Back to Journals » Cancer Management and Research » Volume 12
Circ_0007031 Serves as a Sponge of miR-760 to Regulate the Growth and Chemoradiotherapy Resistance of Colorectal Cancer via Regulating DCP1A
Authors Wang Y, Wang H, Zhang J, Chu Z, Liu P, Zhang X, Li C, Gu X
Received 20 March 2020
Accepted for publication 3 August 2020
Published 14 September 2020 Volume 2020:12 Pages 8465—8479
DOI https://doi.org/10.2147/CMAR.S254815
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Harikrishna Nakshatri
This paper has been retracted.
Yuanyuan Wang, 1, 2,* Hua Wang, 3,* Jian Zhang, 2 Zhifen Chu, 2 Pu Liu, 2 Xing Zhang, 2 Chao Li, 2 Xiaosong Gu 1
1Academy of Medical Engineering and Translational Medicine, Tianjin University, Tianjin, People’s Republic of China; 2Department of General Surgery, Hebei Key Laboratory of Colorectal Cancer Precision Diagnosis and Treatment, The First Hospital of Hebei Medical University, Shijiazhuang, Hebei, People’s Republic of China; 3Department of Pharmacy, Third Hospital of Hebei Medical University, Shijiazhuang, Hebei, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Chao Li
Department of General Surgery, Hebei Key Laboratory of Colorectal Cancer Precision Diagnosis and Treatment, The First Hospital of Hebei Medical University, Shijiazhuang 050031, Hebei, People’s Republic of China
Email [email protected]
Xiaosong Gu
Academy of Medical Engineering and Translational Medicine, Tianjin University, Tianjin 300072, People’s Republic of China
Tel +86-0311-85917000
Email [email protected]
Background: Colorectal cancer (CRC) is a kind of malignant tumor, and the development of chemoradiotherapy resistance (CRR) increases the difficulty of its treatment. The role of circular RNAs (circRNAs) in cancer progression has been well documented. Nevertheless, the function of circ_0007031 in the growth and CRR of CRC has not been well elucidated.
Methods: CRR cell lines were constructed using 5-Fu and radiation. Cell counting kit 8 (CCK8) assay was employed to measure the 5-Fu resistance and proliferation of cells. Clonogenic assay was used to evaluate the radiation resistance of cells. Also, the expression of circ_0007031 and microRNA-760 (miR-760) was determined using quantitative real-time polymerase chain reaction (qRT-PCR). The cell cycle distribution and apoptosis of cells were assessed by flow cytometry. Besides, the levels of apoptosis-related protein and mRNA-decapping enzyme 1a (DCP1A) protein were measured by Western blot (WB) analysis. Further, dual-luciferase reporter assay and RNA immunoprecipitation (RIP) assay were used to confirm the interaction between miR-760 and circ_0007031 or DCP1A. In addition, animal experiments were performed to evaluate the function of silenced circ_0007031 on the 5-Fu and radiation resistance of CRC tumors.
Results: Circ_0007031 expression was markedly increased in CRC tissues and cells, especially in CRC resistant cells. Circ_0007031 knockdown hindered proliferation, induced cell cycle arrest in the G0/G1 phase, enhanced apoptosis, and lowered the CRR of CRC resistant cells. Further, miR-760 could be targeted by circ_0007031, and its inhibitor could reverse the inhibition effect of circ_0007031 knockdown on the growth and CRR of CRC resistant cells. Moreover, DCP1A was a target of miR-760, and its overexpression could invert the suppression effect of miR-760 overexpression on the growth and CRR of CRC resistant cells. Circ_0007031 silencing could enhance the sensitivity of CRC tumors to 5-Fu and radiation to markedly reduce CRC tumor growth in vivo.
Conclusion: Circ_0007031 might play a positive role in the CRR of CRC through regulating the miR-760/DCP1A axis, which might provide a new approach for treating the CRR of CRC.
Keywords: colorectal cancer, CRR, circ_0007031, miR-760, DCP1A
Introduction
Colorectal cancer (CRC) is one of the most common cancers, and the number of CRC patients is increasing year by year.1,2 Because the early symptoms are not obvious, most patients with CRC are already in the advanced stage when diagnosed, so this greatly increases the difficulty of treatment.3 At present, the combination of radiotherapy and chemotherapy is considered the common treatment for locally advanced CRC.4 5-Fluorouracil (5-Fu) is a commonly used chemotherapeutic drug for CRC, but the development of chemoradiotherapy resistance (CRR) is a huge obstacle for the treatment of CRC.5,6 Therefore, it is urgent to clarify the factors affecting the CRR of CRC.
Non-coding RNAs acting as non-protein-coding RNAs have received a lot of attention, while circular RNAs (circRNAs) have been favored by researchers in recent years due to their closed-loop structure.7,8 CircRNAs have been shown to take part in the modulation of cancer progression and may also be involved in the CRR of cancer.9–11 For example, circRNA Cdr1as inhibits the cisplatin resistance of ovarian cancer,12 and circKDM4C participates in the regulation of the tumor progression and doxorubicin resistance of breast cancer.13 In CRC, Abu et al found that there were 773 up-regulated and 732 down-regulated circRNAs in the chemoresistant and chemosensitive HCT116 cells, indicating that the expression levels of circRNAs were crucial to the development of the chemical resistance of CRC.14 The microarray analysis by Xiong et al showed that 71 circRNAs were differentially expressed in the CRR of CRC cells, among which circ_0007031 (TUBGCP3 was its linear mRNA) was significantly highly expressed.15 However, the role of circ_0007031 in the growth and CRR of CRC has not been investigated.
The function of circRNAs as competitive endogenous RNAs (ceRNAs) for microRNAs (miRNAs) is a classic molecular mechanism of circRNAs and has been widely confirmed.16,17 MiR-760 is lower expressed in many cancers and has been shown in many studies to suppress the CRR of many cancers, including pancreatic cancer, hepatocellular carcinoma and breast cancer.18–20 It has been reported that mRNA-decapping enzyme 1a (DCP1A) is a decapping enzyme associated with mRNA degradation and translation inhibition, and may be involved in cell differentiation.21,22 Studies have shown that DCP1A expression is up-regulated in CRC, and its elevated expression is associated with the reduced survival rate in CRC patients.23,24 Therefore, it may be a key target to regulate CRC progress.
Our study was aimed to investigate the role of circ_0007031 in the growth and CRR of CRC and to elucidate its molecular mechanisms. Combined with the results of the background investigation, we focused on the interaction of circ_0007031, miR-760 and DCP1A, which might provide molecular therapeutic strategies for the treatment of CRC with CRR.
Materials and Methods
Cell Culture
CRC cell lines (HCT116 and Caco2) were obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA) and cultured in RPMI-1640 medium (Gibco, Waltham, MA, USA). Human normal colon epithelial cells (NCM460) were purchased from Jining Shiye (Shanghai, China) and cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco). All mediums were contained 10% fetal bovine serum (FBS; Gibco) and 1% penicillin/streptomycin (Invitrogen, Carlsbad, CA, USA), and all cells were incubated at 37°C with 5% CO2 incubator.
Establishment of CRR Cell Model
HCT116 and Caco2 cells were seeded into 6-well plates and cultured until the cells reached 80–90% confluences. Then, the cells were treated with 10 μmol/L 5-Fu (Selleck, Shanghai, China) and simultaneously exposed to a dose of 4 Gy 6 Mv X-ray. After 24 h, the cell medium was replaced with 5-Fu-free medium, and the cells were placed in a normal environment. After 2 days, the surviving cells were transferred to fresh medium and further treated with 5-Fu and X-ray according to the above method. The whole process was repeated 8 times. After that, the last collected cells were HCT116/CRR and Caco2/CRR cell models.
Cell Counting Kit 8 (CCK8) Assay
CCK8 assay was used to evaluate the 5-Fu resistance and proliferation of cells. All cells were seeded in 96-well plates and cultivation for 24 h. For 5-Fu resistance assay, cells were treated with different concentrations of 5-Fu (0, 0.5, 1.0, 1.5, 2.0 and 2.5 μg/mL) for 48 h, followed by incubation with CCK8 solution (Genomeditech, Shanghai, China) for 4 h. The absorbance was detected at 450 nm, and half-maximal inhibitory concentration (IC50) was calculated to evaluate the 5-Fu resistance of cells. For proliferation assay, CCK8 solution was added into each well at a specific time point (0, 24, 48 and 72 h) for 4 h. The absorbance was determined at 450 nm to assess the proliferation ability of cells.
Clonogenic Assay
All cells were seeded in 6-well plates. After 24 h, cells were exposed to graded doses (0, 2, 4, 6 and 8 Gy) of X-ray. After 2 weeks, cells were fixed with methanol, stained with crystal violet, and the colony number (> 50 cells) was counted under a microscope (Novel, Ningbo, China). The survival fraction of cells was calculated as below: survival fraction = the colony number of the treatment group/control group.
Samples Collection
A cohort of 50 CRC patients who underwent surgical resection at The First Hospital of Hebei Medical University, Shijiazhuang were recruited. CRC tumor tissues and adjacent normal tissues were obtained and stored at −80°C. All patients signed informed consent, and our study was approved by the Ethics Committee of The First Hospital of Hebei Medical University, Shijiazhuang.
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
According to the instruction provided by the manufacturer, total RNA was extracted using the RNeasy Mini Kit (Qiagen, Valencia, CA, USA), and cDNA was synthesized using cDNA Synthesis SuperMix (Transgen, Beijing, China). Quantitative analysis was carried out using SYBR Green (Solarbio, Beijing, China) on Real-Time PCR System (Applied Biosystems, Foster City, CA, USA). The thermocycling conditions were as below: 95°C for 10 min, 40 cycles of 95°C for 30 s, 55°C for 30 s, and 72°C for 42 s. The data were analyzed by the 2−ΔΔCt method and normalized using 18S ribosomal RNA (rRNA) or U6. All primers were presented as follows: circ_0007031, F 5ʹ-ATCATTGCTGCACACGAGGT-3ʹ, R 5ʹ-AGGACCTTCCTAGACTGATCCA-3ʹ; TUBGCP3, F 5ʹ-TATGTGGAGCAGATCGAGAAG-3ʹ, R 5ʹ-TTGATGGACCTGAGGTGAG-3ʹ; 18S rRNA, F 5ʹ-ATCGGGGATTGCAATTATTC-3ʹ, R 5ʹ-CTCACTAAACCATCCAATCG-3ʹ; miR-760, F 5ʹ- TCAATCCACCAGAGCATGGATAT-3ʹ, R 5ʹ-CTCTACAGCTATATTGCCAGCCA-3ʹ; U6, F 5ʹ-CTCGCTTCGGCAGCACATATACT-3ʹ, R 5ʹ-CGCTTCACGAATTTGCGTGT-3ʹ.
Subcellular Fractionation and Localization
The cytoplasmic and nuclear RNA of HCT116/CRR and Caco2/CRR cells were isolated and extracted using the Cytoplasmic & Nuclear RNA Purification Kit (Norgen Biotek, Thorold, Ontario, Canada). QRT-PCR was employed to determine the expression of circ_0007031, U6 and 18S rRNA in cytoplasmic and nuclear of cells. U6 and 18S rRNA were used as nuclear control and cytoplasm control, respectively.
RNase R Treatment
Part of extracted RNAs from HCT116/CRR and Caco2/CRR cells were treated with Ribonuclease R (RNase R; Geneseed, Guangzhou, China) for 20 min and then continued for qRT-PCR analysis to detect the circ_0007031 and TUBGCP3 expression. Results without RNase R treatment (RNase R-) were used as negative controls, and TUBGCP3 was used as a representative linear control.
Cell Transfection
Transfection could be performed when the cell density rate reached 50%-60%. Circ_0007031 small interfering RNA and lentiviral short hairpin RNA (si-circ_0007031 and sh-circ_0007031) or their negative controls (si-NC and sh-NC), miR-760 mimic and inhibitor (miR-760 and anti-miR-760) or their negative controls (miR-NC and anti-miR-NC), DCP1A overexpression plasmid (DCP1A) and its negative control (vector) were purchased from RiboBio (Guangzhou, China). Cell transfection was performed using Lipofectamine 3000 (Invitrogen).
Flow Cytometry
Flow cytometry was performed to measure the cell cycle distribution and apoptosis of cells. HCT116/CRR and Caco2/CRR cells were harvested and collected into a centrifuge tube after transfection for 48 h. For cell cycle distribution assay, cells were fixed with 70% ethanol overnight at 4°C. Then, cells were incubated with RNase A (Beyotime, Shanghai, China) for 1 h at 37°C and stained with propidium iodide (PI, Beyotime) for 30 min. The cell cycle was analyzed using FACScan Flow Cytometer (BD Biosciences, San Jose, CA, USA). For apoptosis assay, cells were stained with Annexin V-fluorescein isothiocyanate (FITC) Apoptosis Detection Kit (Beyotime). The apoptosis of cells was also detected using FACScan Flow Cytometer.
Western Blot (WB) Analysis
Tissues and cells were lysed with RIPA lysis buffer (Beyotime) to extract total protein. After quantification with the BCA Protein Assay Kit (Beyotime), the same amount of protein was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel, transferred onto polyvinylidene fluoride (PVDF) membranes (Millipore, Billerica, MA, USA) and blocked with 5% nonfat milk. The membranes were incubated with primary antibodies against B-cell lymphoma-2 (Bcl-2; 1:1000, Abcam, Cambridge, MA, USA), Bcl2-associated X (Bax; 1:5000, Abcam), cleaved-caspase-3 (cleaved-cas-3, 1:1000, Abcam), DCP1A (1:1000, Abcam) or GAPDH (1:5000, Abcam) at 4°C overnight. After that, the membranes were interacted with secondary antibody (1:2000, Abcam) for 1 h and visualized using BeyoECL star (Beyotime).
Dual-Luciferase Reporter Assay
The fragments of circ_0007031-wild type (WT)/mutant type (MUT) or DCP1A 3ʹ-UTR-WT/MUT were inserted into the pGL3 reporter vector (Promega, Madison, WI, USA). HCT116/CRR and Caco2/CRR cells were co-transfected the reporter vectors and miR-760 mimic or miR-NC. After 48 h, the relative luciferase activity was determined using the Dual-Luciferase Reporter Assay Kit (Transgen).
RNA Immunoprecipitation (RIP) Assay
RIP Assay Kit (Millipore) was used to perform this assay. In brief, magnetic beads were incubated with antibodies against argonaute2 (Anti-Ago2) or immunoglobulin G (Anti-IgG) at 4°C overnight. HCT116/CRR and Caco2/CRR cells were lysed using RIP buffer. Then, the cell lysates were added into the magnetic bead-antibody complex at 4°C overnight. After that, the proteinase K buffer was used to purify RNA. The enrichment of circ_0007031 and miR-760 was determined using qRT-PCR.
Mice Xenograft Models
Animal experiments were authorized by the Animal Care Committee of The First Hospital of Hebei Medical University, Shijiazhuang and performed according to the Guide for the Care and Use of Laboratory Animals. Animal studies were performed in compliance with the ARRIVE guidelines and the Basel Declaration. All animals received humane care according to the National Institutes of Health (USA) guidelines. HCT116 cells transfected with sh-circ_0007031 or sh-NC were subcutaneously injected into the flank regions of nude mice (Vital River, Beijing, China). Then, the sh-circ_0007031 or sh-NC group mice were equally divided into 2 groups, one treated with 5-Fu and the other treated with X-ray radiation. For 5-Fu treatment, after 10 days of inoculation, mice in both the sh-NC and sh-circ_0007031 groups were injected with 5-Fu at 50 mg/kg/week, and tumor volume was monitored every 7 days and calculated as follows: volume = 0.5 × length × width2. After 35 days, the mice were sacrificed, and the tumor weight was measured. For X-ray radiation, when the tumor volume was about 300 mm3, the mice had received 8 Gy X-ray radiation. Tumor volume was measured every 7 days until 35 days. The tumor was removed for further testing.
Statistical Analysis
Data were expressed as mean ± standard deviation and analyzed for the statistical difference using Student’s t-test or one-way analysis of variance in GraphPad Prism 5.0 software (GraphPad, La Jolla, CA, USA). P < 0.05 was considered statistically significant. All experiment was performed in triplicate, and all independent experiments were set for 3 times to take the average value.
Results
The Sensitivity of HCT116/CRR and Caco2/CRR Cells to Chemoradiotherapy Was Verified
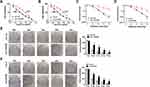
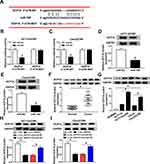
First, we examined the 5-Fu resistance and radiation resistance of HCT116/CRR and Caco2/CRR cell lines using CCK8 and clonogenic assays. As presented in Figure 1A and B, with the increase of 5-Fu concentration, the viabilities of HCT116/CRR and Caco2/CRR cells were significantly higher than that of HCT116 and Caco2 cells, and their IC50 value was also markedly enhanced compared with HCT116 and Caco2 cells, suggesting that the HCT116/CRR and Caco2/CRR cells had a strong resistance to 5-Fu. Besides, clonogenic assay results showed that with the increase of X-ray radiation, the survival fraction of HCT116/CRR and Caco2/CRR cells was increased compared to HCT116 and Caco2 cells (Figure 1C and D), and the statistical results on the colony number of cells also suggested that the colony number of HCT116/CRR and Caco2/CRR cells was remarkably higher than that of HCT116 and Caco2 cells (Figure 1E and F). These results indicated that HCT116/CRR and Caco2/CRR cells had good resistance to radiation. Therefore, our results confirmed the successful construction of 5-Fu-resistant and radiation-resistant HCT116/CRR and Caco2/CRR cells.
The Upregulation of Circ_0007031 in CRC Tissues and Cells
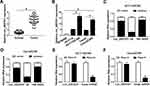
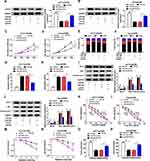
For exploring the role of circ_0007031 in CRC, we first measured its expression in CRC tissues and cells. The results revealed that circ_0007031 had elevated expression in CRC tumor tissues compared with adjacent normal tissues (Figure 2A). The correlation analysis between circ_0007031 expression and the clinicopathologic characteristics of CRC patients showed that the high expression of circ_0007031 was markedly related to the tumor size, TNM stage and CEA of CRC patients (Table 1). Similarly, we also found that compared with NCM460, circ_0007031 was up-regulated in CRC cells, and was significantly higher expressed in HCT116/CRR and Caco2/CRR cells than in HCT116 and Caco2 cells (Figure 2B). By detecting the expression of circ_0007031 in the nucleus and cytoplasm of HCT116/CRR and Caco2/CRR cells, we found that circ_0007031 mainly accumulated in the cytoplasm (Figure 2C and D). Besides, circ_0007031 also showed strong resistance to RNase R digestion in HCT116/CRR and Caco2/CRR cells, indicating that circ_0007031 was a circular transcript (Figure 2E and F).
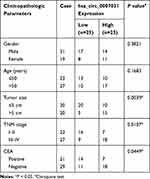 |
Table 1 Correlation Between circ_0007031 Expression and Clinicopathologic Characteristics of CRC Patients |
Silencing of circ_0007031 Repressed the Growth and CRR of CRC Resistant Cells
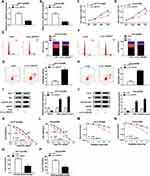
To investigate the function of circ_0007031 in CRC growth and CRR, we silenced its expression using si-circ_0007031. The detection of its expression by qRT-PCR indicated that si-circ_0007031 had a good inhibitory effect on circ_0007031 expression (Figure 3A and B). Subsequently, CCK8 results suggested that the silencing of circ_0007031 could inhibit the proliferation of HCT116/CRR and Caco2/CRR cells (Figure 3C and D). Besides, knockdown of circ_0007031 in HCT116/CRR and Caco2/CRR cells significantly caused the cell cycle arrest in the G0/G1 phase, with an obvious reduction in the number of HCT116/CRR and Caco2/CRR cells at the S phase (Figure 3E and F). Also, the apoptosis of HCT116/CRR and Caco2/CRR cells was promoted by circ_0007031 knockdown (Figure 3G and H). Consistent with this result, WB analysis revealed that the silencing of circ_0007031 suppressed the Bcl-2 protein level, while increased the Bax and Cleaved-cas-3 protein levels in HCT116/CRR and Caco2/CRR cells (Figure 3I and J). Further, silenced circ_0007031 also decreased the resistance of HCT116/CRR and Caco2/CRR cells to 5-Fu (Figure 3K and L). Moreover, the survival fraction of HCT116/CRR and Caco2/CRR cells was also reduced by circ_0007031 knockdown (Figure 3M and N), which could also be confirmed by the decrease in the colony number of HCT116/CRR and Caco2/CRR cells (Figure 3O and P). All data suggested that circ_0007031 might play pro-cancer and pro-resistance roles in CRC.
Circ_0007031 Served as a Sponge of miR-760 in CRC Cells
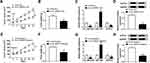
To explore the mechanism by which circ_0007031 inhibited the growth and CRR of CRC resistant cells, we used the starBase v.2.0 tool (http://starbase.sysu.edu.cn/index.php) for bioinformatics analysis and found that miR-760 might directly bind to circ_0007031 (Figure 4A). Dual-luciferase reporter assay determined that miR-760 could restrain the luciferase activity of circ_0007031-WT reporter vector rather than circ_0007031-MUT reporter vector (Figure 4B and C). And RIP assay also revealed that circ_0007031 and miR-760 were enriched in Anti-Ago2 (Figure 4D). Besides, qRT-PCR showed that miR-760 was up-regulated by circ_0007031 knockdown in HCT116/CRR and Caco2/CRR cells (Figure 4E and F). Through detecting the miR-760 expression in CRC, we also found that miR-760 was under-expressed in CRC tumor tissues and cells compared with that in adjacent normal tissues or NCM460 cells, respectively (Figure 4G and H). Moreover, miR-760 was also lower expressed in HCT116/CRR and Caco2/CRR cells than in HCT116 and Caco2 cells (Figure 4H). These results indicated that miR-760 could be sponged by circ_0007031 in CRC.
The Inhibition of circ_0007031 Silencing on the Growth and CRR of CRC Resistant Cells Could Be Partially Reversed by miR-760 Inhibitor
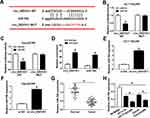
To explore whether the role of circ_0007031 in CRC was mediated by miR-760, we co-transfected si-circ_0007031 and anti-miR-760 into HCT116/CRR and Caco2/CRR cells. The detection results of miR-760 expression showed that si-circ_0007031 had a good promoting effect on the expression of miR-760, while this effect could be reversed by anti-miR-760 (Figure 5A and B). CCK8 assay revealed that miR-760 inhibitor could partially invert the suppression of circ_0007031 knockdown on the proliferation of HCT116/CRR and Caco2/CRR cells (Figure 5C and D). Also, the arresting effect of circ_0007031 knockdown on the G0/G1 phase of HCT116/CRR and Caco2/CRR cells could also be partially recovered by miR-760 inhibitor (Figure 5E and F). Besides, the promotion effect of circ_0007031 silencing on the apoptosis of HCT116/CRR and Caco2/CRR cells also could be reversed by miR-760 inhibitor (Figure 5G and H), which was also reflected by anti-miR-760 changing the protein levels of Bcl-2, Bax and cleaved-cas-3 of HCT116/CRR and Caco2/CRR cells (Figure 5I and J). Meanwhile, we measured the resistance of HCT116/CRR and Caco2/CRR cells to 5-Fu and radiation. The results showed that miR-760 inhibitor partially inverted the reduction effect of silenced circ_0007031 on the 5-Fu resistance and radiation resistance of HCT116/CRR and Caco2/CRR cells (Figure 5K–N). Furthermore, the inhibition effect of circ_0007031 silencing on the colony number of HCT116/CRR and Caco2/CRR cells also could be reversed by miR-760 inhibitor (Figure 5O and P). Therefore, our results confirmed that the effect of circ_0007031 on the growth and CRR of CRC cells was mediated by miR-760.
DCP1A Was a Target of miR-760 in CRC Cells
At the same time, we also used the starBase v.2.0 tool (http://starbase.sysu.edu.cn/index.php) to predict the targets of miR-760. As shown in Figure 6A, DCP1A was predicted to have binding sites for miR-760. Then, miR-760 overexpression could reduce the luciferase activity of DCP1A 3ʹ-UTR-WT reporter vector without affecting that of the DCP1A 3ʹ-UTR-MUT reporter vector, confirming the interaction between them (Figure 6B and C). Further, we also found that the protein level of DCP1A could be suppressed by miR-760 overexpression in HCT116/CRR and Caco2/CRR cells (Figure 6D and E). Moreover, DCP1A expression was up-regulated in CRC tumor tissues compared with that in adjacent normal tissues (Figure 6F). Similarly, its expression was also increased in CRC cells compared to NCM460 cells, and markedly higher in HCT116/CRR and Caco2/CRR cells than that in HCT116 and Caco2 cells (Figure 6G). In addition, we also discovered that DCP1A expression was decreased by circ_0007031 knockdown, while miR-760 inhibitor could reverse this effect (Figure 6H and I). Our data confirmed that DCP1A could be targeted by miR-760.
MiR-760 Regulated the Growth and CRR of CRC Resistant Cells Through Targeting DCP1A
To verify the conclusion of the above, we co-transfected miR-760 mimic and DCP1A overexpression plasmid into HCT116/CRR and Caco2/CRR cells. The inhibition effect of miR-760 mimic on DCP1A protein level and the recovery effect of DCP1A plasmid on DCP1A protein level confirmed the success of both transfection (Figure 7A and B). So, we detected the growth of CRC cells. CCK8 results showed that miR-760 overexpression repressed the proliferation of HCT116/CRR and Caco2/CRR cells, while DCP1A overexpression could invert this effect (Figure 7C and D). Also, miR-760 overexpression could cause the cell cycle arrest in the G0/G1 phase, reducing the proportion of S phase in HCT116/CRR and Caco2/CRR cells, whereas DCP1A overexpression could recover this effect (Figure 7E and F). Besides, the promotion of miR-760 overexpression on the apoptosis of HCT116/CRR and Caco2/CRR cells could be reversed by DCP1A overexpression (Figure 7G and H), which could also be confirmed by reversing the protein levels of Bcl-2, Bax and cleaved-cas-3 in HCT116/CRR and Caco2/CRR cells (Figure 7I and J). On the other hand, DCP1A overexpression inverted the inhibition of miR-760 overexpression on the 5-Fu resistance and radiation resistance of HCT116/CRR and Caco2/CRR cells (Figure 7K–N). In addition, the recovery effect of DCP1A overexpression on the colony number of HCT116/CRR and Caco2/CRR cells also indicated DCP1A overexpression could reverse the inhibitory effect of miR-760 overexpression on the resistance of HCT116/CRR and Caco2/CRR cells to radiation (Figure 7O and P). Hence, we confirmed that the regulation of miR-760 on the growth and CRR of CRC cells was achieved by targeting DCP1A.
Knockdown of circ_0007031 Enhanced the Sensitivity of CRC Tumor to 5-Fu and Radiation in vivo
To further confirm the function of circ_0007031 in CRC, we performed the in vivo experiments. The results showed that under the 5-Fu or radiation condition, the tumor volume of mice in the circ_0007031 knockdown group was markedly smaller than that in the control group (Figure 8A and E). Besides, the tumor weight was also remarkably decreased in the circ_0007031 knockdown group under either 5-Fu treatment or radiation treatment compare with that in the sh-NC group (Figure 8B and F). Through the detection of the circ_0007031 and miR-760 expression, we confirmed that in the sh-circ_0007031 group, circ_0007031 expression was indeed inhibited and miR-760 expression was significantly higher than that in the sh-NC group (Figure 8C and G). At the same time, compared to the sh-NC group, the protein level of DCP1A was also hindered in the sh-circ_0007031 group (Figure 8D and H). Therefore, we confirmed that interference of circ_0007031 could restrain the resistance of CRC tumor to 5-Fu and radiation through regulating the miR-760/DCP1A axis, thereby aggravating the inhibitory effect of 5-Fu and radiation on CRC tumor growth.
Discussion
CRC is a malignant tumor, because it spreads to distant organs.25 Preventing the occurrence of CRR is an important step to improve the CRC treatment response and has important clinical significance. To our knowledge, although many circRNAs have been shown to be differentially expressed before and after CRR, there is no specific evidence for their function in the CRR of CRC. In our system, we discovered that circ_0007031 was remarkably highly expressed in CRC tumor tissues and cells, indicating that circ_0007031 might play a vital function in CRC. Then, we constructed the CRR of CRC cell lines and focused on the role of circ_0007031 in the CRR of CRC. The high expression of circ_0007031 in CRC resistant cells was consistent with previous studies.15 On the other hand, we also confirmed that circ_0007031 was mainly involved in post-transcriptional regulation and had cyclic properties.
Studies have confirmed that the development of CRR is a complex process that may involve DNA damage related to mitochondria and pathways related to mitochondrial metabolism, thus regulating cell survival.26,27 Therefore, the study of cell proliferation and apoptosis can well explain the sensitivity of cells to radiation and drugs. Here, we found that silenced circ_0007031 repressed the proliferation, caused cell cycle arrest in the G0/G1 phase, and accelerated the apoptosis of CRC resistant cells. Also, circ_0007031 knockdown reduced the resistance of CRC resistant cells to 5-Fu and radiation. Further, sh-circ_0007031 also enhanced the sensitivity of CRC tumors to 5-Fu and radiation, thereby notably inhibiting CRC tumor growth in vivo. This suggested that circ_0007031 knockdown might be an important step to improve the chemoradiotherapy sensitivity of CRC.
Previous studies have shown that miR-760 is associated with CRC tumor generation, and its high expression can inhibit the proliferation of CRC.28 Also, miR-760 expression was related to the poor prognosis and malignant clinicopathologic features of CRC patients.29 Besides, Xian et al also reported that miR-760 was involved in the modulation of long-noncoding RNA KCNQ1OT1 on the methotrexate resistance of CRC,30 suggesting that miR-760 might be related to CRC resistance. In our study, we confirmed that miR-760 could be sponged by circ_0007031, its low expression in CRC and the restoring effect of its inhibitor on the silenced circ_0007031 function again confirmed that it could be targeted by circ_0007031. Moreover, we also showed that miR-760 could target DCP1A. Current studies had shown that high expression of DCP1A promoted the development of many cancers, such as gastric cancer and melanoma.31,32 Herein, the reversing effect of DCP1A on the miR-760 function confirmed that DCP1A was also involved in the regulation of circ_0007031 on the growth and CRR of CRC. Therefore, the targeting of DCP1A by miR-760 also helped us better understand the role of circ_0007031 in the growth and CRR of CRC.
Currently, molecular targeted therapy is considered as an effective treatment strategy for cancer.33 The elucidation of the mechanism of CRR also points the way to reduce the occurrence of CRR in cancer treatment. Our study suggests a novel mechanism by which circ_0007031 regulates CRC resistance through the miR-760/DCP1A pathway in vitro and in vivo. However, miR-760 is not the only miRNA that can be targeted by circ_0007031, and there may be other circ_0007031 targets in CRC that are still worthy of further investigation.
In conclusion, we demonstrated that circ_0007031 could increase DCP1A to promote the growth and CRR of CRC via regulating miR-760. The findings of this study might provide a reference for the study of circ_0007031 in other diseases and a solution for the occurrence of CRR in CRC.
Highlights
Abbreviations
CRC, colorectal cancer; CRR, chemoradiotherapy resistance; CCK8, cell counting kit 8; qRT-PCR, quantitative real-time polymerase chain reaction; DCP1A, decapping enzyme 1a; WB, Western blot; RIP, RNA immunoprecipitation; ceRNAs, competitive endogenous RNAs; ATCC, American Type Culture Collection.
Data Sharing Statement
The analyzed data sets generated during the present study are available from the corresponding authors (Chao Li and Xiaosong Gu) upon reasonable request.
Ethics Approval and Consent to Participate
The present study was approved by the ethical review committee of The First Hospital of Hebei Medical University.
Patient Consent for Publication
Not applicable.
Author Contributions
All authors made substantial contribution to conception and design, acquisition of the data, or analysis and interpretation of the data; take part in drafting the article or revising it critically for important intellectual content; gave final approval of the revision to be published; and agree to be accountable for all aspect of the work.
Disclosure
The authors declare that they have no competing interests.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. doi:10.3322/caac.21442
2. Siegel RL, Miller KD, Fedewa SA, et al. Colorectal cancer statistics, 2017. CA Cancer J Clin. 2017;67(3):177–193. doi:10.3322/caac.21395
3. Rundle AG, Lebwohl B, Vogel R, Levine S, Neugut AI. Colonoscopic screening in average-risk individuals ages 40 to 49 vs 50 to 59 years. Gastroenterology. 2008;134(5):1311–1315. doi:10.1053/j.gastro.2008.02.032
4. Appelt AL, Ploen J, Harling H, et al. High-dose chemoradiotherapy and watchful waiting for distal rectal cancer: a prospective observational study. Lancet Oncol. 2015;16(8):919–927. doi:10.1016/S1470-2045(15)00120-5
5. Vodenkova S, Buchler T, Cervena K, Veskrnova V, Vodicka P, Vymetalkova V. 5-fluorouracil and other fluoropyrimidines in colorectal cancer: past, present and future. Pharmacol Ther. 2019;107447. doi:10.1016/j.pharmthera.2019.107447
6. Petrelli F, Cabiddu M, Barni S. 5-fluorouracil or capecitabine in the treatment of advanced colorectal cancer: a pooled-analysis of randomized trials. Med Oncol. 2012;29(2):1020–1029. doi:10.1007/s12032-011-9958-0
7. Li X, Yang L, Chen LL, Biogenesis T. Functions, and challenges of circular RNAs. Mol Cell. 2018;71(3):428–442. doi:10.1016/j.molcel.2018.06.034
8. Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20(11):675–691. doi:10.1038/s41576-019-0158-7
9. Liu J, Li D, Luo H, Zhu X. Circular RNAs: the star molecules in cancer. Mol Aspects Med. 2019;70:141–152. doi:10.1016/j.mam.2019.10.006
10. Yin Y, Long J, He Q, et al. Emerging roles of circRNA in formation and progression of cancer. J Cancer. 2019;10(21):5015–5021. doi:10.7150/jca.30828
11. Yan L, Liu G, Cao H, Zhang H, Shao F. Hsa_circ_0035483 sponges hsa-miR-335 to promote the gemcitabine-resistance of human renal cancer cells by autophagy regulation. Biochem Biophys Res Commun. 2019;519(1):172–178. doi:10.1016/j.bbrc.2019.08.093
12. Zhao Z, Ji M, Wang Q, He N, Li Y. Circular RNA Cdr1as upregulates SCAI to suppress cisplatin resistance in ovarian cancer via miR-1270 suppression. Mol Ther Nucleic Acids. 2019;18:24–33. doi:10.1016/j.omtn.2019.07.012
13. Liang Y, Song X, Li Y, et al. circKDM4C suppresses tumor progression and attenuates doxorubicin resistance by regulating miR-548p/PBLD axis in breast cancer. Oncogene. 2019;38(42):6850–6866. doi:10.1038/s41388-019-0926-z
14. Abu N, Hon KW, Jeyaraman S, et al. Identification of differentially expressed circular RNAs in chemoresistant colorectal cancer. Epigenomics. 2019;11(8):875–884. doi:10.2217/epi-2019-0042
15. Xiong W, Ai YQ, Li YF, et al. Microarray analysis of circular RNA expression profile associated with 5-fluorouracil-based chemoradiation resistance in colorectal cancer cells. Biomed Res Int. 2017;2017:8421614. doi:10.1155/2017/8421614
16. Song W, Fu T. Circular RNA-associated competing endogenous RNA network and prognostic nomogram for patients with colorectal cancer. Front Oncol. 2019;9:1181. doi:10.3389/fonc.2019.01181
17. Yuan W, Peng S, Wang J, et al. Identification and characterization of circRNAs as competing endogenous RNAs for miRNA-mRNA in colorectal cancer. PeerJ. 2019;7:e7602. doi:10.7717/peerj.7602
18. Yang D, Hu Z, Xu J, et al. MiR-760 enhances sensitivity of pancreatic cancer cells to gemcitabine through modulating Integrin beta1. Biosci Rep. 2019;39(11). doi:10.1042/BSR20192358
19. Tian T, Fu X, Lu J, et al. MicroRNA-760 inhibits doxorubicin resistance in hepatocellular carcinoma through regulating Notch1/Hes1-PTEN/Akt signaling pathway. J Biochem Mol Toxicol. 2018;32(8):e22167. doi:10.1002/jbt.22167
20. Hu SH, Wang CH, Huang ZJ, et al. miR-760 mediates chemoresistance through inhibition of epithelial mesenchymal transition in breast cancer cells. Eur Rev Med Pharmacol Sci. 2016;20(23):5002–5008.
21. Chiang PY, Shen YF, Su YL, et al. Phosphorylation of mRNA decapping protein Dcp1a by the ERK signaling pathway during early differentiation of 3T3-L1 preadipocytes. PLoS One. 2013;8(4):e61697. doi:10.1371/journal.pone.0061697
22. Dougherty JD, Reineke LC, Lloyd RE. mRNA decapping enzyme 1a (Dcp1a)-induced translational arrest through protein kinase R (PKR) activation requires the N-terminal enabled vasodilator-stimulated protein homology 1 (EVH1) domain. J Biol Chem. 2014;289(7):3936–3949. doi:10.1074/jbc.M113.518191
23. Wu C, Zhu X, Tao K, et al. MALAT1 promotes the colorectal cancer malignancy by increasing DCP1A expression and miR203 downregulation. Mol Carcinog. 2018;57(10):1421–1431. doi:10.1002/mc.22868
24. Wu C, Liu W, Ruan T, Zhu X, Tao K, Zhang W. Overexpression of mRNA-decapping enzyme 1a affects survival rate in colorectal carcinoma. Oncol Lett. 2018;16(1):1095–1100. doi:10.3892/ol.2018.8730
25. Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet. 2014;383(9927):1490–1502. doi:10.1016/S0140-6736(13)61649-9
26. Lyakhovich A, Lleonart ME. Bypassing mechanisms of mitochondria-mediated cancer stem cells resistance to chemo- and radiotherapy. Oxid Med Cell Longev. 2016;2016:1716341. doi:10.1155/2016/1716341
27. Li L, Zhu T, Gao YF, et al. Targeting DNA damage response in the radio(chemo)therapy of non-small cell lung cancer. Int J Mol Sci. 2016;17(6). doi:10.3390/ijms17060839
28. Cao L, Liu Y, Wang D, et al. MiR-760 suppresses human colorectal cancer growth by targeting BATF3/AP-1/cyclinD1 signaling. J Exp Clin Cancer Res. 2018;37(1):83. doi:10.1186/s13046-018-0757-8
29. Cong K, Li CG, Wei YH, Zhang K, Xu HB. MicroRNA-760 inhibits the biological progression of colorectal carcinoma by directly targeting FOXA1 and regulating epithelial-to-mesenchymal transition and PI3K/AKT signaling pathway. Eur Rev Med Pharmacol Sci. 2019;23(13):5730–5740. doi:10.26355/eurrev_201907_18310
30. Xian D, Zhao Y. LncRNA KCNQ1OT1 enhanced the methotrexate resistance of colorectal cancer cells by regulating miR-760/PPP1R1B via the cAMP signalling pathway. J Cell Mol Med. 2019;23(6):3808–3823. doi:10.1111/jcmm.14071
31. Shi C, Liu T, Chi J, et al. LINC00339 promotes gastric cancer progression by elevating DCP1A expression via inhibiting miR-377-3p. J Cell Physiol. 2019;234(12):23667–23674. doi:10.1002/jcp.28934
32. Tang Y, Xie C, Zhang Y, Qin Y, Zhang W. Overexpression of mRNA-decapping enzyme 1a predicts disease-specific survival in malignant melanoma. Melanoma Res. 2018;28(1):30–36. doi:10.1097/CMR.0000000000000406
33. Lee YT, Tan YJ, Oon CE. Molecular targeted therapy: treating cancer with specificity. Eur J Pharmacol. 2018;834:188–196. doi:10.1016/j.ejphar.2018.07.034
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.