Back to Journals » Cancer Management and Research » Volume 12
Circ_0109046 Promotes the Progression of Endometrial Cancer via Regulating miR-136/HMGA2 Axis
Received 30 July 2020
Accepted for publication 3 October 2020
Published 2 November 2020 Volume 2020:12 Pages 10993—11003
DOI https://doi.org/10.2147/CMAR.S274856
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Yong Teng
Yanping Shi,1 Li Jia,2 Hongli Wen2
1Department of Gynaecology, Henan Provincial People’s Hospital, People’s Hospital of Zhengzhou University, Zhengzhou, Henan 450003, People’s Republic of China; 2Department of Gynaecology, Chongqing Traditional Chinese Medicine Hospital, Chongqing 400021, People’s Republic of China
Correspondence: Li Jia Tel +86 17784307991
Email [email protected]
Background: Endometrial cancer (EC) is one of the most common gynecological malignancies. Circular RNAs (circRNAs) play crucial roles in the occurrence and development of tumors. This research aimed to explore the function and potential mechanism of human serum albumin (hsa)_circ_0109046 in EC.
Materials and Methods: The abundance of circ_0109046, microRNA-136 (miR-136) and high-mobility group AT-hook 2 (HMGA2) was detected by quantitative real-time polymerase chain reaction or Western blot. Cell counting kit-8 (CCK-8) and colony formation assays were employed to assess cell proliferation. Transwell assay was used to measure cell migration and invasion. The levels of E-cadherin, Vimentin and N-cadherin were examined by Western blot. The binding association among circ_0109046, miR-136 and HMGA2 was verified by dual-luciferase reporter assay, RNA pull-down assay and RNA immunoprecipitation assay. Xenograft assay was performed to test tumor growth in vivo.
Results: Circ_0109046 and HMGA2 were up-regulated, and miR-136 was down-regulated in EC tissues and cells. Knockdown of circ_0109046 impeded the proliferation, migration, invasion and epithelial-mesenchymal transition (EMT) of EC cells. Moreover, miR-136 knockdown reversed the suppression of circ_0109046 silencing on EC development. HMGA2 overexpression abolished the inhibition of miR-136 on EC progression. Besides, depletion of circ_0109046 inhibited EC growth in vivo.
Conclusion: Circ_0109046 accelerated EC progression via modulating miR-136/HMGA2 axis, indicating that circ_0109046 might be a promising therapeutic target for EC.
Keywords: circ_0109046, miR-136, HMGA2, endometrial cancer
Introduction
Endometrial cancer (EC) is a common gynecological tumor, especially in developed countries.1 The incidence of EC is increasing year by year, and the 5-year overall survival rate of patients without metastatic EC is 74~91%.2 In contrast, the 5-year overall survival rate for recurrent or metastatic EC is only 10~20%.3 Despite significant progress in the research of EC, proper early screening methods are still lacking. Hence, exploring EC pathogenesis to seek new biomarkers for diagnosis and treatment is urgent.
Circular RNAs (circRNAs) are endogenous molecules with covalently closed loops without 5ʹ to 3ʹ polarity.4,5 Mounting evidence has confirmed that circRNAs exert significant effects on various diseases.6 Synchronously, more and more studies have corroborated that dysregulated circRNAs can mediate the occurrence and development of tumors in gynecological cancers by regulating a range of biological functions.7 For instance, hsa_circ_0000520 suppressed cell growth and mobility and induced apoptosis in cervical cancer by absorbing microRNA-146b-3p.8 Also, hsa_circ_0009910 triggered cell proliferation and motor phenotypes in ovarian cancer via targeting microRNA-145.9 Furthermore, many circRNAs participated in EC progression, including hsa_circ_0061140,10 circ_PUM111 and hsa_circ_0002577.12 Xu et al revealed that hsa_circ_0109046 was prominently increased in EC by using RNA-seq analysis.13 Nevertheless, the exact role and molecular basis of circ_0109046 in EC are unknown.
Emerging evidence has demonstrated that circRNAs can sponge microRNAs (miRNAs) and regulate gene expression by competitively combining with miRNA binding sites.14 Besides, numerous studies have verified that aberrantly expressed miRNAs are strongly related to EC development.15 For example, miR-21-5p facilitated epithelial-mesenchymal transition (EMT) in EC cells through inhibiting SOX17.16 In addition, miR-214-3p suppressed EC cell metastasis via binding to TWIST1.17 Moreover, up-regulation of miR-320a hindered cell proliferation in EC by negatively modulating IGF-1R.18 A previous research unveiled that miR-136 was overtly down-regulated in EC and affected EC progression by combining with circ_PUM1.11 However, the association between circ_0109046 and miR-136 has not been studied.
In this research, we ascertained the effect of circ_0109046 in EC development. Meanwhile, we investigated the potential relationship between circ_0109046 and miR-136/high-mobility group AT-hook 2 (HMGA2) regulatory axis in EC, which might provide a new biomarker for EC therapy.
Materials and Methods
Clinical Specimens
All EC tissues (n=44) and adjacent normal tissues (n=44) were obtained from EC patients recruited at Henan Provincial People’s Hospital. None of patients received any radiotherapy or chemotherapy before resection. All participants signed written informed consent. The protocol was ratified by the Ethics Committee of Henan Provincial People’s Hospital. Some clinical pathological features of patients are listed in Table 1.
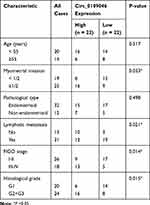 |
Table 1 Correlation Between Circ_0109046 Expression and the Clinical Pathological Features of 44 Endometrial Cancer Patients |
Cell Culture
Human normal endometrial epithelial cells (HEECs) and three EC cells (HEC1-A, KLE and Ishikawa) were purchased from Tongpai Biotech (Shanghai, China) and incubated in RPMI-1640 medium (Hyclone, Logan, UT, USA) supplemented with 10% fetal bovine serum (FBS; Hyclone). The other two EC cells (HEC1-B and AN3CA) were bought from American Type Culture Collection (ATCC, Manassas, VA, USA) and cultured in Dulbecco’s Modified Eagle Medium (DMEM; Hyclone, Logan, UT, USA) containing 10% FBS. All cells were maintained in an incubator with 5% CO2 at 37°C.
Cell Transfection
Short hairpin RNA (shRNA) against circ_0109046 (sh-circ#1, sh-circ#2 and sh-circ#3) and the control (sh-NC), miR-136 mimics (miR-136) and negative control (miR-NC), miR-136 inhibitor (anti-miR-136) and negative control (anti-NC), HMGA2 overexpression vector (HMGA2) and the empty pcDNA3.1 (vector) were bought from Genechem (Shanghai, China). The oligonucleotides or vectors were introduced into HEC1-A and Ishikawa cells via Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA).
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
After isolating RNA using Trizol reagent (Solarbio, Beijing, China), the specific reverse transcription kit (Vazyme, Nanjing, China) was utilized to obtain complementary DNA. Then, the RNA levels were monitored via SYBR Green PCR Master Mix (Vazyme) and quantified using the 2−ΔΔCt method. Glyceraldehyde 3-phosphate dehydrogenase (GADPH) or U6 was taken as an internal control. The primer sequences were shown in Table 2.
 |
Table 2 Quantitative Real-Time PCR Primer |
Cell Counting Kit-8 (CCK-8) Assay
Transfected HEC1-A and Ishikawa cells were injected into 96-well plates. After incubating for 24 h, 48 h or 72 h, the fresh medium containing 10% CCK-8 solution (Beyotime, Shanghai, China) was added, followed by culture for 4 h. Finally, the optical density was measured using a Microplate Reader (BioTek, Burlington, VT, USA).
Colony Formation Assay
Transfected HEC1-A and Ishikawa cells (3×103) were grown in six-well plates and maintained for 2 weeks at 37°C. After fixing with methanol and staining with crystal violet (Solarbio), the colonies were calculated under a microscope in three independent replicates.
Transwell Assay
For cell migration test, HEC1-A and Ishikawa cells were plated into the upper chamber (Corning, Corning, NY, USA). Synchronously, the lower chamber was filled with fresh medium harboring 10% FBS. After culturing for 24 h, the cells were fixed with methanol and stained with crystal violet (Solarbio). Subsequently, the migrated cells were photographed and counted under a microscope at 100× magnification. Additionally, the difference is that Transwell used in cell invasion assay is pre-coated with Matrigel (Corning).
Western Blot Assay
Protein samples extracted with RIPA buffer (Solarbio) were separated by polyacrylamide gel electrophoresis and transferred onto polyvinylidene fluoride (PVDF) membranes (Millipore, Billerica, MA, USA). After sealing with 5% skimmed milk, the membranes were incubated with primary antibodies against E-cadherin (ab15148, 1:500, Abcam, Cambridge, UK), Vimentin (ab137321, 1:2000, Abcam), N-cadherin (ab76057, 1:1000, Abcam), HMGA2 (ab97276, 1:3000, Abcam) or GAPDH (ab9485, 1:2500, Abcam) at 4°C. Afterwards, the membranes were probed with a secondary antibody (ab205718, 1:20,000, Abcam). The signal intensity was examined with ECL reagent (Millipore).
Nuclear and Cytoplasmic Fraction Assay
The subcellular localization of circ_0109046 was evaluated via mirVanaTM PARIS Kit (Invitrogen). U6 and GAPDH were regarded as positive controls for nuclear and cytoplasmic fractions, respectively.
Dual-Luciferase Reporter Assay
The sequences of circ_0109046 or HMGA2-3ʹUTR containing the predicted miR-136 binding site and the mutant sequence were inserted into the pmirGLO vector (LMAI Bio, Shanghai, China) to form circ_0109046-WT, circ_0109046-MUT, HMGA2-3ʹUTR-WT or HMGA2-3ʹUTR-MUT. Subsequently, the constructed reporter and miR-136 or miR-NC were co-transfected into HEC1-A and Ishikawa cells. The luciferase intensity was determined using Dual-Lucy Assay Kit (Solarbio).
RNA Pull-Down Assay
Biotinylated miR-136 (Bio-miR-136) and the control (Bio-NC) were purchased from Genechem. HEC1-A and Ishikawa cells were lysed and then incubated with streptavidin-coated magnetic beads (Invitrogen). Afterwards, qRT-PCR was used to measure the abundance of circ_0109046 and HMGA2 in the isolated RNA.
RNA Immunoprecipitation (RIP) Assay
RIP analysis was conducted using EZ-Magna RIP Kit (Millipore). HEC1-A and Ishikawa cells were lysed in RIP lysis buffer, and then cell lysates were incubated with magnetic beads conjugated with Ago2 antibody or IgG antibody (Abcam). Finally, the precipitated RNAs were purified and measured using qRT-PCR.
Xenograft Assay
Ishikawa cells (5×106) stably transfected with sh-NC or sh-circ#1 were subcutaneously injected into the right-back of five-week-old BALB/c female nude mice (Vital River Laboratory, Beijing, China; n=5 per group). Then, tumor volume was monitored every 7 days. At 35 days, the xenograft tumors were weighed after the mice were killed. Additionally, the levels of circ_0109046, miR-136, HMGA2 and EMT-related markers were examined in xenograft tissues by qRT-PCR or Western blot. The experiment was approved by the Animal Ethics Committee of Henan Provincial People’s Hospital. Animal studies were performed in compliance with the ARRIVE guidelines and the Basel Declaration. All animals received humane care according to the National Institutes of Health (USA) guidelines.
Statistical Analysis
All data were displayed as mean ± standard deviation by Graphpad Prism 7.0 software (GraphPad, San Diego, CA, USA). The relationship between circ_0109046 level and overall survival was tested using Kaplan-Meier survival analysis. Student’s t-test and one-way analysis of variance were utilized to assess the differences. P <0.05 was considered statistically significant.
Results
Circ_0109046 is Up-Regulated in EC Tissues and Cells
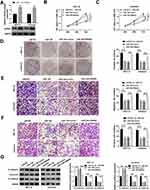
To evaluate the role of circ_0109046 in EC, we first detected circ_0109046 expression in EC tissues (n=44) and adjacent normal tissues (n=44) using qRT-PCR. The results showed that circ_0109046 level in EC tissues was remarkably increased compared with normal tissues (Figure 1A). Additionally, circ_0109046 expression in EC cells (HEC1-A, HEC1-B, KLE, AN3CA and Ishikawa) was significantly higher than that in normal endometrial epithelial cells (HEECs) (Figure 1B). Besides, EC patients were divided into circ_0109046 high expression group (n=22) and circ_0109046 low expression group (n=22) according to the median of circ_0109046 expression level in EC tissues. As illustrated in Figure 1C, circ_0109046 high expression was closely related to poor overall survival. We also studied some clinical pathological features of EC patients. The data suggested that circ_0109046 expression was not related to age and pathological type, but was associated with myometrial invasion, lymphatic metastasis, FIGO stage and histological grade (Table 1). These data hinted that circ_0109046 might play a carcinogenic role in EC.
Circ_0109046 Silencing Inhibits the Proliferation, Migration, Invasion and EMT of EC Cells
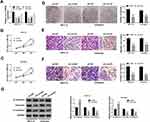
Next, we performed loss-of-function experiments by transfecting circ_0109046 shRNA to HEC1-A and Ishikawa cells. Firstly, transfection with sh-circ_0109046 (sh-circ#1, sh-circ#2 and sh-circ#3) markedly reduced circ_0109046 level compared to the sh-NC group, and sh-circ#1 had the highest knockdown efficiency (Figure 2A). CCK-8 and colony formation assays suggested that down-regulation of circ_0109046 decreased the viability (Figure 2B and C) and colony number (Figure 2D) of HEC1-A and Ishikawa cells. Transwell analysis showed that silencing of circ_0109046 reduced the migration and invasion of HEC1-A and Ishikawa cells (Figure 2E and F). In addition, knockdown of circ_0109046 led to a marked elevation in E-cadherin level and significant decreases in Vimentin and N-cadherin levels (Figure 2G). Collectively, these data indicated that circ_0109046 depletion suppressed EC cell progression.
Circ_0109046 Directly Targets miR-136
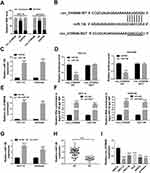
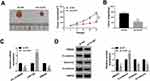
To determine circ_0109046 distribution in EC cells, nuclear and cytoplasmic fraction assay was carried out. The results showed that circ_0109046 was mainly distributed in cytoplasm (Figure 3A). Subsequently, the circinteractome online database illustrated that circ_0109046 and miR-136 had a putative binding site (Figure 3B). As displayed in Figure 3C, miR-136 was strikingly up-regulated in HEC1-A and Ishikawa cells after transfection with miR-136. Next, dual-luciferase reporter assay was conducted to verify the binding relationship, and the results suggested that miR-136 mimics remarkably reduced the luciferase activity of circ_0109046-WT reporter (Figure 3D). Moreover, RNA pull-down assay revealed that circ_0109046 was pulled down by Bio-miR-136 (Figure 3E). Meanwhile, RIP analysis showed that circ_0109046 and miR-136 were enriched in anti-Ago2 group instead of anti-lgG group (Figure 3F). In addition, qRT-PCR showed that circ_0109046 silencing promoted miR-136 expression (Figure 3G). Also, miR-136 level was overtly decreased in EC tissues and cells relative to the control group (Figure 3H and I). These results evidenced that circ_0109046 negatively regulated miR-136 in EC cells.
Inhibition of miR-136 Alleviates the Effect of Circ_0109046 Knockdown on EC Cells
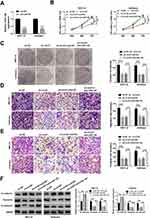
To illuminate whether circ_0109046 affected EC development by modulating miR-136, sh-circ#1 and anti-miR-136 were simultaneously transfected into EC cells. First of all, qRT-PCR revealed that miR-136 knockdown efficiency was significant (Figure 4A). Next, CCK-8 and colony formation assays showed that circ_0109046 knockdown suppressed cell proliferation in HEC1-A and Ishikawa cells, while this change was abolished by down-regulating miR-136 (Figure 4B and C). Simultaneously, co-transfection of sh-circ#1 and anti-miR-136 reversed the inhibitory effect of circ_0109046 depletion on cell migration and invasion (Figure 4D and E). Additionally, Western blot assay showed that down-regulation of circ_0109046 inhibited EMT in HEC1-A and Ishikawa cells, whereas this impact was abrogated after introduction of anti-miR-136 (Figure 4F). Overall, these data indicated that circ_0109046 facilitated EC development by sponging miR-136.
HMGA2 is a Target of miR-136
Moreover, starBase prediction software revealed that miR-136 had a putative binding site in HMGA2-3ʹUTR (Figure 5A). As shown in Figure 5B, miR-136 mimics markedly decreased the luciferase activity of HMGA2-3ʹUTR-WT reporter. Next, RNA pull-down and RIP assays were performed to validate the binding relationship between miR-136 and HMGA2. The results suggested that HMGA2 was pulled down by Bio-miR-136 (Figure 5C). Synchronously, HMGA2 and miR-136 were enriched in anti-Ago2 group (Figure 5D). Furthermore, miR-136 overexpression suppressed HMGA2 protein expression, and miR-136 knockdown induced HMGA2 protein expression (Figure 5E). To clarify the interaction between circ_0109046 and miR-136/HMGA2 axis, HMGA2 protein level was examined in HEC1-A and Ishikawa cells transfected with sh-NC, sh-circ#1, sh-circ#1+anti-NC or sh-circ#1+anti-miR-136. The results indicated that introduction of anti-miR-136 mitigated the reduction of HMGA2 protein level caused by circ_0109046 silencing (Figure 5F). These data evidenced that circ_0109046 regulated HMGA2 expression by targeting miR-136.
HMGA2 Overexpression Attenuates the Effect of miR-136 on EC Cells
To investigate the role of miR-136/HMGA2 axis in EC progression, HEC1-A and Ishikawa cells were introduced with miR-NC, miR-136, miR-136+vector or miR-136+HMGA2 to perform rescue experiments. Firstly, Western blot showed that HMGA2 overexpression efficiency was significant (Figure 6A). Then, rescue experiments revealed that up-regulation of miR-136 resulted in marked inhibition of proliferation (Figure 6B–D), migration (Figure 6E), invasion (Figure 6F) and EMT (Figure 6G) in HEC1-A and Ishikawa cells, while these effects were abolished after co-transfection with miR-136 and HMGA2. These data evidenced that miR-136 hindered EC cell progression via binding to HMGA2.
Knockdown of Circ_0109046 Inhibits EC Growth in vivo
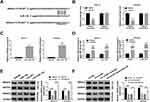
To explore the role of circ_0109046 in tumorigenesis, we constructed xenograft models by injecting transfected Ishikawa cells into the nude mice. The results showed that tumor volume and weight were strikingly decreased in the sh-circ#1 group compared to the sh-NC group (Figure 7A and B). Next, qRT-PCR and Western blot were performed in xenograft tissues. As displayed in Figure 7C, knockdown of circ_0109046 overtly decreased circ_0109046 and HMGA2 levels and elevated miR-136 level. In addition, Western blot analysis showed that silence of circ_0109046 led to a marked decrease in HMGA2, Vimentin and N-cadherin levels, while an increase in E-cadherin level (Figure 7D). These data unveiled that circ_0109046 knockdown blocked tumor growth in vivo.
Discussion
The competing endogenous RNA (ceRNA) hypothesis has received widespread attention, indicating that transcripts containing miRNA binding sites compete for post-transcriptional regulation.19 Compelling evidence has highlighted that circRNAs play critical roles in tumorigenesis and development via serving as ceRNAs or miRNA sponges.20 In the present research, we selected circ_0109046 as the research candidate based on the previous high-throughput sequencing results.13 We validated that circ_0109046 was remarkably up-regulated in EC. In addition, loss-of-function experiments revealed that circ_0109046 down-regulation suppressed EC cell growth and metastasis, suggesting that circ_0109046 might be a potential biomarker for EC treatment.
In terms of mechanism, we first confirmed that miR-136 combined with circ_0109046 and HMGA2, and circ_0109046 indirectly elevated HMGA2 expression via modulating miR-136, indicating that circ_0109046 sponged miR-136 to regulate HMGA2 through the ceRNA mechanism. Furthermore, recent investigations have manifested that circRNAs mediate EC progression via intervening ceRNA network. For instance, hsa_circ_0002577 expedited the proliferation and metastasis of EC cells via sponging miR-197 to up-regulate CTNND1.12 Liu et al disclosed that circWHSC1 aggravated the malignant behaviors of EC by absorbing miR-646 and increasing NPM1 expression.21 Moreover, hsa_circ_0061140 contributed to EC development through functioning as a ceRNA for miR-149-5p to activate STAT3.10 This research discovered a new ceRNA regulatory mechanism.
EMT confers malignant properties to cancer cells, including migration and invasion.22 EMT is characterized by down-regulation of E-cadherin and up-regulation of N-cadherin and Vimentin.23 In this report, we chose miR-136 as a possible target for circ_0109046 based on its anti-tumor effect. In triple-negative breast cancer, miR-136 up-regulation restrained tumor metastasis via combining with RASAL2.24 Additionally, augmentation of miR-136 reduced cancer stem cell activity and sensitized paclitaxel-resistant ovarian cancer cells in ovarian cancer by repressing Notch3.25 MiR-136 overexpression decelerated cell proliferation and increased radiosensitivity in cervical carcinoma via inhibiting E2F1 and inactivating NF-κB pathway.26 Besides, Zong et al unveiled that miR-136 was an anti-proliferative and anti-metastatic microRNA in EC.11 In the current research, miR-136 level was dramatically declined in EC. More importantly, miR-136 knockdown abrogated the effect of circ_0109046 depletion on EC development.
Moreover, mounting evidence has demonstrated that miRNAs facilitate post-transcriptional silencing of genes via base-pairing with 3ʹUTR of mRNAs.27 Our research discovered that miR-136 directly targeted HMGA2. HMGA2 is a common oncogenic factor and participates in various cellular processes, including cell proliferation, apoptosis and differentiation.28 Meanwhile, HMGA2 accelerates tumor progression in gynecological cancers, including cervical carcinoma,29 breast cancer30 and ovarian cancer.31 Ma et al suggested that HMGA2 mediated by miR-302a-5p/367-3p promoted the malignancy of EC cells.32 In this research, HMGA2 was identified as a target for miR-136. Importantly, augmentation of HMGA2 abolished the inhibition of miR-136 mimics on EC development.
In short, our findings revealed that circ_0109046 sponged miR-136 to up-regulate HMGA2 through the ceRNA mechanism. Furthermore, circ_0109046 facilitated EC progression via regulating miR-136/HMGA2 signaling (Figure S1), indicating that circ_0109046 might be a promising therapeutic marker for EC.
Funding
There is no funding to report.
Disclosure
The authors declare that they have no financial or non-financial conflicts of interest for this work.
References
1. Martin-Hirsch PP, Bryant A, Keep SL, Kitchener HC, Lilford R. Adjuvant progestagens for endometrial cancer. Cochrane Database Syst Rev. 2011;6:CD001040. doi:10.1002/14651858.CD001040.pub2
2. Morice P, Leary A, Creutzberg C, Abu-Rustum N, Darai E. Endometrial cancer. Lancet. 2016;387(10023):1094–1108. doi:10.1016/S0140-6736(15)00130-0
3. Post CCB, Westermann AM, Bosse T, Creutzberg CL, Kroep JR. PARP and PD-1/PD-L1 checkpoint inhibition in recurrent or metastatic endometrial cancer. Crit Rev Oncol Hematol. 2020;152:102973. doi:10.1016/j.critrevonc.2020.102973
4. Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20(11):675–691. doi:10.1038/s41576-019-0158-7
5. Qian L, Yu S, Chen Z, Meng Z, Huang S, Wang P. The emerging role of circRNAs and their clinical significance in human cancers. Biochim Biophys Acta Rev Cancer. 2018;1870(2):247–260. doi:10.1016/j.bbcan.2018.06.002
6. Su M, Xiao Y, Ma J, et al. Circular RNAs in cancer: emerging functions in hallmarks, stemness, resistance and roles as potential biomarkers. Mol Cancer. 2019;18(1):90. doi:10.1186/s12943-019-1002-6
7. Dong P, Xu D, Xiong Y, et al. The expression, functions and mechanisms of circular RNAs in gynecological cancers. Cancers. 2020;12(6):1472. doi:10.3390/cancers12061472
8. Zhang J, Cai R, Zhang Y, Wang X. Involvement of a novel circularRNA, hsa_circ_0000520, attenuates tumorigenesis of cervical cancer cell through competitively binding with miR-146b-3p. J Cell Mol Med. 2020. doi:10.1111/jcmm.15414
9. Li Y, Lin S, An N. Hsa_circ_0009910: oncogenic circular RNA targets microRNA-145 in ovarian cancer cells. Cell Cycle. 2020;1–12. doi:10.1080/15384101.2020.1731650
10. Liu Y, Chang Y, Cai Y. Hsa_circ_0061140 promotes endometrial carcinoma progression via regulating miR-149-5p/STAT3. Gene. 2020;745:144625. doi:10.1016/j.gene.2020.144625
11. Zong ZH, Liu Y, Chen S, Zhao Y. Circ_PUM1 promotes the development of endometrial cancer by targeting the miR-136/NOTCH3 pathway. J Cell Mol Med. 2020;24(7):4127–4135. doi:10.1111/jcmm.15069
12. Shen Q, He T, Yuan H. Hsa_circ_0002577 promotes endometrial carcinoma progression via regulating miR-197/CTNND1 axis and activating Wnt/beta-catenin pathway. Cell Cycle. 2019;18(11):1229–1240. doi:10.1080/15384101.2019.1617004
13. Xu H, Gong Z, Shen Y, Fang Y, Zhong S. Circular RNA expression in extracellular vesicles isolated from serum of patients with endometrial cancer. Epigenomics. 2018;10(2):187–197. doi:10.2217/epi-2017-0109
14. Li J, Sun D, Pu W, Wang J, Peng Y. Circular RNAs in cancer: biogenesis, function, and clinical significance. Trends Cancer. 2020;6(4):319–336. doi:10.1016/j.trecan.2020.01.012
15. Vasilatou D, Sioulas VD, Pappa V, Papageorgiou SG, Vlahos NF. The role of miRNAs in endometrial cancer. Epigenomics. 2015;7(6):951–959. doi:10.2217/epi.15.41
16. Wang C, Li Q, He Y. MicroRNA215p promotes epithelial to mesenchymal transition by targeting SRYbox 17 in endometrial cancer. Oncol Rep. 2020;43(6):1897–1905. doi:10.3892/or.2020.7556
17. Fang YY, Tan MR, Zhou J, et al. miR-214-3p inhibits epithelial-to-mesenchymal transition and metastasis of endometrial cancer cells by targeting TWIST1. Onco Targets Ther. 2019;12:9449–9458. doi:10.2147/OTT.181037
18. Shu S, Liu X, Xu M, et al. MicroRNA-320a acts as a tumor suppressor in endometrial carcinoma by targeting IGF-1R. Int J Mol Med. 2019;43(3):1505–1512. doi:10.3892/ijmm.2019.4051
19. Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet. 2016;17(5):272–283. doi:10.1038/nrg.2016.20
20. Cui X, Wang J, Guo Z, et al. Emerging function and potential diagnostic value of circular RNAs in cancer. Mol Cancer. 2018;17(1):123. doi:10.1186/s12943-018-0877-y
21. Liu Y, Chen S, Zong ZH, Guan X, Zhao Y. CircRNA WHSC1 targets the miR-646/NPM1 pathway to promote the development of endometrial cancer. J Cell Mol Med. 2020;24(12):6898–6907. doi:10.1111/jcmm.15346
22. Lu W, Kang Y. Epithelial-mesenchymal plasticity in cancer progression and metastasis. Dev Cell. 2019;49(3):361–374. doi:10.1016/j.devcel.2019.04.010
23. Loh CY, Chai JY, Tang TF, et al. The E-Cadherin and N-Cadherin switch in epithelial-to-mesenchymal transition: signaling, therapeutic implications, and challenges. Cells. 2019;8:10. doi:10.3390/cells8101118
24. Yan M, Li X, Tong D, et al. miR-136 suppresses tumor invasion and metastasis by targeting RASAL2 in triple-negative breast cancer. Oncol Rep. 2016;36(1):65–71. doi:10.3892/or.2016.4767
25. Jeong JY, Kang H, Kim TH, et al. MicroRNA-136 inhibits cancer stem cell activity and enhances the anti-tumor effect of paclitaxel against chemoresistant ovarian cancer cells by targeting Notch3. Cancer Lett. 2017;386:168–178. doi:10.1016/j.canlet.2016.11.017
26. Lu HJ, Jin PY, Tang Y, et al. microRNA-136 inhibits proliferation and promotes apoptosis and radiosensitivity of cervical carcinoma through the NF-kappaB pathway by targeting E2F1. Life Sci. 2018;199:167–178. doi:10.1016/j.lfs.2018.02.016
27. Fabian MR, Sonenberg N, Filipowicz W. Regulation of mRNA translation and stability by microRNAs. Annu Rev Biochem. 2010;79:351–379. doi:10.1146/annurev-biochem-060308-103103
28. Young AR, Narita M. Oncogenic HMGA2: short or small? Genes Dev. 2007;21(9):1005–1009. doi:10.1101/gad.1554707
29. Wang WY, Cao YX, Zhou X, Wei B, Zhan L, Fu LT. HMGA2 gene silencing reduces epithelial-mesenchymal transition and lymph node metastasis in cervical cancer through inhibiting the ATR/Chk1 signaling pathway. Am J Transl Res. 2018;10(10):3036–3052.
30. Mansoori B, Duijf PHG, Mohammadi A, et al. Overexpression of HMGA2 in breast cancer promotes cell proliferation, migration, invasion and stemness. Expert Opin Ther Targets. 2020:1–11. doi:10.1080/14728222.2020.1736559.
31. Han W, Zhang Y, Niu C, et al. BTB and CNC homology 1 (Bach1) promotes human ovarian cancer cell metastasis by HMGA2-mediated epithelial-mesenchymal transition. Cancer Lett. 2019;445:45–56. doi:10.1016/j.canlet.2019.01.003
32. Ma J, Li D, Kong FF, Yang D, Yang H, Ma XX. miR-302a-5p/367-3p-HMGA2 axis regulates malignant processes during endometrial cancer development. J Exp Clin Cancer Res. 2018;37(1):19. doi:10.1186/s13046-018-0686-6
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.