-383 signaling in CRC cells. The glycolysis-related proteins (HK2 and LDHA) were modulated by circ_0136666/miR-383/CREB1 axis in CRC cells. circ_0136666 accelerated the growth of CRC tumors via circ_0136666/miR-383/CREB1 axis in vivo.
Conclusion: circ_0136666 deteriorated CRC through miR-383/CREB1 axis. circ_0136666/miR-383/CREB1 axis might be an underlying therapeutic target for CRC therapy.
Keywords: colorectal cancer, circ_0136666, miR-383, CREB1, proliferation, apoptosis, glycolysis
Introduction
There were 551,269 deaths of colorectal cancer (CRC) globally in 2018, accounting for 5.8% of all cancers.1 The therapeutic strategies for early-stage CRC patients were surgical resection, chemotherapy and radiotherapy. Nevertheless, the prognosis of CRC patients remains dismal due to the metastasis and recurrence of CRC tumors.2,3 Therefore, it is urgent to investigate the pathogenesis and explore effective markers for CRC treatment.
Circular RNAs (circRNAs) are a class of non-coding RNAs (ncRNAs), and they are characterized by the covalently closed loop structure. circRNAs have been reported to participate in the pathogenesis of many diseases and cancers.4–7 circRNAs could serve as microRNAs (miRNAs) sponges and down-regulate the levels of target miRNAs.8 circ_0136666 is generated from protein kinase, DNA-activated, catalytic subunit (PRKDC) gene, which is involved in the process of DNA damage repair.9,10 circ_0136666 was implicated in the progression of CRC, and it accelerated the development of CRC through miR-136/SH2B1 signaling.11 Herein, we intended to uncover the signal network behind circ_0136666 in the modulation of CRC progression.
miRNAs are involved in the growth, migration, invasion and apoptosis of cancer cells through negatively modulating the levels of their target messenger RNAs (mRNAs) or inhibiting their translation.12–15 Cui et al claimed that miR-383 suppressed the development of CRC through targeting APRIL.16 Li et al reported that miR-383 was a tumor suppressor in CRC, and it inhibited the progression of CRC through regulating the abundance of CREPT/RPRD1B.17 However, the molecular mechanism of miR-383 in CRC is not fully addressed.
The cAMP response element binding protein 1 (CREB1) gene encodes a pivotal transcription factor that modulates a variety of stress signals and growth factors.18,19 CREB1 has been conformed to function as an oncogene in multiple cancers,20 including CRC. For instance, Lu et al found that circ_0079993 promoted the progression of CRC through miR-203a-3p.1/CREB1 axis.21 Yang et al reported that miR-101 restrained the proliferation and migration of CRC cells through down-regulating CREB1.22 Nevertheless, the underlying mechanism by which CREB1 accelerating carcinogenesis of CRC is largely unknown.
In this study, we analyzed the expression of circ_0136666 in CRC tissues and cells, and we investigated the molecular mechanism by which circ_0136666 accelerating the progression of CRC.
Materials and Methods
Patients
A total of 37 pairs of CRC tumor tissues and adjacent normal tissues were obtained from patients in China-Japan Union Hospital of Jilin University. The tissues were frozen in liquid nitrogen immediately and stored at −80°C. We have received written informed consents from all patients, and this study was approved by the ethics committee of China-Japan Union Hospital of Jilin University. The correlation between circ_0136666 expression and clinical clinicopathological parameters of CRC patients is displayed in Table 1.
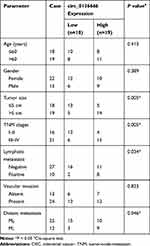 |
Table 1 Correlation Between circ_0136666 Expression and Clinical Clinicopathological Parameters of CRC Patients |
Cell Culture
Human normal colon epithelial cell line NCM460, CRC cell lines SW480 and LOVO and human embryonic kidney cell line 293T were purchased from Bena Culture Collection (Beijing, China). All cells were cultured in Dulbecco’s Modified Eagle Medium (DMEM; Gibco, Carlsbad, CA, USA) added with 10% fetal bovine serum (FBS; Gibco), 100 units/mL penicillin and 100 μg/mL streptomycin in a 37°C, 5% CO2 humidified incubator.
Real-Time Quantitative Polymerase Chain Reaction (RT-qPCR)
Total RNA from tissues and cells was isolated using TRIzol solution (Invitrogen, Carlsbad, CA, USA). Complementary DNA (cDNA) was acquired using a High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA). SYBR Green PCR Master Mix (Applied Biosystems) was utilized for PCR with the ABI 7900 thermocycler (Applied Biosystems), and U6 (for miR-383) or β-actin (circ_0136666, PRKDC or CREB1) served as the internal control. The abundance of circ_0136666, PRKDC, miR-383 and CREB1 was analyzed by 2−ΔΔCt method.23 The primer sequences were as follows: circ_0136666 (Forward, 5ʹ-TGAACACCTGGACAAACAGA-3ʹ; Reverse, 5ʹ-CAGCTCACCAGCCAATCGTC-3ʹ), PRKDC (Forward, 5ʹ-CCTGGGGCAGGAATGCGTCC-3ʹ; Reverse, 5ʹ-CCCATTTTTTCTAAGAAAAT-3ʹ), miR-383 (Forward, 5ʹ-CACGAAAGATCAGAAGGTGATTG-3ʹ; Reverse, universal reverse primer), CREB1 (Forward, 5ʹ-CTGCCTCTGGAGACGTACAA-3ʹ; Reverse, 5ʹ-CAAGCACTGCCACTCTGTTT-3ʹ), U6 (Forward, 5ʹ-CTCGCTTCGGCAGCACA-3ʹ; Reverse, 5ʹ-AACGCTTCACGAATTTGCGT-3ʹ), β-actin (Forward, 5ʹ-AGCCTCGCCTTTGCCGA-3ʹ; Reverse, 5ʹ-CTGGTGCCTGGGGCG-3ʹ).
Cell Transfection
Lipofectamine 3000 (Invitrogen) was used to conduct transfection. Small interfering RNA negative control (si-NC), circ_0136666 specific siRNA (si-circ_0136666), pcDNA empty vector (pcDNA-Control), CREB1 overexpression plasmid (pcDNA-CREB1), circ_0136666 specific short hairpin RNA (sh-circ_0136666) and sh-NC were obtained from Genepharma (Shanghai, China). miR-383 inhibitor and its control (inhibitor NC), miR-383 mimic and its control (miRNA NC) were synthesized from Ribobio (Guangzhou, China). The specific siRNA sequences were listed as below. Si-NC (5ʹ-3ʹ): UUCUCCGAACGUGUCACGUTT, si-circ_0136666 (5ʹ-3ʹ): ACAAAGAGACUGUUUUCAGCA.
3-(4,5-Dimethylthiazol-2-Yl)-2,5-Diphenyltetrazolium Bromide (MTT) Assay
The proliferation of CRC cells was detected by using MTT assay. CRC cells were plated in 96-well cell culture plates overnight. After transfection for 0 h, 24 h, 48 h or 72 h, 10 µL MTT reagent (Invitrogen) was pipetted into each well of 96-well plates and incubated for a further 4 h. The optical density at 490 nm was detected by a microplate reader.
Cell Apoptosis Analysis
Transfected CRC cells were resuspended using phosphate buffer saline (PBS). The CRC cells were stained with Annexin V combined fluorescein isothiocyanate (FITC) and propidine iodide (PI; Solarbio, Beijing, China) in dark. After washing with PBS, the apoptotic cells (FITC+/PI±) and non-apoptotic cells were subjected to the analysis by the flow cytometer (BD Biosciences, San Jose, CA, USA).
Glucose Uptake and Lactate Production Assay
SW480 and LOVO cells were cultivated in glucose-free DMEM medium for 16 h. And then the medium was replaced with high-glucose DMEM medium, and the CRC cells were cultured for a further 24 h. The glucose uptake and lactate production were detected using Fluorescence-based glucose assay kit (BioVision, Milpitas, California, USA) and lactate oxidase-based colorimetric assay.
Dual-Luciferase Reporter Assay
Dual-luciferase reporter assay was implemented to analyze the target relationship between miR-383 and circ_0136666 or CREB1.
The wild-type or mutant type binding sites with miR-383 in circ_0136666 sequences were cloned by PCR and inserted into pmirGLO vector (Promega, Madison, WI, USA), designated as WT-circ_0136666 or MUT-circ_0136666. GeneArtTM Site-Directed Mutagenesis System Kit (Invitrogen) was used to alter the specific binding sites in circ_0136666. The luciferase activity was measured in 293T cells co-transfected with 20 nM miRNA NC or miR-383 mimic and 50 ng WT-circ_0136666 or MUT-circ_0136666 through Dual-Luciferase Reporter Assay System (Promega) via the luminometer (Plate Chameleon V, Hidex, Finland). Firefly luciferase activity was normalized to Renilla fluorescence intensity.
The 3ʹ untranslated region (3ʹ UTR) of CREB1 sequences including wild-type or mutant type binding sites were amplified by PCR and subcloned into pmirGLO vector (Promega), named as WT-CREB1 or MUT-CREB1. Luciferase activity was determined in 293T cells after transfection for 48 h through Dual-Luciferase Reporter Assay System (Promega).
Western Blot Assay
The proteins were isolated from CRC cells. Protein quantification was conducted with the BCA assay kit (Bio-Rad, Hercules, CA, USA). Protein samples (25 μg) were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gel and transferred onto a polyvinylidene fluoride (PVDF) membrane. The PVDF membrane was blocked with 5% non-fat milk for 1 h followed by incubation with the specific primary antibodies at 4°C overnight. The antibodies of CREB1 (ab31387), hexokinase 2 (HK2; ab104836), lactate dehydrogenase A (LDHA; ab226016) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH; ab37168) were purchased from Abcam (Cambridge, MA, USA). The membrane was then incubated with the horseradish peroxidase-labeled secondary antibody (ab205718, Abcam) at for 1 h at room temperature. The protein signal was visualized through the enhanced chemiluminescent (ECL) system (Beyotime, Shanghai, China). Protein quantification was performed using ImageJ software.
Murine Xenograft Assay
Animal experiments were approved by China-Japan Union Hospital of Jilin University. All animal procedures were conducted in accordance with the Guide for the Care and Use of Laboratory Animals. SW480 cells were stably transfected with sh-NC or sh-circ_0136666. The above cells (2 × 106 cells/200 μL PBS) were collected and re-suspended. The cell suspension was subcutaneously injected to the right side of the flank area of the mice (n=6). Tumor volume was measured every 4 d after inoculation for 7 d with the formula of width2 × length × 0.5. The mice were killed following inoculation for 27 d, and the tumors were weighed. The enrichment of circ_0136666, miR-383 and CREB1 was detected by RT-qPCR or Western blot assay.
Statistical Analysis
All data were showed as mean±standard deviation. The significance of differences was evaluated by Student’s t-test or one-way analysis of variance (ANOVA) followed by Tukey’s test as appropriate. P<0.05 was considered statistically significant.
Results
The Abundance of circ_0136666 is Markedly Increased in CRC Patients and Cells
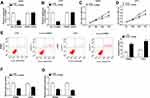
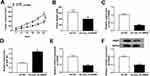
To clarify the role of circ_0136666 in CRC, we first measured the abundance of circ_0136666 in CRC tissues and cells. As showed in Figure 1A and B, the expression of circ_0136666 was significantly up-regulated in CRC tissues and cells compared with that in corresponding normal tissues and normal colon epithelial NCM460 cells. circ_0136666 expression was positively associated with tumor size, TNM stages, lymphatic metastasis and distant metastasis in CRC patients (Table 1). These findings suggested that circ_0136666 might play an oncogenic role in CRC.
circ_0136666 Promotes the Proliferation and Glycolysis While Represses the Apoptosis of CRC Cells
We conducted loss-of-function experiments to explore the function of circ_0136666 in CRC cells. As indicated in Figure 2A and B, the expression of circ_0136666 was prominently reduced in si-circ_0136666 transfected group compared with that in si-NC group of CRC cells. The proliferation of CRC cells was inhibited by si-circ_0136666 transfection (Figure 2C and D). Besides, circ_0136666 depletion also promoted the apoptosis of SW480 and LOVO cells (Figure 2E). Apart from this, we measured the glycolysis of CRC cells transfected with si-circ_0136666 through detecting the glucose uptake and lactate production. As exhibited in Figure 2F and G, the glucose uptake and lactate production were repressed with circ_0136666 intervention in CRC cells, demonstrating that circ_0136666 knockdown suppressed the glycolysis of CRC cells. Taken together, circ_0136666 promoted the progression of CRC through accelerating the proliferation and glycolysis and inhibiting the apoptosis of CRC cells.
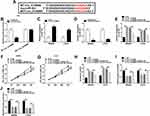
miR-383 is a Direct Target of circ_0136666 in 293T Cells
Circular RNA Interactome software was used to explore the downstream components by which circ_0136666 contributing to the progression of CRC. The complementary sequence between miR-383 and circ_0136666 is shown in Figure 3A. Subsequently, we conducted dual-luciferase reporter assay to confirm this combination. WT-circ_0136666 or MUT-circ_0136666 and miRNA NC or miR-383 mimic were co-transfected into 293T cells, and the luciferase activity was decreased in miR-383 mimic and WT-circ_0136666 co-transfected group (Figure 3B), suggesting that miR-383 was a direct target of circ_0136666 in 293T cells. To illustrate the regulatory relationship between miR-383 and circ_0136666 in CRC cells, we measured the abundance of miR-383 in si-circ_0136666 or si-NC transfected CRC cells. As mentioned in Figure 3C, the level of miR-383 was conspicuously up-regulated with si-circ_0136666 transfection in CRC cells, suggesting that miR-383 was negatively regulated by circ_0136666 in CRC cells.
To test whether circ_0136666 exerted its function through miR-383 in CRC cells, we first measured the knockdown efficiency of miR-383 inhibitor in CRC cells. As indicated in Figure 3D, the transfection of miR-383 inhibitor notably reduced the expression of miR-383 in SW480 and LOVO cells. SW480 and LOVO cells were transfected with si-NC, si-circ_0136666, si-circ_0136666 + inhibitor NC or si-circ_0136666 + miR-383 inhibitor. The abundance of miR-383 was notably increased by circ_0136666 knockdown, and the co-transfection of si-circ_0136666 and miR-383 inhibitor abolished this promoting effect in CRC cells (Figure 3E). The depletion of miR-383 abated the suppressive effects of circ_0136666 intervention on the proliferation and glycolysis and the promoting impact on the apoptosis of CRC cells (Figure 3F-J), demonstrating that miR-383 acted as a downstream gene of circ_0136666 to exert its function in CRC cells.
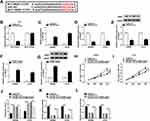
CREB1 Could Bind to miR-383 in 293T Cells
CREB1 was predicted as a target of miR-383 by Starbase bioinformatic software (Figure 4A). And this target relationship between CREB1 and miR-383 in 293T cells was verified by dual-luciferase reporter assay. As mentioned in Figure 4B, the accumulation of miR-383 in WT-CREB1 group notably reduced the luciferase activity in CRC cells, demonstrating that CREB1 was a target of miR-383 in 293T cells. miRNAs could down-regulate the abundance of target mRNAs by directly binding to their 3ʹ UTR.14,15 We wondered whether miR-383 could negatively regulate the mRNA and protein expression of CREB1 in CRC cells. Firstly, we assessed the overexpression efficiency of miR-383 mimic in CRC cells. As indicated in Figure 4C, the level of miR-383 was increased by the transfection of miR-383 mimic in CRC cells. The accumulation of miR-383 down-regulated the mRNA and protein expression of CREB1 in CRC cells (Figure 4D and E). The mRNA and protein levels of CREB1 were conspicuously elevated by the transfection of pcDNA-CREB1 in CRC cells (Figure 4F and G). SW480 and LOVO cells were transfected with miRNA NC, miR-383 mimic, miR-383 mimic + pcDNA-Control, miR-383 mimic + pcDNA-CREB1. As mentioned in Figure 4H-L, CREB1 accumulation counteracted the inhibitory effects of miR-383 overexpression on the proliferation and glycolysis and the promoting impact on the apoptosis of CRC cells, demonstrating that miR-383 functioned as a cancer suppressor through negatively regulating CREB1 in CRC cells.
CREB1 Accumulation Reverses the Inhibitory Effects of circ_0136666 Knockdown on the Proliferation and Glycolysis and the Promoting Impact on the Apoptosis of CRC Cells
SW480 and LOVO cells were transfected with si-NC, si-circ_0136666, si-circ_0136666 + pcDNA-Control or si-circ_0136666 + pcDNA-CREB1. As showed in Figure 5A and B, circ_0136666 depletion down-regulated the mRNA and protein levels of CREB1, and the addition of pcDNA-CREB1 recovered the abundance of CREB1 mRNA and protein in CRC cells. CREB1 overexpression reversed the suppressive effects of circ_0136666 interference on the proliferation and glycolysis and the promoting impact on the apoptosis of CRC cells (Figure 5C-G).
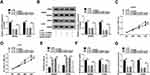
CREB1 is Modulated by circ_0136666/miR-383 Axis in CRC Cells
To further elucidate the regulatory relationship between circ_0136666, miR-383 and CREB1 in CRC cells, we transfected si-NC, si-circ_0136666, si-circ_0136666 + inhibitor NC or si-circ_0136666 + miR-383 inhibitor into SW480 and LOVO cells. The abundance of CREB1 mRNA and protein was notably decreased by the transfection of si-circ_0136666, and the co-transfection of si-circ_0136666 and miR-383 inhibitor recovered the mRNA and protein expression of CREB1 in CRC cells (Figure 6A and B).
The Glycolysis of CRC Cells is Regulated by circ_0136666/miR-383/CREB1 Axis
HK2 is the first rate-limiting enzyme of glycolysis, and LDHA is a vital oxidoreductase in the glycolytic pathway,24,25 thus these two proteins were used as glycolysis indicators in the following experiments. As indicated in Figure 7A-C, circ_0136666 intervention reduced the levels of HK2 and LDHA, while the addition of miR-383 inhibitor attenuated these inhibitory effects caused by circ_0136666 depletion in CRC cells. As mentioned in Figure 7D-F, the accumulation of CREB1 counteracted the suppressive effects of miR-383 overexpression on the levels of HK2 and LDHA in CRC cells. Collectively, the glycolysis of CRC cells was regulated by circ_0136666/miR-383/CREB1 signaling.
circ_0136666 Depletion Inhibits the Growth of CRC Tumors Through miR-383/CREB1 Axis in vivo
To investigate the function of circ_0136666/miR-383/CREB1 axis in vivo, we built the murine xenograft model using SW480 cells stably transfected with sh-NC or sh-circ_0136666. As showed in Figure 8A and B, circ_0136666 silencing inhibited the growth of the CRC tumors in vivo. Meanwhile, we detected the expression of circ_0136666, miR-383 and CREB1 in resected tumor tissues. The expression of circ_0136666 was notably reduced in sh-circ_0136666 group, while the level of miR-383 showed an opposite phenomenon (Figure 8C and D). circ_0136666 intervention also reduced the abundance of CREB1 mRNA and protein compared with that in the sh-NC group (Figure 8E and F). Taken together, circ_0136666 promoted the growth of CRC tumors by elevating the level of CREB1 via sponging miR-383 in vivo.
Discussion
circRNAs were implicated in the initiation and development of CRC. For example, Guo et al demonstrated that circ_0000069 accelerated the growth and metastasis of CRC cells.26 Fang et al claimed that circ_100290 accelerated the development of CRC through miR-516b/FZD4 axis and Wnt/β-catenin signal pathway.27 circRNAs have also emerged as crucial regulators in the glycolytic metabolism of CRC cells. circ_0007534 silencing was found to suppress the malignant behaviors and glycolysis of CRC cells through targeting miR-613/SLC25A22 axis.28 Herein, we concentrated on the roles of circ_0136666 in the proliferation, apoptosis and glycolysis of CRC.
circ_0136666 contributed to the progression of breast cancer through miR-1299/CDK6 axis.29 Besides, Jin et al found that circ_0136666 played an oncogenic role in CRC, and it facilitated the growth and metastasis of CRC cells via miR-136/SH2B1 axis.11 Consistent with the above findings, we found that circ_0136666 was abnormally up-regulated in CRC tissues and cells, and it promoted the proliferation and inhibited the apoptosis of CRC cells. Cancer cells gain energy through aerobic glycolysis rather than oxidative phosphorylation, and this phenomenon is called the Warburg effect.30 Increased aerobic glycolysis has emerged as a hallmark for many malignancies. A large amount of lipids, proteins, and nucleotides are generated in glycolytic metabolism, thereby promoting the proliferation of cancer cells.31,32 Accumulating works have pointed out the significance of aerobic glycolysis in CRC progression.33,34 However, the potential regulatory mechanism behind the Warburg effect of cancer cells remains to be revealed. Herein, we found that circ_0136666 contributed to the glycolysis of CRC cells for the first time.
miR-383 was predicted as a target of circ_0136666 by Circular RNA Interactome software, and this target relationship was then validated by dual-luciferase reporter assay. miR-383 was a tumor suppressor in ovarian cancer, hepatocellular carcinoma and gastric cancer.35–37 Subsequently, we found that the depletion of miR-383 counteracted the inhibitory effects of circ_0136666 depletion on the proliferation and glycolysis and the promoting effect on the apoptosis of CRC cells, demonstrating that circ_0136666 promoted the progression of CRC via miR-383.
To clarify the modulatory mechanism by which miR-383 inhibiting the progression of CRC, we aimed to explore the downstream components of miR-383. CREB1 was predicted as a potential target of miR-383 by Starbase software. Subsequently, the combination of miR-383 and CREB1 in 293T cells was confirmed by dual-luciferase reporter assay. Yan et al showed that CREB1 promoted the proliferation and metastasis of CRC cells.38 Consistent with the above findings, we found that CREB1 served as an oncogene in CRC. CREB1 accumulation reversed the inhibitory effects of miR-383 overexpression on the proliferation and glycolysis and the promoting effect on the apoptosis of CRC cells. Therefore, we concluded that miR-383 suppressed the development of CRC by negatively regulating CREB1. Based on the results that miR-383 was a functional target of circ_0136666, we speculated that circ_0136666 facilitated the proliferation and glycolysis while impeded the apoptosis of CRC cells through miR-383/CREB1 axis.
CREB1 overexpression ameliorated the suppressive effects of circ_0136666 interference on the proliferation and glycolysis and the promoting effect on the apoptosis of CRC cells. Besides, CREB1 could be modulated by circ_0136666/miR-383 axis in CRC cells. These two findings supported our former hypothesis.
To further explore the role of circ_0136666/miR-383/CREB1 signaling in the glycolysis of CRC cells, we determined the abundance of glycolysis-related proteins (HK2 and LDHA) by Western blot assay. circ_0136666 promoted the glycolysis of CRC cells through elevating the abundance of glycolysis-related proteins via miR-383/CREB1 axis. The effect of circ_0136666 on the growth of CRC tumors was validated by murine xenograft model. circ_0136666 promoted the growth of CRC tumors through up-regulating CREB1 via miR-383.
Conclusion
In summary, circ_0136666 promoted the development of CRC through accelerating the proliferation and glycolysis and restraining the apoptosis of CRC cells via miR-383/CREB1 axis in vitro and in vivo.
Highlights
- The level of circ_0136666 is conspicuously enhanced in CRC tissues and cells, and circ_0136666 promotes the proliferation and glycolysis while impedes the apoptosis of CRC cells.
- circ_0136666 could bind to miR-383 in 293T cells.
- CREB1 is a direct target of miR-383 in 293T cells.
- circ_0136666 accelerates the progression of CRC through up-regulating the abundance of CREB1 through sponging miR-383.
Data Sharing Statement
The analyzed data sets generated during the present study are available from the corresponding author on reasonable request.
Ethics Approval
The present study was approved by the ethical review committee of China-Japan Union Hospital of Jilin University.
Author Contributions
All authors made substantial contribution to conception and design, acquisition of the data, or analysis and interpretation of the data; take part in drafting the article or revising it critically for important intellectual content; gave final approval of the revision to be published; and agree to be accountable for all aspect of the work.
Disclosure
The authors report no funding and no conflicts of interest for this work.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
2. Weinberg BA, Marshall JL. Colon cancer in young adults: trends and their implications. Curr Oncol Rep. 2019;21(1):3. doi:10.1007/s11912-019-0756-8
3. Gerner EW, Bruckheimer E, Cohen A. Cancer pharmacoprevention: targeting polyamine metabolism to manage risk factors for colon cancer. J Biol Chem. 2018;293(48):18770–18778. doi:10.1074/jbc.TM118.003343
4. Qian L, Yu S, Chen Z, Meng Z, Huang S, Wang P. The emerging role of circRNAs and their clinical significance in human cancers. Biochim Biophys Acta Rev Cancer. 2018;1870(2):247–260. doi:10.1016/j.bbcan.2018.06.002
5. Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12(4):381–388. doi:10.1080/15476286.2015.1020271
6. Fu L, Jiang Z, Li T, Hu Y, Guo J. Circular RNAs in hepatocellular carcinoma: functions and implications. Cancer Med. doi:2018;doi:10.1002/cam4.1574
7. Zhou B, Yu JW. A novel identified circular RNA, circRNA_010567, promotes myocardial fibrosis via suppressing miR-141 by targeting TGF-beta1. Biochem Biophys Res Commun. 2017;487(4):769–775. doi:10.1016/j.bbrc.2017.04.044
8. Chan JJ, Tay Y. Noncoding RNA:RNA regulatory networks in cancer. Int J Mol Sci. 2018;19(5):1310. doi:10.3390/ijms19051310
9. Holgersson A, Erdal H, Nilsson A, Lewensohn R, Kanter L. Expression of DNA-PKcs and Ku86, but not Ku70, differs between lymphoid malignancies. Exp Mol Pathol. 2004;77(1):1–6. doi:10.1016/j.yexmp.2004.02.001
10. Sun G, Yang L, Dong C, Ma B, Shan M, Ma B. PRKDC regulates chemosensitivity and is a potential prognostic and predictive marker of response to adjuvant chemotherapy in breast cancer patients. Oncol Rep. 2017;37(6):3536–3542. doi:10.3892/or.2017.5634
11. Jin C, Wang A, Liu L, Wang G, Li G. Hsa_circ_0136666 promotes the proliferation and invasion of colorectal cancer through miR-136/SH2B1 axis. J Cell Physiol. 2019;234(5):7247–7256. doi:10.1002/jcp.27482
12. Croce CM, Calin GA. miRNAs, cancer, and stem cell division. Cell. 2005;122(1):6–7. doi:10.1016/j.cell.2005.06.036
13. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi:10.1016/s0092-8674(04)00045-5
14. Ambros V. The functions of animal microRNAs. Nature. 2004;431(7006):350–355. doi:10.1038/nature02871
15. Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6(11):857–866. doi:10.1038/nrc1997
16. Cui Y, Chen LG, Yao HB, Zhang J, Ding KF. Upregulation of microRNA-383 inhibits the proliferation, migration and invasion of colon cancer cells. Oncol Lett. 2018;15(1):1184–1190. doi:10.3892/ol.2017.7409
17. Li J, Smith AR, Marquez RT, et al. MicroRNA-383 acts as a tumor suppressor in colorectal cancer by modulating CREPT/RPRD1B expression. Mol Carcinog. 2018;57(10):1408–1420. doi:10.1002/mc.22866
18. Shaywitz AJ, Greenberg ME. CREB: a stimulus-induced transcription factor activated by a diverse array of extracellular signals. Annu Rev Biochem. 1999;68(1):821–861. doi:10.1146/annurev.biochem.68.1.821
19. Shankar DB, Cheng JC, Kinjo K, et al. The role of CREB as a proto-oncogene in hematopoiesis and in acute myeloid leukemia. Cancer Cell. 2005;7(4):351–362. doi:10.1016/j.ccr.2005.02.018
20. Sakamoto KM, Frank DA. CREB in the pathophysiology of cancer: implications for targeting transcription factors for cancer therapy. Clin Cancer Res. 2009;15(8):2583–2587. doi:10.1158/1078-0432.ccr-08-1137
21. Lu X, Yu Y, Liao F, Tan S. Homo Sapiens circular RNA 0079993 (hsa_circ_0079993) of the POLR2J4 gene acts as an oncogene in colorectal cancer through the microRNA-203a-3p.1 and CREB1 axis. Med Sci Monitor. 2019;25:6872–6883. doi:10.12659/msm.916064
22. Yang Q, Yu W, Han X. Overexpression of microRNA101 causes antitumor effects by targeting CREB1 in colon cancer. Mol Med Rep. 2019;19(4):3159–3167. doi:10.3892/mmr.2019.9952
23. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–408. doi:10.1006/meth.2001.1262
24. Akram M. Mini-review on glycolysis and cancer. J Cancer Educ. 2013;28(3):454–457. doi:10.1007/s13187-013-0486-9
25. Kim S, Lee E, Jung J, et al. microRNA-155 positively regulates glucose metabolism via PIK3R1-FOXO3a-cMYC axis in breast cancer. Oncogene. 2018;37(22):2982–2991. doi:10.1038/s41388-018-0124-4
26. Guo JN, Li J, Zhu CL, et al. Comprehensive profile of differentially expressed circular RNAs reveals that hsa_circ_0000069 is upregulated and promotes cell proliferation, migration, and invasion in colorectal cancer. Onco Targets Ther. 2016;9:7451–7458. doi:10.2147/ott.s123220
27. Fang G, Ye BL, Hu BR, Ruan XJ, Shi YX. CircRNA_100290 promotes colorectal cancer progression through miR-516b-induced downregulation of FZD4 expression and Wnt/beta-catenin signaling. Biochem Biophys Res Commun. 2018;504(1):184–189. doi:10.1016/j.bbrc.2018.08.152
28. Ding DY, Wang D, Shu ZB. Hsa_circ_0007534 knockdown represses the development of colorectal cancer cells through regulating miR-613/SLC25A22 axis. Eur Rev Med Pharmacol Sci. 2020;24(6):3004–3022. doi:10.26355/eurrev_202003_20665
29. Liu LH, Tian QQ, Liu J, Zhou Y, Yong H. Upregulation of hsa_circ_0136666 contributes to breast cancer progression by sponging miR-1299 and targeting CDK6. J Cell Biochem. 2019;120(8):12684–12693. doi:10.1002/jcb.28536
30. Koppenol WH, Bounds PL, Dang CV. Otto Warburg’s contributions to current concepts of cancer metabolism. Nat Rev Cancer. 2011;11(5):325–337. doi:10.1038/nrc3038
31. Gatenby RA, Gillies RJ. Why do cancers have high aerobic glycolysis? Nat Rev Cancer. 2004;4:891–899. doi:10.1038/nrc1478
32. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi:10.1016/j.cell.2011.02.013
33. Kawada K, Toda K, Sakai Y. Targeting metabolic reprogramming in KRAS-driven cancers. Int J Clin Oncol. 2017;22(4):651–659. doi:10.1007/s10147-017-1156-4
34. Brown RE, Short SP, Williams CS. Colorectal cancer and metabolism. Curr Colorectal Cancer Rep. 2018;14(6):226–241. doi:10.1007/s11888-018-0420-y
35. Zhu C, Huang Q, Zhu H. miR-383 inhibited the cell cycle progression of gastric cancer cells via targeting cyclin E2. DNA Cell Biol. 2019;38(8):849–856. doi:10.1089/dna.2019.4624
36. Chen L, Guan H, Gu C, Cao Y, Shao J, Wang F. miR-383 inhibits hepatocellular carcinoma cell proliferation via targeting APRIL. Tumour Biol. 2016;37(2):2497–2507. doi:10.1007/s13277-015-4071-1
37. Han RL, Wang FP, Zhang PA, Zhou XY, Li Y. miR-383 inhibits ovarian cancer cell proliferation, invasion and aerobic glycolysis by targeting LDHA. Neoplasma. 2017;64(2):244–252. doi:10.4149/neo_2017_211
38. Yan L, You WQ, Sheng NQ, et al. A CREB1/miR-433 reciprocal feedback loop modulates proliferation and metastasis in colorectal cancer. Aging (Albany NY). 2018;10(12):3774–3793. doi:10.18632/aging.101671
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.