Back to Journals » OncoTargets and Therapy » Volume 14
Circ_0007444 Inhibits the Progression of Ovarian Cancer via Mediating the miR-570-3p/PTEN Axis
Authors Wu X, Liu D, Wang S, Liu J
Received 4 June 2020
Accepted for publication 11 November 2020
Published 7 January 2021 Volume 2021:14 Pages 97—110
DOI https://doi.org/10.2147/OTT.S266186
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Gaetano Romano
Xinyu Wu,* Daoyan Liu,* Shuzhen Wang, Jie Liu
Department of Laboratory Medicine, The Affiliated Maternity and Child Health Care Hospital, Xuzhou Medical University, Xuzhou 221009, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Jie Liu
Department of Laboratory Medicine, The Affiliated Maternity and Child Health Care Hospital, Xuzhou Medical University, NO. 46 Heping Road, Xuzhou 221009, People’s Republic of China
Tel +86-516-83907033
Email [email protected]
Shuzhen WangEmail Email [email protected]
Purpose: Some circular RNAs have been found to be effective therapeutic targets for OC. However, the biological function of circ_0007444 in OC is still unknown. Thus, this study investigated the role of circ_0007444 in OC progression.
Methods: circ_0007444 expression was monitored in 87 OC patients and OC cells by quantitative real-time polymerase chain reaction. An in vitro study was performed to research the biological function of circ_0007444, including cell counting kit-8 assay, flow cytometry, wound healing assay, and transwell experiment. Luciferase reporter gene assay and RNA immunoprecipitation assay were used to reveal the interaction between circ_0007444, miR-570-3p, and PTEN. PTEN protein expression was determined by Western blot. In vivo study was performed using nude mice. Ki67, PTEN expression, and apoptosis in xenograft tumors was respectively researched by immunohistochemistry and Tunel assay.
Results: circ_0007444 was down-regulated in 87 OC patients, which was related to advanced tumor stage and grade, large tumor size, and low 60-month percent survival (P< 0.05 or P< 0.01). circ_0007444 inhibited proliferation, migration, and invasion, and promoted apoptosis of OC cells (P< 0.01). circ_0007444 promoted PTEN expression via sponging miR-570-3p. miR-570-3p up-regulation and PTEN down-regulation reversed the inhibitory effect of circ_0007444 on OC cells malignant phenotype (P< 0.01). circ_0007444 inhibited OC growth in vivo. In xenograft tumor, circ_0007444 decreased Ki67 expression but increased PTEN expression and apoptosis.
Conclusion: circ_0007444 is a tumor suppressor in OC, which inhibits OC progression by mediating the miR-570-3p/PTEN. circ_0007444 can be a potential candidate for targeted therapy of OC.
Keywords: OC, circ_0007444, miR-570-3p, PTEN, progression
Introduction
Ovarian cancer (OC) is one of the most common malignant tumors that occur in the female reproductive system. The number of OC incidence increases at a rate of 200,000 cases per year.1 Even worse, 70% of ovarian cancer patients will relapse, posing a serious threat to their lives.2 Traditional treatments, including surgery, radiotherapy, and chemotherapy, are effective for most patients with early OC. However, the high postoperative recurrence rate and insensitivity to radiotherapy and chemotherapy still poses great adverse effects on the prognosis of OC patients. In addition, for patients with advanced OC, these traditional treatment strategies cannot save their lives in most cases.3 These challenges urgently require us to explore the pathogenesis of OC, so as to provide effective therapeutic targets for the diagnosis and treatment of OC and to design more effective OC targeted therapeutic drugs.
Circular RNAs (circRNAs) are a class of endogenous non-coding RNA which has been found to be commonly expressed in eukaryotic genes.4 In recent years, circRNAs have been demonstrated to play an important role in biological processes including proliferation, invasion and apoptosis of malignant tumors.5,6 Of course, the functions of several circRNAs in OC have also been elucidated. In general, circ-CSPP1 acted as a cancer-promoting gene in OC, which exacerbated OC cells proliferation, migration, and invasion via sponging miR-1236-3p.7 Pei et al8 revealed the carcinogenic role of circ_0013958 in OC. They declared that circ_0013958 facilitated the malignant development of OC through promotion of epithelial-mesenchymal transition and inhibition of the apoptotic signaling pathways. On the contrary, circPLEKHM3 was proved to be a tumor suppressor in OC. It inhibited OC progression through sponging miR-9.9 Moreover, circ-ITCH was discovered to weaken the malignant progression of OC both in vitro and in vivo via sponging miR-145.10 The discovery of more circRNAs that regulates the development of OC will provide a more effective choice for OC targeted therapy.
Recently, Zhu et al11 revealed that circ_0007444 expression was declined in colorectal cancer. It was suggested to be used as a prognostic indicator of colorectal cancer. As a newly discovered circRNA, the function of circ_0007444 in the development of other tumors, including OC, has not yet been reported. Hence, this paper studied the role of circ_0007444 in OC progression for the first time. circRNAs are usually found to be involved in tumor regulation by sponging miRNAs.12 Through online analysis of bioinformatics, we noticed that miR-570-3p possessed binding sites for circ_0007444 and PTEN might be a target gene for miR-570-3p. Based on this, this article fully reported the internal molecular mechanism of circ_0007444 in regulating OC progression with miR-570-3p/PTEN as the axis. The findings of this study will provide a novel target for the treatment of OC.
Methods
Patients and Tissues
OC patients (n=87) who were admitted to our hospital from March 2011 to September 2014 were enrolled in this research. All patients were diagnosed with OC for the first time and underwent surgical resection in our hospital. The inclusion criteria of patients were as follows: 1) Patients who volunteered to participate in this study; 2) Patients diagnosed with OC for the first time; and 3) Patients without a previous treatment history of cancer-related diseases. The exclusion criteria for patients were as follows: 1) Patients who were not willing to join the study; and 2) Patients with previous treatment history of cancer-related diseases. During surgery, tumor tissues and adjacent normal tissues were collected and quickly frozen in liquid nitrogen. Clinical features, including ages, FIGO stage, grade, distant metastasis and tumor size, were recorded. After surgery, all patients were followed up for 60 months.
All subjects voluntarily participated in this study and written informed consent was obtained from all patients. This research was approved by the ethics committee of Xuzhou Medical University in line with the Declaration of Helsinki.
Detection of Circular Structure Stability of Circ_0007444
The circular structure stability of circ_0007444 in OC was monitored using Ribonuclease R (RNase R, Geneseed Biotech, Guangzhou, China). Tumor tissues of OC patients were incubated with TRIzol reagent (Thermo Fisher Scientific, Waltham, MA, USA) in order to extract total RNA. For each total RNA sample, 3.0 μg was collected for 30 minutes of incubation with 10 U RNase R at 37 °C. Thereafter, circ_0007444 and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression was detected by quantitative real-time polymerase chain reaction (qRT-PCR). Notably, 3.0 μg of each total RNA sample without any treatment was also collected to detect the expression of circ_0007444 and GAPDH expression (served as Mock) by qRT-PCR.
Cell Lines
Human ovarian epithelial cell line (HOSEpiC) and OC cell lines (SKOV3, OV420, A2780, CAOV3, and OVCAR3) were commercially provided by the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). Cells were separately cultured in Dulbecco’s modified eagle’s medium (DMEM) and maintained at 37°C, 5% CO2. It was worth noting that 10% (v/v) fetal bovine serum (FBS), and 100 U/mL penicillin/streptomycin were supplemented in the DMEM. The medium was changed every 3 days.
Transfection
SKOV3 and OVCAR3 cells were separately dispersed in serum free DMEM to a density of 2×106 cells/mL. Then a total of 1 mL cell suspension was added into each well of 6-well plates. The circ_0007444 sequence was designed and synthesized by Gene Pharma (Shanghai, China), followed by being cloned into pcDNA3.1 vectors (Thermo Fisher Scientific, Waltham, MA, USA) according to the instructions. pcDNA3.1-circ_0007444 vectors and empty pcDNA3.1 vectors were respectively transfected into OVCAR3 cells (set as the oe-circ_0007444 group and oe-NC group). In addition, shRNA targeting circ_0007444 and corresponding negative control (Gene Pharma, Shanghai, China) were used to transfect SKOV3 cells (set as sh-circ_0007444 group and sh-NC group). miR-570-3p mimic and corresponding negative control were obtained (Gene Pharma, Shanghai, China), which were applied for the transfection of SKOV3 and OVCAR3 cells (set as miR-570-3p mimic group and miR-NC group).
For OVCAR3 cells transfected by pcDNA3.1-circ_0007444 vectors, they were further experienced cotransfection with miR-570-3p mimic or corresponding negative control (set as oe-circ_0007444 + miR-570-3p mimic group and oe-circ_0007444 + miR-NC group). In addition, PTEN siRNA or corresponding negative control (Gene Pharma, Shanghai, China) was also used to cotransfect OVCAR3 cells which have been transfected by pcDNA3.1-circ_0007444 vectors previously (set as oe-circ_0007444 + si-PTEN group and oe-circ_0007444 + si-NC group). All transfection operations were performed according to the instructions of Lipofectamine 2000 reagent (Thermo Fisher Scientific, Waltham, MA, USA). After 8 hours of incubation at 37°C, 5% CO2, cells were cultured with DMEM containing 10% FBS.
Cell Counting Kit (CCK)-8 Assay
The proliferative capacity of SKOV3 and OVCAR3 cells was monitored by CCK-8 assay. Cells were dispersed in DMEM (with 10% FBS) to a density of 1×106 cells/mL. A total of 100 μL cell suspension was added into each well of 96-well plates. DMEM containing 10% FBS (100 μL) was added into each well for the culture of cells. Cells were kept at 37°C, 5% CO2. The proliferation of cells were monitored every 24 hours. Briefly, CCK-8 regent (10 μL, Solarbio, Beijing, China) was added into wells to incubate cells for 4 hours at 37°C. The optical density (OD) value of wells was monitored using a microplate reader (Bio-Tek Instruments Inc., Winooski, VT, USA) at 450 nm wavelength.
Flow Cytometry
SKOV3 and OVCAR3 cells were harvested after 48 hours of transfection, followed by being centrifuged for 5 minutes at 1,000 rpm. A total of 1×106 cells were collected and washed with pre-cooled phosphate buffered saline (PBS) 3-times. Thereafter, 1× binding buffer (500 μL) was used to disperse these cells. Annexin V-FITC and propidium iodide (PI) were added into cells. After 15 minutes of incubation in the dark at room temperature, apoptotic cells were detected using flow cytometry.
Wound Healing Assay
The migration ability of cells was evaluated by wound healing assay. SKOV3 and OVCAR3 cells were seeded in 6-well plates with 1 m of DMEM containing 10% FBS at 37°C, 5% CO2. After cells were attached to the bottom of the well plate, a scratch was made using a sterile pipette tip. The width of this original scratch was recorded. Afterward, residual liquid in the wells was replaced by fresh DMEM containing 10% FBS. Cells were maintained at 37°C, 5% CO2 for 24 hours. The width of the scratch at 24 hours was then measured and recorded. The relative wound width was then calculated with the formula of (the scratch at 24 hours/the original scratch).
Transwell Experiment
Cell invasion ability was assessed by transwell experiment. The transwell chamber was inserted into 6-well plates containing 600 μL of DMEM (with 10% FBS). Matrigel (Sigma-Aldrich, St Louis, MO, USA) was pre-spread on the upper chamber. SKOV3 and OVCAR3 cells were collected and seeded in the upper chamber with 500 μL of serum-free DMEM. After 24 hours of culture at 37°C, 5% CO2, cells on the upper surface of the membrane was gently scraped off with a cotton swab. Cells on the lower surface were fixed with 4% paraformaldehyde and stained with 0.1% crystal violet for 10 minutes. The invasion cells were observed under a microscope (Olympus, Tokyo, Japan). The number of invasion cells was counted with five non-overlapping random fields.
Luciferase Reporter Gene Assay
According to the predictions of Circular RNA Interactome and miRDB online software, circ_0007444 possessed binding sites for miR-570-3p in the 3ʹ-UTR region. In addition, TargetScan online software indicated that PTEN had binding sites for miR-570-3p in the 3ʹ-UTR region. In order to verify the targeting relationship between two genes, a luciferase reporter gene assay was performed. Briefly, based on the prediction results from these online software, the wild type (WT) and mutant type (Mut) sequences of circ_0007444, as well as WT and Mut sequences of PTEN was designed and synthesized (Gene Pharma, Shanghai, China). These sequences were cloned and loaded into the pmirGLO luciferase vectors. SKOV3 and OVCAR3 cells dispersed in serum-free DMEM were transfected by miR-570-3p mimic (miR-570-3p mimic group) and corresponding negative control (miR-NC group), as well as miR-570-3p inhibitor (miR-570-3p inhibitor group) and corresponding negative control (NC-inh group). Subsequently, WT-circ_0007444-pmirGLO luciferase vectors, Mut-circ_0007444-pmirGLO luciferase vectors, WT-PTEN-pmirGLO luciferase vectors, and Mut-PTEN-pmirGLO luciferase vectors were used respectively to cotransfected SKOV3 and OVCAR3 cells of the above four groups. Cells were cultured at 37°C, 5% CO2 for 8 hours, followed by being cultured with DMEM containing 10% FBS for 48 hours. Dual-Glo luciferase assay kit (Promega, Madison, WI, USA) was responsible for the detection of relative luciferase activity.
RNA Immunoprecipitation (RIP) Assay
RIP assay was conducted by using the IgG and AGO2 antibody. Briefly, the lysate of SKOV3 cells of miR-570-3p mimic group and miR-NC group were obtained. Cell lysate sample with a volume of 10 μL was used as the input. The magnetic beads were incubated with IgG and AGO2 antibody, respectively, at 4°C, followed by being mixed with the lysate. After being purified, RNA was dissolved in 20 μL of diethylpyrocarbonate-treated water. The expression of circ_0007444 in each sample was then detected by qRT-PCR.
qRT-PCR
Total RNA extraction in tissues and cells were carried out using Trizol reagent. Based on the instructions, PrimeScript RT reagent Kit (TaKaRa, Shiga, Japan) was responsible for the synthesis of cDNA template with 5 μg of total RNA sample. Subsequently, the cDNA was quantified by SYBR Green reagent (TaKaRa, Shiga, Japan) using the ABI 7500 fast real-time PCR System (Applied Biosystems, Foster City, CA, USA). The reaction conditions were as follows: 94°C for 5 minutes, followed by 40 cycles of 94°C for 45 seconds, 56°C for 45 seconds, and 72°C for 40 seconds. The expression of circ_0007444, miR-570-3p, and PTEN mRNA was determined by 2−ΔΔCt method. U6 was used as the control for circ_0007444 and miR-570-3p, while β-actin was used as the control for PTEN. Primers were synthesized by GenePharma (Shanghai, China) and listed as follows: circ_0007444, forward, 5′-AAGTTGAAAGATTCTGGGGATG-3′, reverse, 5′-TGTGACGCTTCAGCCTTT-3′, miR-570-3p, forward, 5′-CGAAAACAGCAATTACCTTTGC-3′, reverse, 5′-TGGTGTCGTGGAGTCG-3′. U6, forward, 5′-CTCGCTTCGGCAGCACA-3′, reverse, 5′-AACGCTTCACGAATTTGCGT-3′. PTEN, forward, 5′-CGGCAGCATCAAATGTTTCAG-3′, reverse, 5′-AACTGGCAGGTAGAAGGCAACTC-3′. β-actin, forward, 5′-CCAAGGCCAACCGCG AGAAGATGAC-3′, reverse, 5′-AGGGTACATGGTGGTGCCGCCAGAC-3′.
Western Blot
Western blot was responsible for the detection of PTEN protein expression in cells. Briefly, SKOV3 and OVCAR3 cells were collected and incubated with RIPA lysis buffer (Solarbio, Beijing, China) on ice for 30 minutes. BCA kit (Beyotime, Shanghai, China) was used to determine the concentration of total proteins. Protein extract (10 μL) was collected and experienced separation with 10% sodium dodecyl sulphate-polyacrylamide gel electrophoresis (SDS-PAGE). Thereafter, proteins were transferred onto PVDF membrane, followed by being blocked with 5% skimmed milk for 30 minutes at room temperature. Specific primary antibody was used to incubate the membrane overnight at 4°C. β‑actin was set as control. The specific primary antibodies used in this study were mouse monoclonal anti-PTEN antibody (1:1,000, Cascade Biosciences, Portland, OR, USA) and mouse anti-β‑actin (1:1,000, Novus Biologicals, Littleton, CO, USA). Subsequently, horseradish peroxidase (HRP)-conjugated goat-anti mouse secondary antibody (1:2,000, Solarbio, Beijing, China) was added onto the membrane for 2 hours incubation at room temperature. The protein bands were visualized using the enhanced chemiluminescence system (Pierce Biotechnology, Rockford, IL, USA).
In vivo Study
In this study, animal experiments have been approved by the Animal Ethics Committee of Xuzhou Medical University. The animal experiments were performed in accordance with relevant guidelines and regulations of the Animal Care and Use Committees at the Xuzhou Medical University, and a signed document issued by the Animal Care and Use Committees that granted approval was obtained. Female nude mice at 5-weeks-old (n=24) were commercially obtained from the Shanghai Experimental Animal Center, Chinese Academy of Sciences (Shanghai, China). Mice were kept in a room without specific pathogens. The cycle for day and night was 12 hours. All mice had free access to food and water.
OVCAR3 cells of the oe-circ_0007444 group and oe-NC group were collected and dispersed in PBS (2×106 cells/mL). Moreover, SKOV3 cells of the sh-circ_0007444 group and sh-NC group were also prepared as cell suspensions with PBS to a density of 2×106 cells/mL. Cells of each group were injected subcutaneously into the back of six nude mice. The injection volume of each cell suspension was 100 μL. The tumor volume was measured every 7 days with the equation of V=0.5×a×b2. Notably, “a” represented the longitudinal diameter and “b” represented the latitudinal diameter. On the 28th day of post-injection, mice were sacrificed by rapid neck dislocation to collect the xenograft tumors. The weight of xenograft tumors was then measured.
Immunohistochemistry
Xenograft tumors were fixed with 10% formaldehyde and embedded in paraffin, followed by being sliced, dewaxed, and hydrated. Antigen retrieval was performed using 0.01 M citrate buffer. H2O2 solution (3%) was applied to incubate sections for 15 minutes at room temperature. Goat serum (5%) was added onto sections for 30 minutes incubation at room temperature. Thereafter, sections were sequentially subjected to incubation with primary antibody (1:100, mouse anti-Ki67 and anti-PTEN, Geneseed Biotech, Guangzhou, China) overnight at 4°C and with HRP labeled secondary antibody (1:300, Boster, Wuhan, China) for 30 minutes at room temperature. After being stained with diaminobenzidine (DAB) and hematoxylin, sections were sealed with neutral resin. The positive signals of Ki67 and PTEN were observed under a microscope (Olympus, Tokyo, Japan).
Tunel Assay
Xenograft tumor sections were dewaxed and hydrated, followed by being incubated with proteinase K for 20 minutes at 37°C. Then, H2O2 (3%) and 0.1% TritonX-100 were added onto sections to incubate for 10 minutes. TUNEL reaction solution was used to incubate sections for 60 minutes at 37°C. After being washed with PBS, DAB and hematoxylin weres used to stain sections for 5 minutes at room temperature. Ethanol was used for the dehydration and xylene was responsible for transparency. Sections were sealed with neutral resin, and the Tunel positive cells were observed under a microscope (Olympus, Tokyo, Japan).
Statistical Analysis
SPSS 19.0 software (SPSS, Inc., Chicago, IL, USA) was used for the statistical analysis. GraphPad Prism 6 was responsible for the process of graph. The differences between groups were analyzed by Student’s t-test. One-way analysis of variance was the method used for the comparison of more than two groups. Survival curves were made based on Kaplan-Meier survival analysis and Log rank tests. Pearson’s correlation analysis was applied for the analysis of the expression correlation between two genes. P<0.05 was considered statistically significant.
Results
Low Expression of Circ_0007444 in OC Patients Predicted Poor Prognosis
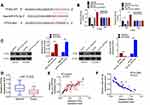
In 87 cases with OC, circ_0007444 expression in their tumor tissues was reduced severely relative to adjacent normal tissues, with statistically significant differences (P<0.01) (Figure 1A). According to analysis of clinical features, it could be noted that OC cases with low circ_0007444 expression exhibited advanced tumor stage and grade (P<0.05), and large tumor size (P<0.01) (Figure 1B, Table 1). Meanwhile, patients with low circ_0007444 expression presented distinctly lower 60-month percent survival (P<0.05) (Figure 1C). Hence, circ_0007444 expression was abnormally reduced in OC cases, which was significantly associated with poor prognosis.
 |
Table 1 Relationship Between hsa_circ_0007444 Expression and Clinical Features of OC Patients |
In addition, the circular structure stability of circ_0007444 was monitored. As presented in Figure 1D, RNase R treatment prominently decreased the expression of GAPDH (P<0.01). By contrast, circ_0007444 expression was not obviously changed by treatment of RNase R. It indicated the stable circular structure of circ_0007444, and circ_0007444 could be stably expressed in OC. In vitro research further indicated the down-regulated circ_0007444 in OC, because remarkably lower circ_0007444 expression was observed in OC cells than that in HOSEpiC cells (P<0.01) (Figure 1E).
Circ_0007444 Inhibited Proliferation, Migration, Invasion, and Promoted Apoptosis of OC Cells
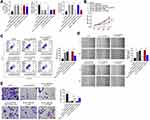
From Figure 2A, the expression of circ_0007444 in SKOV3 cells of sh-circ_0007444 group was markedly lower than that of sh-NC group (P<0.01). Meanwhile, circ_0007444 expression in OVCAR3 cells of the oe-circ_0007444 group was significantly higher than that of oe-NC group (P<0.01). Thus, the expression of circ_0007444 in SKOV3 and OVCAR3 cells was successfully regulated by transfection.
After transfection, the malignant phenotype of SKOV3 and OVCAR3 cells was studied in vitro. The proliferation was monitored by CCK-8 assay. As a result, SKOV3 cells of sh-circ_0007444 group presented dramatically higher OD value than that of the sh-NC group (P<0.01). On the contrary, OVCAR3 cells of the oe-circ_0007444 group exhibited remarkably lower OD value than that of the oe-NC group (P<0.01) (Figure 2B). Flow cytometry results showed that, relative to the sh-NC group, much lower apoptosis was found in SKOV3 cells of the sh-circ_0007444 group (P<0.01). However, compared with the oe-NC group, OVCAR3 cells of the oe-circ_0007444 group had obviously higher apoptosis (P<0.01) (Figure 2C). Wound healing assay was responsible for cells migration ability detection. The relative wound width of SKOV3 cells in the sh-circ_0007444 group was prominently narrower than that of the sh-NC group (P<0.01). Oppositely, markedly wider relative wound width was observed in OVCAR3 cells of the oe-circ_0007444 group in comparison with the oe-NC group (P<0.01) (Figure 2D). According to transwell experiment, a higher invasive cell number was found in SKOV3 cells in the sh-circ_0007444 group when relative to the sh-NC group (P<0.01). Conversely, a less invasive cell number was observed in OVCAR3 cells of the oe-circ_0007444 group when compared with the oe-NC group (P<0.01) (Figure 2E). Therefore, circ_0007444 attenuated the malignant phenotype of OC cells in vitro.
Circ_0007444 Acted as a Sponge for miR-570-3p
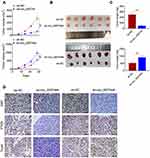
Prediction of Circular RNA Interactome and miRDB illustrated that miR-570-3p and circ_0007444 had common binding sites. Based on this, circ_0007444-WT and circ_0007444-Mut fragments containing the binding sites for miR-570-3p was designed and synthesized (Figure 3A). These fragments were loaded into luciferase reporter vectors to further verify the relationship between circ_0007444 and miR-570-3p through luciferase reporter gene assay. As a result, relative to the miR-NC group, SKOV3 and OVCAR3 cells of the miR-570-3p mimic group exhibited much lower relative luciferase activity of the circ_0007444-WT reporter (P<0.01). Conversely, in comparison with the NC-inh group, SKOV3 and OVCAR3 cells of the miR-570-3p inhibitor group presented markedly higher relative luciferase activity of the circ_0007444-WT reporter (P<0.01). Interestingly, no statistically significant difference was found in the relative luciferase activity of circ_0007444-Mut reporter between the miR-NC group and miR-570-3p mimic group. Meanwhile, the difference of the relative luciferase activity between the NC-inh group and miR-570-3p inhibitor group was not obvious (Figure 3B). In addition, RIP for AGO2 was used in SKOV3 cells and qRT-PCR was applied to detect the expression of endogenous pulled-down circ_0007444. The results are shown in Figure 3C. Notably, circ_0007444 was highly precipitated by AGO2 antibody when relative to the input. Meanwhile, the expression of circ_0007444 was obviously elevated in the miR-570-3p mimic group in comparison with the miR-NC group (P<0.01). All of these results revealed that circ_0007444 acted as a sponge for miR-570-3p.
miR-570-3p expression in OC cells and patients were further monitored by qRT-PCR. SKOV3 cells of the sh-circ_0007444 group exhibited significantly higher miR-570-3p expression than those of the sh-NC group (P<0.01). On the contrary, OVCAR3 cells of the oe-circ_0007444 group showed prominently lower miR-570-3p expression when compared with the oe-NC group (P<0.01) (Figure 3D). In OC patients, pronounced higher miR-570-3p expression was found in their tumor tissues than in adjacent normal tissues (P<0.01) (Figure 3E). At the same time, miR-570-3p expression was negatively correlated with circ_0007444 expression in tumor tissues of OC patients (P<0.01) (Figure 3F). Thus, miR-570-3p expression was inhibited by circ_0007444 in OC.
PTEN Expression Was Regulated by miR-570-3p and Circ_0007444
According to the online prediction of TargetScan, PTEN possessed the binding sites for miR-570-3p. Therefore, the fragments of PTEN-WT and PTEN-Mut were designed and synthesized (Figure 4A), followed by being loaded into luciferase reporter vectors. Luciferase reporter gene assay was carried out to verify the relationship between PTEN and miR-570-3p. As shown in Figure 4B, the relative luciferase activity of the PTEN-WT reporter in SKOV3 and OVCAR3 cells of the miR-570-3p mimic group was severely lower than that of the miR-NC group (P<0.01). Conversely, a significantly higher relative luciferase activity of PTEN-WT reporter occurred in SKOV3 and OVCAR3 cells of the miR-570-3p inhibitor group when relative to the NC-inh group (P<0.01). In terms of the relative luciferase activity of the PTEN-Mut reporter, no obvious changes were found between the miR-NC group and the miR-570-3p mimic group, or between the NC-inh group and the miR-570-3p inhibitor group. Hence, PTEN was a target gene of miR-570-3p.
In vitro studies exhibited that, for SKOV3 and OVCAR3 cells of the miR-570-3p mimic group, much higher PTEN protein expression was observed when compared with the miR-NC group (P<0.01). Furthermore, SKOV3 cells of the sh-circ_0007444 group presented significantly lower PTEN protein expression than that of the sh-NC group (P<0.01). Conversely, OVCAR3 cells of the oe-circ_0007444 group showed a markedly higher PTEN protein expression than that of the oe-NC group (P<0.01) (Figure 4C). In OC patients, prominently lower PTEN expression was found in tumor tissues than that in adjacent normal tissues (P<0.01) (Figure 4D). Pearson’s correlation analysis revealed a positive correlation between PTEN and circ_0007444 expression (P<0.01), but a negative correlation between PTEN and miR-570-3p expression in tumor tissues of OC patients (P<0.01) (Figure 4E and F). These results illustrated that PTEN expression was regulated by miR-570-3p and circ_0007444.
Circ_0007444 Inhibited OC Cells Malignant Phenotype by Mediating the miR-570-3p/PTEN Axis
To investigate whether circ_0007444 affected OC cells, the malignant phenotype via mediating the miR-570-3p/PTEN axis, rescue experiments were subsequently conducted using OVCAR3 cells. Cells were subjected to cotransfection and qRT-PCR was used for the detection of transfection efficiency. As presented in Figure 5A, relative to the oe-NC group, remarkably higher circ_0007444 expression occurred in OVCAR3 cells of the oe-circ_0007444 group, oe-circ_0007444 + miR-NC group, oe-circ_0007444 + miR-570-3p mimic group, oe-circ_0007444 + si-NC group and oe-circ_0007444 + si-PTEN group (P<0.01). In terms of miR-570-3p expression, it was much lower in OVCAR3 cells of the oe-circ_0007444 group when compared with the oe-NC group (P<0.01). Meanwhile, significantly higher miR-570-3p expression was found in OVCAR3 cells of the oe-circ_0007444 + miR-570-3p mimic group when relative to the oe-circ_0007444 + miR-NC group (P<0.01). In addition, the expression of PTEN was obviously higher in OVCAR3 cells of the oe-circ_0007444 group in comparison with the oe-NC group (P<0.01). Relative to the oe-circ_0007444 + miR-NC group, markedly lower PTEN expression was observed in OVCAR3 cells of the oe-circ_0007444 + miR-570-3p mimic group (P<0.01). At the same time, OVCAR3 cells of the oe-circ_0007444 + si-PTEN group exhibited prominently lower PTEN expression than that of the oe-circ_0007444 + si-NC group (P<0.01). Therefore, OVCAR3 cells were successfully transfected.
Regarding the proliferation ability, a pronounced lower OD value was presented in OVCAR3 cells in the oe-circ_0007444 group when compared with the oe-NC group (P<0.01). In comparison with the oe-circ_0007444 + miR-NC group, OVCAR3 cells of the oe-circ_0007444 + miR-570-3p mimic group showed much higher OD value (P<0.01). Meanwhile, the OD value of the OVCAR3 cells in the oe-circ_0007444 + si-PTEN group was prominently higher than that in the oe-circ_0007444 + si-PTEN group (P<0.01) (Figure 5B). Apoptosis was researched by flow cytometry. Relative to OVCAR3 cells of the oe-NC group, the apoptosis was significantly increased in the oe-circ_0007444 group (P<0.01). However, much reduced apoptosis was found in OVCAR3 cells of the oe-circ_0007444 + miR-570-3p mimic group when compared with the oe-circ_0007444 + miR-NC group (P<0.01). Meanwhile, the apoptosis of OVCAR3 cells in the oe-circ_0007444 + si-PTEN group was prominently lower than that of the oe-circ_0007444 + si-NC group (P<0.01) (Figure 5C). Wound healing assay and transwell experiment were carried out to explore migration and invasion, respectively. As exhibited in Figure 5D and E, dramatically wider relative wound width and less invasive cell number was observed in OVCAR3 cells of the oe-circ_0007444 group when compared with the oe-NC group (P<0.01). On the contrary, compared with the oe-circ_0007444 + miR-NC group, OVCAR3 cells of the oe-circ_0007444 + miR-570-3p mimic group showed a markedly narrower relative wound width and more invasive cell number (P<0.01). In comparison with the oe-circ_0007444 + si-NC group, OVCAR3 cells of the oe-circ_0007444 + si-PTEN group exhibited a narrower relative wound width and more invasive cell number (P<0.01). All of these results revealed that circ_0007444 inhibited the OC cells malignant phenotype by mediating the miR-570-3p/PTEN axis.
Circ_0007444 Inhibited OC Growth in vivo
The effect of circ_0007444 on OC progression was monitored by in vivo experiment using nude mice. Distinctly diminished xenograft tumor volume and weight were found in mice of the oe-circ_0007444 group when compared to the oe-NC group (P<0.01). In contrast, relative to the sh-NC group, the xenograft tumor volume and weight were pronounced elevated in mice of the sh-circ_0007444 group (P<0.01) (Figure 6A–C). Ki67 and PTEN expression in the xenograft tumor was detected by immunohistochemistry. Xenograft tumors of the oe-circ_0007444 group exhibited less Ki67 positive signals and more PTEN positive signals than those of the oe-NC group. However, more Ki67 positive signals and less PTEN positive signals were observed in xenograft tumors of the sh-circ_0007444 group relative to the sh-NC group. Tunel assay was then performed for apoptosis analysis. It could be noted that, in comparison with the oe-NC group, xenograft tumors of the oe-circ_0007444 group showed more Tunel positive signals. Conversely, less Tunel positive signals were observed in xenograft tumors of the sh-circ_0007444 group when compared with the sh-NC group (Figure 6D). Thus, all of these data illustrated that circ_0007444 inhibited the growth of OC in vivo.
Discussion
circRNAs are a type of endogenous RNA with the function of regulating genes. The covalently closed loop structure of circRNAs has good stability which can be resistant to degradation by exonuclease RNase R.13 Accumulated data indicated that many circRNAs were abnormally expressed in malignant tumors and were related to malignant development or patients’ prognosis.14 Concretely, one of the main ways for circRNAs regulating tumors progression are through serving as sponges for microRNAs (miRNAs) and then further regulating the expression of mRNAs. The circRNAs–miRNAs–mRNAs network ultimately mediates the proliferation and metastasis of tumors via intervening in cell cycle, epigenetic modulation, or signal transduction, etc.15 In this study, we proposed a novel target for OC treatment, namely circ_0007444, which served as an oncogene in OC. Lower expression of circ_0007444 was prominently associated with poor outcome of OC patients, such as advanced tumor stage and grade, large tumor size, and lower 60-month percent survival. Interestingly, this research still demonstrated that circ_0007444 was resistant to the degradation by RNase R. Thus, circ_0007444 could be stably expressed in OC, and could be used as a potential target for OC therapy. Importantly, this study revealed that circ_0007444 acted as a sponge for miR-570-3p. It suppressed the progression of OC through regulating the miR-570-3p/PTEN axis.
The function of miR-570-3p in regulating the progression of human cancers has been reported previously. However, according to available data, the function of miR-570-3p was differently among human tumors. For instance, miR-570-3p was found to be down-regulated in triple negative breast cancer. It could inhibit proliferation and induce apoptosis of triple negative breast cancer cells. Thus, miR-570-3p acted as a tumor suppressor in triple negative breast cancer.16 Meanwhile, Martha et al17 reported that, miR-570-3p greatly reduced the risk of death in colorectal cancer patients. On the contrary, in bladder cancer, miR-570-3p was served as a cancer-promoting gene, which exacerbated the migration and invasion of tumor cells.18 Bao et al19 suggested that miR-570-3p could be used as a potential biomarker of the metastasis and prognosis of osteosarcoma. They discovered that, in comparison with non-metastatic osteosarcoma tissues, the expression of miR-570-3p in metastatic osteosarcoma tissues was distinctly diminished. Currently, the function of miR-570-3p has not been reported in OC. In this research, miR-570-3p was found to be sponged by circ_0007444 based on the results of luciferase reporter gene assay and RIP experiment. More importantly, miR-570-3p was aberrantly up-regulated in OC patients. Up-regulation of miR-570-3p reversed the inhibitory effect of circ_0007444 on OC cells malignant phenotype. Thus, miR-570-3p served as a cancer-promoting gene in OC.
According to bioinformatics analysis and luciferase reporter gene assay, PTEN was identified as a target gene of miR-570-3p. In OC, the expression of PTEN was negatively regulated by miR-570-3p, but positively regulated by circ_0007444. PTEN is considered as a tumor suppressor in most human tumors, such as pancreatic carcinoma, hepatocellular carcinoma, and breast cancer.20–22 Generally, the decreased PTEN expression predicts more aggressive tumors and a worse prognosis of patients. In OC, the abnormally reduced PTEN expression is also reported to predict the malignant progression. Zhao et al23 illustrated that PTEN expression in OC was declined by the aberrantly up-regulated miR-552. This enhanced the growth and metastasis capacity of OC cells. In addition, researchers revealed that the elevated PTEN expression in OC cells could induce apoptosis and weaken OC cells invasion and proliferation.24 In terms of the mechanism, there was research which illustrated that PTEN might inhibit the OC malignant phenotype via inhibiting the PI3K/Akt signaling pathway.25 It is well known that the activation of the PI3K/Akt signaling pathway is one of the main reasons for the tumorigenesis and progression of OC.26 As a tumor suppressor, PTEN expression in OC was discovered to be abnormally down-regulated in this research. Knockdown of PTEN reversed the inhibitory effect of circ_0007444 on OC cells malignant phenotype. Regarding the mechanism, circ_0007444 could facilitate the expression of PTEN via sponging miR-570-3p, thereby inhibiting the malignant progression of OC.
Last but not the least, this study conducted in vivo experiments by using nude mice. Results revealed that circ_0007444 could attenuate the growth of OC in vivo. circ_0007444 reduced Ki67 expression and elevated PTEN expression and apoptosis in xenograft tumors. This discovery provided a more solid theoretical basis for the application of circ_0007444 in target treatment of OC.
There was a limitation in this study. For circ_0007444 knockdown or overexpression study in vitro, at least two cell lines should be used. However, due to the limitations of the laboratory, we are currently unable to study this experiment. This point will be the focus in our future research.
In summary, this was the first report of circ_0007444 expression in OC. Results illustrated that circ_0007444 expression was aberrantly reduced in OC and circ_0007444 acted as a tumor suppressor in OC. It could inhibit the malignant progression of OC through enhancing PTEN expression via sponging miR-570-3p. Therefore, circ_0007444 was proposed as a potential candidate target for the treatment of OC.
Highlights
- Low expression of circ_0007444 in OC patients predicted poor prognosis.
- circ_0007444 inhibited proliferation, invasion, and promoted apoptosis of OC cells.
- circ_0007444 promoted PTEN expression by sponging miR-570-3p.
- circ_0007444 inhibited OC cells malignant phenotype by mediating miR-570-3p/PTEN.
- circ_0007444 inhibited OC growth in vivo.
Acknowledgment
We thank Dr Guilin Li for helping us complete the Luciferase reporter gene assay and perform the data analysis.
Disclosure
The authors report no conflicts of interest for this work.
References
1. Torre LA, Trabert B, DeSantis CE, et al. Ovarian cancer statistics. CA Cancer J Clin. 2018;68:284–296. doi:10.3322/caac.21456
2. Zhang M, Xia B, Xu Y, et al. Circular RNA (hsa_circ_0051240) promotes cell proliferation, migration and invasion in ovarian cancer through miR-637/KLK4 axis. Artif Cells Nanomed Biotechnol. 2019;47:1224–1233. doi:10.1080/21691401.2019.1593999
3. Holmes D. Ovarian cancer: beyond resistance. Nature. 2015;527:S217. doi:10.1038/527S217a
4. Guo N, Liu XF, Pant OP, et al. Circular RNAs: novel promising biomarkers in ocular diseases. Int J Med Sci. 2019;16:513–518. doi:10.7150/ijms.29750
5. Li X, Diao H. Circular RNA circ_0001946 acts as a competing endogenous RNA to inhibit glioblastoma progression by modulating miR1 and CDR1. J Cell Physiol. 2019;234:13807–13819. doi:10.1002/jcp.28061
6. Liu L, Yang X, Li NF, et al. Circ_0015756 promotes proliferation, invasion and migration by microRNA-7-dependent inhibition of FAK in hepatocellular carcinoma. Cell Cycle. 2019;18:2939–2953. doi:10.1080/15384101.2019.1664223
7. Li QH, Liu Y, Chen S, et al. circ-CSPP1 promotes proliferation, invasion and migration of ovarian cancer cells by acting as a miR-1236-3p sponge. Biomed Pharmacother. 2019;114:108832. doi:10.1016/j.biopha.2019.108832
8. Pei C, Wang H, Shi C, et al. CircRNA hsa_circ_0013958 may contribute to the development of ovarian cancer by affecting epithelial-mesenchymal transition and apoptotic signaling pathways. J Clin Lab Anal. 2020;34:e23292. doi:10.1002/jcla.23292
9. Zhang L, Zhou Q, Qiu Q, et al. CircPLEKHM3 acts as a tumor suppressor through regulation of the miR-9/BRCA1/DNAJB6/KLF4/AKT1 axis in ovarian cancer. Mol Cancer. 2019;18:144. doi:10.1186/s12943-019-1080-5
10. Hu J, Wang L, Chen J, et al. The circular RNA circ-ITCH suppresses ovarian carcinoma progression through targeting miR-145/RASA1 signaling. Biochem Biophys Res Commun. 2018;505:222–228. doi:10.1016/j.bbrc.2018.09.060
11. Zhu M, Dang Y, Yang Z, et al. Comprehensive RNA sequencing in adenoma-cancer transition identified predictive biomarkers and therapeutic targets of human CRC. Mol Ther Nucleic Acids. 2020;20:25–33. doi:10.1016/j.omtn.2020.01.031
12. Ng WL, Mohd Mohidin TB, Shukla K. Functional role of circular RNAs in cancer development and progression. RNA Biol. 2018;15:995–1005. doi:10.1080/15476286.2018.1486659
13. Xie L, Mao M, Xiong K, et al. Circular RNAs: A novel player in development and disease of the central nervous system. Front Cell Neurosci. 2017;11:354. doi:10.3389/fncel.2017.00354
14. Yang C, Yuan W, Yang X, et al. Circular RNA circ-ITCH inhibits bladder cancer progression by sponging miR-17/miR-224 and regulating p21, PTEN expression. Mol Cancer. 2018;17:19. doi:10.1186/s12943-018-0771-7
15. Zhao ZJ, Shen J. Circular RNA participates in the carcinogenesis and the malignant behavior of cancer. RNA Biol. 2017;14:514–521. doi:10.1080/15476286.2015.1122162
16. Wang LL, Huang WW, Huang J, et al. Protective effect of hsa-miR −570-3p targeting CD274 on triple negative breast cancer by blocking PI3K /AKT/mTOR signaling pathway. Kaohsiung J Med Sci. 2020;36:581–591. doi:10.1002/kjm2.12212
17. Slattery ML, Herrick JS, Mullany LE, et al. An evaluation and replication of miRNAs with disease stage and colorectal cancer-specific mortality. Int J Cancer. 2015;137:428–438. doi:10.1002/ijc.29384
18. He Q, Yan D, Dong W, et al. circRNA circFUT8 upregulates krüpple-like factor 10 to inhibit the metastasis of bladder cancer via sponging miR-570-3p. Mol Ther Oncolytics. 2020;16:172–187. doi:10.1016/j.omto.2019.12.014
19. Bao X, Zhao L, Guan H, et al. Inhibition of LCMR1 and ATG12 by demethylation-activated miR-570-3p is involved in the anti-metastasis effects of metformin on human osteosarcoma. Cell Death Dis. 2018;9:611. doi:10.1038/s41419-018-0620-z
20. Zhang H, Liu A, Feng X, et al. MiR-132 promotes the proliferation, invasion and migration of human pancreatic carcinoma by inhibition of the tumor suppressor gene PTEN. Prog Biophys Mol Biol. 2019;148:65–72. doi:10.1016/j.pbiomolbio.2017.09.019
21. Wu Q, Yu S, Chen J, et al. Downregulation of STRAP promotes tumor growth and metastasis in hepatocellular carcinoma via reducing PTEN level. IUBMB Life. 2018;70:120–128. doi:10.1002/iub.1707
22. Li S, Shen Y, Wang M, et al. Loss of PTEN expression in breast cancer: association with clinicopathological characteristics and prognosis. Oncotarget. 2017;8:32043–32054. doi:10.18632/oncotarget.16761
23. Zhao W, Han T, Li B, et al. miR-552 promotes ovarian cancer progression by regulating PTEN pathway. J Ovarian Res. 2019;12:121. doi:10.1186/s13048-019-0589-y
24. Wang J, Xu W, He Y, et al. LncRNA MEG3 impacts proliferation, invasion, and migration of ovarian cancer cells through regulating PTEN. Inflamm Res. 2018;67:927–936. doi:10.1007/s00011-018-1186-z
25. Liu J, Chen W, Zhang H, et al. miR-214 targets the PTEN-mediated PI3K/Akt signaling pathway and regulates cell proliferation and apoptosis in ovarian cancer. Oncol Lett. 2017;14:5711–5718. doi:10.3892/ol.2017.6953
26. Shen F, Zong ZH, Liu Y, et al. CEMIP promotes ovarian cancer development and progression via the PI3K/AKT signaling pathway. Biomed Pharmacother. 2019;114:108787. doi:10.1016/j.biopha.2019.108787
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.