Back to Journals » OncoTargets and Therapy » Volume 13
Circ_0057553/miR-515-5p Regulates Prostate Cancer Cell Proliferation, Apoptosis, Migration, Invasion and Aerobic Glycolysis by Targeting YES1
Authors Zhang Y, Shi Z, Li Z, Wang X, Zheng P, Li H
Received 15 July 2020
Accepted for publication 28 September 2020
Published 4 November 2020 Volume 2020:13 Pages 11289—11299
DOI https://doi.org/10.2147/OTT.S272294
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Federico Perche
Yang Zhang, Zhenguo Shi, Zhijun Li, Xiaohui Wang, Pengyi Zheng, Huibing Li
Department of Urology Surgery, The First Affiliated Hospital, and College of Clinical Medicine of Henan University of Science and Technology, Luoyang, 471003, People’s Republic of China
Correspondence: Zhijun Li Tel +86 379-64830626
Email [email protected]
Background: Prostate cancer (PCa) is one of the most common malignant cancer in males worldwide. Circular RNAs (CircRNAs) are novel type of non-coding RNAs. Recently, circRNAs have been reported participating in various cancers, including prostate cancer. However, the function and mechanism of circ_0057553 remain to be elucidated.
Methods and Materials: The RNA expression levels of circ_0057553, miR-515-5p, YES proto-oncogene 1 (YES1) and glycolytic genes mRNA were detected by qRT-PCR in PCa tissues or cells. Western blotting was performed to analyze YES1 protein level. Cell viability, migration and invasion and cell apoptosis were assessed by cell counting kit-8 (CCK-8) assay, transwell assay and flow cytometry. In addition, the effects of cell glycolysis were evaluated by measuring lactate production, glucose consumption and adenosine triphosphate (ATP) level. Moreover, dual-luciferase reporter assay was used to detect the target sites of circ_0057553 and miR-515-5p, miR-515-5p and YES1. RNA immunoprecipitation (RIP) was conducted to evaluate the target relationship between circ_0057553 and miR-515-5p. Xenograft mouse model was conducted to measure tumor formation in vivo.
Results: Circ_0057553 was significantly up-regulated in PCa tissues and cells. Knockdown of circ_0057553 inhibited cell viability, migration, invasion and glycolysis and facilitated apoptosis in PCa cells. Furthermore, circ_0057553 bound to miR-515-5p and miR-515-5p directly targeted YES1. Interestingly, miR-515-5p inhibitor partially rescued the function of circ_0057553 knockdown, while YES1 restored the effects of miR-515-5p overexpression. Circ_0057553 down-regulation remarkably decreased tumor volume and weight in vivo.
Conclusion: Circ_0057553 affected PCa cell viability, migration, invasion, apoptosis and glycolysis through miR-515-5p/YES1 axis.
Keywords: circ_0057553, miR-515-5p, YES1, prostate cancer
Introduction
Even though current advances in treatments and diagnoses, prostate cancer (PCa) remains the second most frequent cancer and the third leading cause of death from cancer in males worldwide.1 For the treatments, advanced-stage prostate cancer patients need to face the huge economic burden.2 Furthermore, the novel screening strategy prostate-specific antigen (PSA) had a limitation of over-diagnosis.3 Therefore, it is urgent to develop more effective clinical therapeutic targets which can improve the treatments and diagnoses of PCa.
Circular RNAs (CircRNAs) comprise a type of non-coding RNAs which are produced by back-splicing. Different from the linear structure, the covalent ring structure has high stability and is not easily decomposed by exonuclease.4 For the biological functions of circRNAs, most circRNAs acted as the miRNA sponges.5 Moreover, emerging studies have shown that circRNAs can also directly interact with RNA-binding proteins (RBPs).6–8 In different tumor types, aberrant expression of circRNAs was associated with tumor cell biological activities.9 For example, circ_0000096 accelerated gastric cancer cells proliferation and migration.10 In addition, Hsiao et al proved that circRNA CCDC66 had the function of promoting colon cancer proliferation and Metastasis.11 Furthermore, a recent study showed that circ_103908 was suggested relating to stages and metastasis of lung cancer.12,13 These studies suggested that circRNA had significant potential in therapeutic targets. Previous study showed that circ_0057553 was significantly up-regulated in PCa.14 Nevertheless, the regulatory mechanisms of circ_0057553 in PCa remain unclear.
MiR-515-5p was reported to repress cancer cell migration through targeting MARK4 signal pathway.15 Moreover, one study proved that miR-515-5p was a tumor inhibitor in PCa through targeting TRIP13.16 YES proto-oncogene 1 (YES1) is a member of a non-receptor protein tyrosine kinase which can be found in many tumor formation process. In lung cancer, YES1 has been shown to be a tumor promoter driving cell growth.17 Meanwhile, YES1 can act as a targetable oncogene in breast cancer.18 Moreover, the expression of YES1 was significantly up-regulated in prostate cancer.19 In our present study, according to the starBase 2.0 and TargetScan software, we were surprised to predict that there were binding sites between circ_0057553 and miR-515-5p, and between miR-515-5p and YES1. Therefore, we speculated that circ_0057553 regulated prostate cancer progression through miR-515-5p/YES1 axis.
In this research, we explored the relationships among circ_0057553, miR-515-5p and YES1 in PCa and uncovered that circ_0057553/miR-515-5p/YES1 axis regulated cell viability, apoptosis, migration and invasion and glycolysis process in vitro. In addition, the tumor formation was detected in vivo. Thus, the purpose of our study was to find novel clinical targets for PCa and further understand the regulatory mechanisms of circ_0057553.
Methods and Materials
Patient Sample
After obtaining the informed consents from patients, thirty-seven PCa tissues and normal nearby tissues were gathered from PCa patients at The First Affiliated Hospital, and College of Clinical Medicine of Henan University of Science and Technology. All procedures were approved by the Ethics Committee of The First Affiliated Hospital, and College of Clinical Medicine of Henan University of Science and Technology.
Cell Culture and Transfection
The human PCa cell lines (22RV1, PC3, DU145, LNCap) and the normal human prostatic epithelial cell line (RWPE-1) were obtained from Cell Bank of Chinese Academy of Sciences (Shanghai, China). All cell lines were cultured in RPMI-1630 medium (Sigma-Aldrich, St. Louis, MO, USA) at 37°C. Ten percent fetal bovine serum (FBS), penicillin and streptomycin were added in culture medium. For the procedure of transfection, small interfering RNA against circ_0057553#1, circ_0057553#2 and circ_0057553#3 (si-circ_0057553#1, si-circ_0057553#2 and si-circ_0057553#3) and their control (si-NC), short-hairpin against circ_0057553 (sh-circ_0057553) and its control (sh-NC), circ_0057553 overexpression vector (circ_0057553) and Vector, miR-515-5p mimics and its scramble miR-NC, miR-515-5p inhibitor (in-miR-515-5p) and its control (in-miR-NC), and transfection plasmid vectors pcDNA and YES1 were bought from GenePharma Company (Shanghai, China). These experiments utilized the instructions of Lipofectamine 3000 (Thermo, Carlsbad, CA, USA).
Quantitative Real-Time PCR (qRT-PCR)
Total RNAs were isolated by TRIzol reagent (Invitrogen, Carlsbad, CA, USA). Then, cDNA Synthesis Kit (Vazyme, Nanjing, China) was utilized to reverse-transcribe under the manufacturer’s instructions. In addition, quantitative real-time PCR was performed using SYBR Green PCR Master Mix (Ambion, Carlsbad, CA, USA) and then processed by method of 2−ΔΔCt. The primers as follows: circ_0057553 (forward: 5ʹ- CGATCCATGTGGAATCCGAA-3ʹ and reverse: 5ʹ-ACGCTCACTGGGAGTAATGG-3ʹ), linear SLC39A10 (forward: 5ʹ- TCGCATACCCACCAGCATTT-3ʹ and reverse: 5ʹ- TCACTGTGAGCAACGGAGTC-3ʹ), miR-515-5p (forward: 5ʹ-TTCTCCAAAAGAAAGCACTTTCTG-3ʹ and reverse 5ʹ-CTCGCTTCGGCAGCACA-3ʹ), YES1 (forward: 5ʹ-AAAAGTTGGCATGGGCTGC-3ʹ and reverse 5ʹ-ACACTTCGCCGAAACATCCT-3ʹ), U6 (forward: 5ʹ-CTCGCTTCGGCAGCACATATACT-3ʹ and reverse: 5ʹ-ACGCTTCACGAATTT-GCGTGTC-3ʹ), GAPDH (forward: 5ʹ-TGTTCGTCATGGGTGTGAAC-3ʹ and reverse: 5ʹ-ATGGCATGGACTGTGGTCAT-3ʹ).
RNase R Digestion
CircRNAs stability was assessed by RNase R experiment. The extracted RNAs from PCa cells were separated with two parts. One part was added with RNase R digestion, the control was digested with digestion buffer. Then, the expression level of circ_0057553 and linear SLC39A10 were measured by qRT-PCR.
Western Blotting Assay
The protein extraction kit (Beyotime, Shanghai, China) was conducted for protein isolation. Protein concentration was examined by Pierce BCA Protein Assay Kit (Beyotime). The total proteins were loaded on 10% SDS-PAGE. And then proteins were transferred onto the PVDF membrane (GE Healthcare, Piscataway, NJ, USA). In addition, the membranes were blocked with 5% milk and added with primary antibodies at room temperature for 10 min, and left at 4°C overnight. Then, the second antibody was used for identification. Finally, we used eyoECL Plus Kit (Beyotime) for chemiluminescence intensity analysis. Primary antibodies were anti-YES1 (1:1000, Abcam, Cambridge, MA, USA) and anti-GAPDH (1:1000; Cell Signaling Technology, Danvers, MA, USA).
Cell Counting Kit-8 Assay
DU145 and LNCap cells (1×105 cells/well) were incubated in a 96-well plate for 0 h, 24 h, 48 h, and 72 h. Then, we added 10 μL CCK-8 solution. The optical density value was observed by Microplate Reader ELx808 (Bio-Tek, Winooski, Vermont, USA) at 450 nm.
Cell Migration and Invasion Assay
Cell migration was examined by transwell experiment. Transfected DU145 and LNCap cells cultured with serum-free medium and then seeded them into upper chamber. And the lower chamber containing the complete medium. The ability to migrate could be demonstrated by observing the number of cells stained by 0.1% crystal violet through the membrane. The invasion assay was added with the matrigel (BD Biosciences, San Jose, CA, USA) in upper chamber and the other steps were same with migration assay. All the experiments were carried out under a 100× optical microscope.
Cell Apoptosis Experiment
Cell apoptosis was detected by flow cytometry. In brief, transfected cells were washed with PBS twice and then resuspended in binding buffer. The Annexin V/propidium iodide (PI) kit (BD Pharmingen, San Diego, CA, USA) was used to double stain the cells to better observe the apoptosis cells. The cells were detected by flow cytometer (Cytek Biosciences, Shanghai, China).
Glycolysis Level
Glycolysis metabolism was analyzed through measuring lactate production, glucose consumption and adenosine triphosphate (ATP) level in DU145 and LNCap cells. After transfection for 48 h, we collected EU145, LNCap cells and their culture medium. Then, Lactate Assay Kit (BioVision, Milpitas, CA, USA) was used to measure the Lactate production in culture medium. In addition, Glucose Colorimetric Assay Kit (BioVision) was used to calculate the Glucose uptake in PCa cells, and finally, we used a specific ATP assay kit (Promega, Madison, Wisconsin, USA) to measure the change of ATP level in cells.
Dual-Luciferase Reporter Assay
The sequences of wild type circ_0057553, mutant circ_0057553, wild type YES1 (3ʹ UTR) and mutant YES1 (3ʹUTR) were cloned into the pGL3 luciferase vector (Promega, Madison, Wl, USA). Above vectors and miR-515-5p or miR-NC were cotransfected in the DU145 and LNCap cells. The transfection procedure was performed by Lipofectamine 3000 (Invitrogen). The luciferase activities were detected by Dual-luciferase Reporter Assay Kit (Promega).
RIP Assay
With the manufacturer’s instructions, RNA Immunoprecipitation experiment was conducted using Dynabeads Protein G Immunoprecipitation Kit (Thermo). In brief, transfected DU145 and LNCap cells were lysed with RIP lysis buffer. And then we used magnetic beads conjugated with anti-Argonaute 2 (Ago2) antibody (Abcam) or control anti-IgG (Abcam) to enrich RNA-protein complexes. In addition, RNA fragments isolated from the mixture with TRIzol Reagent were detected by qRT-PCR.
Mouse Model
Ten 4-week-old male nude mice were used. And LNCap cells containing negative control and circ_0057553 knockdown were injected subcutaneously. The tumor volumes were detected every 7 days and tumor weights were measured after the animals were sacrificed. All the animal studies were approved by the Animal Care committee of The First Affiliated Hospital, and College of Clinical Medicine of Henan University of Science and Technology, Animal studies were performed in compliance with the ARRIVE guidelines and the Basel Declaration. All animals received humane care according to the National Institutes of Health (USA) guidelines.
Statistical Analysis
For statistical analysis, we used GraphPad Prism 7.0 and SPSS 20.0 software. The difference between two groups was analyzed by the Student’s t-test. Furthermore, the difference among multiple groups was measured by one-way Analysis of Variance (ANOVA). The linear correlation was analyzed by Pearson correlation analysis. All the experimental data were showed as mean ± standard deviation and detected at least three times. P value <0.5 was considered significant.
Results
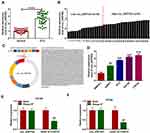
circ_0057553 Was Up-Regulated in PCa Tissues and Cells
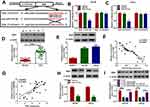
In order to investigate the expression level of circ_0057553 in PCa, we first measured circ_0057553 expression in PCa tissues and adjacent normal tissues using qRT-PCR. Compared with control, the expression level of circ_0057553 was significantly increased in PCa tissues (Figure 1A). And of these 37 expressions, 21 were higher and 16 were lower (Figure 1B). Furthermore, the biological structure and full sequence of circ_0057553 are shown in Figure 1C. The total length of circ_0057553 was 2348 nt, including 8 exons (2–9). The expression level of circ_0057553 was remarkably increased in PCa cell lines 22RV1, PC3, DU145 and LNCap with a comparison to control RWPE-1 cell line (Figure 1D). In addition, circ_0057553 was more difficult to decompose under the function of RNase R enzyme than linear SLC39A10 in DU145 and LNCap cells (Figure 1E and F). We speculated that circ_0057553 played a critical role in PCa progression and development.
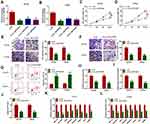
Knockdown of circ_0057553 Inhibited Cell Viability, Migration, Invasion and Glycolysis Process and Promoted Cell Apoptosis
To explore the cellular functions of circ_0057553 in PCa cell lines, we used loss-of-function experiments. The expression level of circ_0057553 was reduced after transfection with si-circ_0057553#1, si-circ_0057553#2 and si-circ_0057553#3 in DU145 and LNCap cell lines (Figure 2A and B). Knockdown of circ_0057553 inhibited cell viability in DU145 and LNCap cell lines (Figure 2C and D). Moreover, the transwell assay proved that the cell migration and invasion abilities were significantly reduced in DU145 and LNCap cells after transfection with si-circ_00575533#1 (Figure 2E and F). Meanwhile, circ_0057553 down-regulation promoted cell apoptosis in PCa cell lines (Figure 2G). Circ_0057553 silencing group decreased lactate production, glucose consumption and ATP level (Figure 2H–J). The glycolytic genes HIF-1A, SLC2A1, HK2, PFKFB2, PFKFB3, PGK1, TPI1 and PKM2 also were significantly decreased in circ_0057553 knockdown group in DU145 and LNCap cells (Figure 2K and L). These data suggested that downregulation of circ_0057553 affected cellular functions in PCa cell lines.
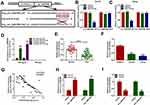
circ_0057553 Directly Targeted miR-515-5p
To confirm the potential targets of circ_0057553 in PCa, starbase 2.0 first predicted the binding sites of circ_0057553 and miR-515-5p (Figure 3A). In order to verify the targeting relationship between circ_0057553 and miR-515-5p, we used dual-luciferase reporter assay. The results showed that overexpression of miR-515-5p remarkably decreased luciferase activity in wild type circ_0057553 group, whereas mutant circ_0057553 group had no change (Figure 3B and C). So there was a correlation relationship between circ_0057553 and miR-515-5p in DU145 and LNCap cell lines. Besides, RNA immunoprecipitation (RIP) experiments indicated that enrichment of circ_0057553 was dramatically up-regulated when we performed antibody against Ago2 compared with anti-lgG (Figure 3D). Furthermore, the expression of miR-515-5p was evidently decreased in PCa tissues and cells with a comparison to normal control (Figure 3E and F). Therefore, there was a negative correlation between circ_0057553 and miR-515-5p (Figure 3G). QRT-PCR results indicated that overexpression of circ_0057553 up-regulated circ_0057553 expression, while greatly down-regulated miR-515-5p expression (Figure 3H and I). As expected, miR-515-5p was the direct target of circ_0057553 and there was a negative correlation between circ_0057553 and miR-515-5p.
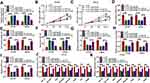
miR-515-5p Inhibitor Rescued the Effects of circ_0057553 Knockdown
We speculated that the interaction between circ_0057553 and miR-515-5p can affect the functional activity of the PCa cell lines. We first tested the expression level of miR-515-5p after transfection with si-circ_0057553#1 and miR-515-5p inhibitor. As presented in Figure 4A, miR-515-5p was significantly increased after transfection with circ_0057553 inhibitor and restored when addition with in-miR-515-5p. CCK-8 assay suggested that miR-515-5p inhibitor could rescue the effect of circ_0057553 knockdown on cell viability in DU146 and LNCap cells (Figure 4B and C). We subsequently tested cell migration and invasion abilities using transwell assay. The results demonstrated that knockdown of circ_0057553 reduced cell migration and invasion ability in PCa cells while addition with miR-515-5p inhibitor recovered the function of circ_0057553 knockdown in DU145 and LNCap cells (Figure 4D and E). In addition, the cell apoptosis was measured by flow cytometry and showed that cell apoptosis was up-regulated after transfection with si-circ_0057553#1, while inhibition of miR-515-5p reverted the impact of circ_0057553 knockdown on cell apoptosis in PCa cells (Figure 4F). For the glycolysis process, miR-515-5p inhibitor played a critical role to rescue the lactate production, glucose consumption and ATP production caused by circ_0057553 knockdown in PCa cells (Figure 4G–I). We further investigated glycolysis-related genes, and miR-515-5p inhibitor relieved the decrease in glycolytic gene expression caused by the knockdown of circ_0057553 in DU145 and LNCap cells (Figure 4J and K). From these data, it was confirmed that miR-515-5p inhibitor resuced the cellular activities caused by circ_0057553 knockdown in PCa cells.
YES1 Acted as the Direct Target of miR-515-5p
In order to find downstream target genes of miR-515-5p, we first used starbase 2.0 prediction and found binding sites between 3ʹUTR of YES1 and miR-515-5p (Figure 5A). Furthermore, the dual-luciferase reporter assay was conducted to analyze correlation between miR-515-5p and YES1 and confirmed that miR-515-5p directly targeted YES1 in DU145 and LNCap cells (Figure 5B and C). In addition, the Western blotting assay showed that the expression of YES1 protein in PCa tissues and cells was significantly up-regulated compared with normal controls (Figure 5D and E). As shown in Figure 5F and G, miR-515-5p was negatively correlated with YES1, whereas circ_0057553 was positively correlated with YES1. Moreover, YES1 expression was evidently decreased by overexpression of miR-515-5p (Figure 5H). Then, we used Western blotting assay to detect YES1 expression after transfection with si-circ_0057553#1 and in-miR-515-5p. Interestingly, when inhibition of circ_0057553, the YES1 expression was significantly down-regulated, while this change would recover by inhibition of miR-515-5p (Figure 5I). These data suggested that miR-515-5p directly targeted YES1 and had an inverse correlation with YES1.
YES1 Rescued the Function of miR-515-5p
We have demonstrated that miR-515-5p directly targeted YES1, and in order to further investigate the potential function of miR-515-5p and YES1 in PCa, we conducted the following studies. Western blotting assay found that the protein expression level of YES1 was significantly decreased after transfection with miR-515-5p, while this change was alleviated by overexpression of YES1 (Figure 6A). In addition, the cell viability was markedly reduced when addition with miR-515-5p, whereas this effect was recovered by transfecting YES1 in DU145 and LNCap cells (Figure 6B and C). Transwell assay revealed that cell migration and cell invasion were incredibly decreased in miR-515-5p overexpression group; however, transfection with YES1 reversed the effects of miR-515-5p in DU145 and LNCap cell lines (Figure 6D and E). For the cell apoptosis experiment, the apoptosis rate of miR-515-5p overexpression group was remarkably increased, while the effects were rescued by addition with YES1 (Figure 6F). Furthermore, miR-515-5p could rescue the impact of YES1 overexpression on lactate production level, glucose consumption and ATP production (Figure 6G–I). Moreover, the expression level of glycolytic genes was evidently decreased after transfection with miR-515-5p, while addition of YES1 could reverse the expression of glycolysis-related genes in DU145 and LNCap cell lines (Figure 6J and K). These results indicated that YES1 could rescue cell viability, cell migration and invasion, cell apoptosis and glycolysis process caused by miR-515-5p.
circ_0057553 Knockdown Affected Tumor Volume and Weight in vivo
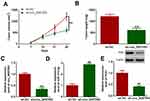
To further understand the function of circ_0057553 in PCa, we conducted in vivo experiments. Tumor volume was significantly decreased in circ_0057553 knockdown group (Figure 7A) and silencing circ_0057553 greatly suppressed tumor weight (Figure 7B). Moreover, the expression levels of circ_0057553 and YES1 were significantly decreased after transfection with sh-circ_0057553, while knockdown of circ_0057553 up-regulated miR-515-5p expression in vivo (Figure 7C–E). These results suggest that circ_0057553 affected tumor growth in vivo through miR-515-5p/YES1 axis.
Discussion
PCa is a common cancer in men and the number of PCa associated deaths worldwide was 359,000 in 2018.1 We found that circ_0057553 was up-regulated in PCa tissues, DU145 and LNCap cell lines. In addition, compared with linear SLC39A10, circ_0057553 which had the conversely ring structure was less likely to be decomposed by RNase R enzyme. Moreover, circ_0057553 facilitated cell viability, migration and invasion and restrained cell apoptosis in PCa cell lines. Glycolysis metabolism is needed to meet the rapid growth of tumor cells.20 We further studied three variables in the glycolysis process. Lactate production, glucose consumption and ATP level were markedly decreased after transfection of circ_0057553 knockdown.
MiR-515-5p acted as a tumor suppressor in multiple cancers. For example, miR-515-5p impeded lung squamous cell carcinoma development by acting on YAP1.21 Zhang et al proposed that miR-515-5p suppressed proliferation in breast cancer cells.16 Li et al demonstrated that down-regulation of miR-515-5p promoted non-small cell lung cancer cell viability and metastasis.22 In our present study, miR-515-5p expression was significantly down-regulated in PCa tissues and cells. Starbase 2.0 was used to predict the binding sites of circ_0057553 and miR-515-5p. In addition, there was an inverse correlation between circ_0057553 and miR-515-5p. Overexpression of circ_0057553 down-regulated the expression of miR-515-5p. Interestingly, we displayed that miR-515-5p inhibitor could partially rescue the function of circ_0057553 knockdown in cell viability, migration, invasion, and apoptosis. Furthermore, the changes in glycolysis caused by down-regulation of circ_0057553 can also be recovered by miR-515-5p inhibitors.
YES1 is a non-receptor tyrosine kinase that consists with the SRC family kinases (SFKs). Previous studies indicated that YES1 acted as a tumor promoter in lung cancer,17 gastric cancer23 and thyroid cancer.24 Our results were in agreement with the previously described YES1 functions in tumor cells. The expression of YES1 was remarkably increased in PCa tissues as well as DU145 and LNCap cells. Furthermore, we applied TargetScan software to predict the binding sites between miR-515-5p and YES1. We found that there was an inverse correlation between YES1 and miR-515-5p. Interestingly, cell viability, migration and invasion experiments demonstrated that YES1 could restore the effects of miR-515-5p in PCa cells. For glycolysis process, YES1 had the role of up-regulating the lactate production, glucose consumption and ATP level to rescue the effects of miR-515-5p. Moreover, we used xenograft mouse model to further prove the mechanism of circ_0057553/miR-515-5p/YES1 axis in vivo.
In conclusion, we exhibited that circ_0057553 was increased in PCa tissues and cells. In addition, circ_0057553 promoted cell viability, migration, invasion and glycolysis process and suppressed cell apoptosis through miR-515-5p/YES1 axis. Furthermore, circ_0057553 affected tumor growth in vivo. Thus, our study suggested that circ_0057553/miR-515-5p/YES1 axis would provide potential clinic targets in the future.
Funding
There is no funding to report.
Disclosure
The authors declare that they have no financial or non-financial conflicts of interest for this work.
References
1. Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
2. Crawford ED, Black L, Eaddy M, et al. A retrospective analysis illustrating the substantial clinical and economic burden of prostate cancer. Prostate Cancer Prostatic Dis. 2010;13(2):162–167. doi:10.1038/pcan.2009.63
3. Loeb S, Bjurlin MA, Nicholson J, et al. Overdiagnosis and overtreatment of prostate cancer. Eur Urol. 2014;65(6):1046–1055. doi:10.1016/j.eururo.2013.12.062
4. Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. Rna. 2013;19(2):141–157. doi:10.1261/rna.035667.112
5. Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi:10.1038/nature11993
6. Holdt LM, Stahringer A, Sass K, et al. Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans. Nat Commun. 2016;7:12429. doi:10.1038/ncomms12429
7. Dudekula DB, Panda AC, Grammatikakis I, et al. CircInteractome: a web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. 2016;13(1):34–42. doi:10.1080/15476286.2015.1128065
8. Wang Z, Lei X, Wu FX. Identifying cancer-specific circRNA-RBP binding sites based on deep learning. Molecules. 2019;24(22).
9. Zhang HD, Jiang LH, Sun DW, et al. CircRNA: a novel type of biomarker for cancer. Breast Cancer. 2018;25(1):1–7. doi:10.1007/s12282-017-0793-9
10. Li P, Chen H, Chen S, et al. Circular RNA 0000096 affects cell growth and migration in gastric cancer. Br J Cancer. 2017;116(5):626–633. doi:10.1038/bjc.2016.451
11. Hsiao KY, Lin YC, Gupta SK, et al. Noncoding effects of circular RNA CCDC66 promote colon cancer growth and metastasis. Cancer Res. 2017;77(9):2339–2350. doi:10.1158/0008-5472.CAN-16-1883
12. Cheng XY, Shen H. [Circular RNA in lung cancer research: biogenesis, functions and roles]. Zhongguo Fei Ai Za Zhi. 2018;21(1):50–56. Chinese.
13. Liu W, Ma W, Yuan Y, et al. Circular RNA hsa_circRNA_103809 promotes lung cancer progression via facilitating ZNF121-dependent MYC expression by sequestering miR-4302. Biochem Biophys Res Commun. 2018;500(4):846–851. doi:10.1016/j.bbrc.2018.04.172
14. Shan G, Shao B, Liu Q, et al. circFMN2 sponges miR-1238 to promote the expression of LIM-homeobox gene 2 in prostate cancer cells. Mol Ther Nucleic Acids. 2020;21:133–146. doi:10.1016/j.omtn.2020.05.008
15. Pardo OE, Castellano L, Munro CE, et al. miR-515-5p controls cancer cell migration through MARK4 regulation. EMBO Rep. 2016;17(4):570–584. doi:10.15252/embr.201540970
16. Zhang X, Zhou J, Xue D, et al. MiR-515-5p acts as a tumor suppressor via targeting TRIP13 in prostate cancer. Int J Biol Macromol. 2019;129:227–232. doi:10.1016/j.ijbiomac.2019.01.127
17. Garmendia I, Pajares MJ, Hermida-Prado F, et al. YES1 drives lung cancer growth and progression and predicts sensitivity to dasatinib. Am J Respir Crit Care Med. 2019;200(7):888–899. doi:10.1164/rccm.201807-1292OC
18. Takeda T, Yamamoto H, Kanzaki H, et al. Yes1 signaling mediates the resistance to Trastuzumab/Lap atinib in breast cancer. PLoS One. 2017;12(2):e0171356. doi:10.1371/journal.pone.0171356
19. Chen L, Cao H, Feng Y. MiR-199a suppresses prostate cancer paclitaxel resistance by targeting YES1. World J Urol. 2018;36(3):357–365. doi:10.1007/s00345-017-2143-0
20. Bose S, Le A. Glucose metabolism in cancer. Adv Exp Med Biol. 2018;1063:3–12.
21. Ye P, Lv X, Aizemaiti R, et al. H3K27ac-activated LINC00519 promotes lung squamous cell carcinoma progression by targeting miR-450b-5p/miR-515-5p/YAP1 axis. Cell Prolif. 2020;e12797.
22. Li J, Tang Z, Wang H, et al. CXCL6 promotes non-small cell lung cancer cell survival and metastasis via down-regulation of miR-515-5p. Biomed Pharmacother. 2018;97:1182–1188. doi:10.1016/j.biopha.2017.11.004
23. Fang Z, Yin S, Sun R, et al. miR-140-5p suppresses the proliferation, migration and invasion of gastric cancer by regulating YES1. Mol Cancer. 2017;16(1):139. doi:10.1186/s12943-017-0708-6
24. Liu L, Yang J, Zhu X, et al. Long noncoding RNA H19 competitively binds miR-17-5p to regulate YES1 expression in thyroid cancer. FEBS J. 2016;283(12):2326–2339. doi:10.1111/febs.13741
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.