Back to Journals » Cancer Management and Research » Volume 13
Circ_0005927 Inhibits the Progression of Colorectal Cancer by Regulating miR-942-5p/BATF2 Axis
Authors Yu C, Li D, Yan Q, Wang Y, Yang X, Zhang S, Zhang Y, Zhang Z
Received 10 December 2020
Accepted for publication 16 January 2021
Published 11 March 2021 Volume 2021:13 Pages 2295—2306
DOI https://doi.org/10.2147/CMAR.S281377
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Eileen O'Reilly
Chao Yu,* Deguan Li,* Qiang Yan, Yigao Wang, Xiaodong Yang, Shangxin Zhang, Yonghong Zhang, Zhen Zhang
Department of General Surgery, The First Affiliated Hospital of Anhui Medical University, Hefei, Anhui, 230022, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Zhen Zhang
Department of General Surgery, The First Affiliated Hospital of Anhui Medical University, No. 218, Jixi Road, Shushan District, Hefei, Anhui, 230022, People’s Republic of China
Tel +86-055162923887
Email [email protected]
Background: Colorectal cancer (CRC) is one of the most common aggressive neoplasms worldwide. Circular RNAs (circRNAs) have been involved in the biological process of CRC. This study aimed to explore the effects of circ_0005927 on CRC progression and underneath mechanism.
Materials and Methods: The expression of circ_0005927, microRNA-942-5p (miR-942-5p) and basic leucine zipper ATF-like transcription factor 2 (BATF2) was detected by quantitative real time polymerase chain reaction (qRT-PCR). The protein expression of BATF2 was determined by Western blot. The effects among circ_0005927, miR-942-5p and BATF2 on cell colony-forming ability, apoptosis and migratory and invasive abilities were revealed by cell colony formation, flow apoptosis and transwell assays, respectively. The associated relationship between miR-942-5p and circ_0005927 or BATF2 was predicted by Circinteractome or TargetScan online database, and identified by dual-luciferase reporter or RNA immunoprecipitation (RIP) assay. The impacts of circ_0005927 overexpression on tumor growth in vivo were investigated by in vivo tumor formation assay.
Results: Circ_0005927 expression and the mRNA and protein expression of BATF2 were dramatically downregulated, while miR-942-5p expression was obviously upregulated in CRC tissues or cells compared with control groups. Circ_0005927 overexpression repressed cell colony-forming ability, migration and invasion, whereas induced cell apoptosis in CRC; however, these impacts were hindered by miR-942-5p mimic or BATF2 knockdown. Furthermore, circ_0005927 was a sponge of miR-942-5p, and miR-942-5p bound to BATF2. In addition, circ_0005927 overexpression repressed tumor growth in vivo.
Conclusion: Circ_0005927 suppressed cell colony-forming ability, migration and invasion, and promoted cell apoptosis by sponging miR-942-5p to induce BATF2 in CRC. The possible mechanism provides a theoretical basis for the study of circRNA-directed therapy for CRC.
Keywords: circular RNA, circ_0005927, miR-942-5p, BATF2, CRC
Introduction
Colorectal cancer (CRC), a neoplasm in colon or rectum, is threatening the health of human.1–3 CRC is the second major cause of cancer-linked death, and the patients with CRC account for about 9% of total cancer patients.4–6 Although the great progress has been achieved in the treatment of CRC, some cases still have a poor prognosis. Therefore, profoundly understanding the etiopathogenesis of CRC is essential to seek reliable drug.
Circular RNA (circRNA) is a non-coding RNA with closed loop structure.7,8 CircRNA is more stable than linear RNA and constantly acts as a sponge of microRNA (miRNA).9 Accumulating data showed circRNAs participated in cancer progression, including CRC.10–13 Circ_0006332 was indicated to promote cell invasion via repressing miR-143 in bladder cancer;14 circ_001783 silencing suppressed the proliferative ability of breast cancer by binding to miR-200c-3p.15 In addition, yang et al illustrated circ_0137008 could suppress cell proliferation via associating with miR-338-5p in CRC.16 Du et al also explained circ_0038646 accelerated cell proliferative and invasive abilities of CRC via modulating miR-331-3p and glutamate receptor ionotropic kainate 3 (GRIK3).17 However, the effects of circ_0005927 on CRC development remain unknown.
MiRNAs are a flock of RNAs with about 19 nucleotides in length without protein coding capacity.18,19 MiRNA is a key ingredient of non-coding RNA and has been revealed to serve as a tumor promoter or suppressor.20–22 For instance, miR-942-5p suppressed cell proliferation and invasion in cervical cancer;23 miR-942-5p also participated in tumor metastasis in lung cancer.24 However, there was little data on the regulatory of CRC by miR-942-5p. Basic leucine zipper ATF-like transcription factor 2 (BATF2) takes part in the early process induced by type I IFN and acts as an antioncogene.25 BEAT2 has been related to CRC progression and poor clinical characteristic of CRC cases,26,27 which suggests that BATF2 plays an essential part in CRC progression.
In this study, the expression of circ_001783, miR-942-5p and BATF2 was determined in CRC cells. The effects among circ_001783, miR-942-5p and BATF2 on CRC progression were revealed. Furthermore, the mechanism by which circ_001783 mediated CRC malignancy progression was unveiled. In addition, the impacts of circ_001783 on tumor growth in vivo were disclosed.
Materials and Methods
Specimen Collection
Eighty-seven pairs of human CRC tissues and paracancerous normal intestinal tissues were acquired from CRC patients from the First Affiliated Hospital of Anhui Medical University during operation. Tissues were kept in liquid nitrogen and stored in a refrigerator at −80°C. This study was approved by the Ethics Committee of the First Affiliated Hospital of Anhui Medical University. The relevant patients signed the written informed consent.
Cell Acquirement and Culture
Human CRC cell lines (SW620, SW480, Caco-2 and LoVo) and human normal intestinal epithelial cell line NCM460 were purchased from Otwobiotech (Shenzhen, China). Cells were grown in Roswell Park Memorial Institute-1640 (RPMI-1640) (Thermo Fisher Scientific, Waltham, MA, USA) containing 10% fetal bovine serum (FBS; Thermo Fisher) and 1% streptomycin/penicillin (Thermo Fisher) in an incubator at 37°C with 5% CO2.
Cell Transfection
The overexpression plasmid of circ_0005927 (Lv-circ_0005927), small interfering RNA against circ_0005927 (si-circ_0005927), miR-942-5p mimic (miR-942-5p), miR-942-5p inhibitor (anti-miR-942-5p), small interfering RNA targeting BATF2 (si-BATF2) and their controls (Lv-NC, si-NC, miR-NC, anti-miR-NC and siRNA-NC) were synthesized by Ribobio Co., Ltd. (Guangzhou, China). Lipofectamine 2000 (Thermo Fisher) was employed for cell transfection. The sequences in transfection were si-circ_0005927 5ʹ-GCTAAGTACATGCTGGATT-3ʹ; miR-942-5p mimic 5ʹ-UCUUCUCUGUUUUGGCCAUGUG-3ʹ; miR-942-5p inhibitor 5ʹ-CACAUGGCCAAAACAGAGAAGA-3ʹ; si-BATF2 5ʹ-CCTCTGCTCAAGTCCACTT-3ʹ; si-NC 5ʹ-GCTCATGCGTAGGTAAATT; miR-NC 5ʹ-UUUGUACUACACAAAAGUACUG-3ʹ; anti-miR-NC 5ʹ-CAGUACUUUUGUGUAGUACAAA-3ʹ; siRNA-NC 5ʹ-CCTCTCGTGAAACCTCCTT-3ʹ.
Quantitative Real Time Polymerase Chain Reaction (qRT-PCR)
CRC tissues and cells were lysed with trizol reagent (Takara, Dalian, China). RNA concentration was measured via NanoDrop-1000 apparatus (Thermo Fisher). cDNA was amplified using PrimeScript RT Master Mix (Takara). For determining the expression of circ_0005927, miR-942-5p, BATF2 and glyceraldehyde 3-phosphate dehydrogenase (GAPDH), SYBR Green SuperMix (Roche, Basel, Switzerland) was employed. Data were analyzed using the 2−∆∆Ct method. U6 and GAPDH were selected as references. The forward and reverse primers in this part were circ_0005927 5ʹ-TCTCTCTCTAAAGAAGAGTGGGAA-3ʹ and 5ʹ-CTGATAGCCAGCAAGCCACC-3ʹ; GAPDH 5ʹ-TCGGAGTCAACGGATTTGGT-3ʹ and 5ʹ-TTCCCGTTCTCAGCCTTGAC-3ʹ; miR-942-5p 5ʹ-CTCACGTCTTCTCTGTTTT-3ʹ and 5ʹ-ACCTCAAGAACAGTATTTCCAGG-3ʹ; BATF2 5ʹ-GACAGACCCCAAGGAGCAAC-3ʹ and 5ʹ-GGCGAGGTTGTCTTTTTCCAG; U6 5ʹ-TGCGGGTGCTCGCTTCGGCAGC-3ʹ and 5ʹ-GTGCAGGGTCCGAGGT-3ʹ.
RNase R Treatment Assay
LoVo and SW620 cells were harvested and washed with phosphate buffer solution (PBS; Thermo Fisher). Total RNA was extracted with trizol (Takara) and divided into two groups. Following that, RNA was incubated with or without RNase R (Epicentre, Madison, WI, USA) at 37°C for 30 min, respectively. The enrichment of circ_0005927 and GAPDH was determined by qRT-PCR.
Cell Colony Formation Assay
LoVo and SW620 cells were grown in 6-well plate (500 cells per well) for 2 weeks after various treatments. The proliferating colonies were immobilized using 4% paraformaldehyde (Beyotime, Shanghai, China) and dyed with crystal violet (Beyotime). Finally, cell colony-forming ability was assessed by calculating the colonies under microscope (Olympus, Tokyo, Japan). More than 50 cells per colony were considered as a colony.
Flow Cytometry Analysis
Annexin V-fluorescein isothiocyanate (Annexin V-FITC) apoptosis detection kit (Beyotime) was performed to reveal the apoptosis of LoVo and SW620 cells. In short, trypsin (Beyotime) was added into plate to digest cells at 48 h after treatment, and cells were washed using PBS (Thermo Fisher). Then, cells were centrifuged at 800 rpm for 8 min. Cells were suspended in 195 μL binding buffer. Annexin V-FITC and propidium iodide (PI) were used to fix and stain cells, respectively. Results were assessed by flow cytometry (BD Biosciences, San Diego, CA, USA).
Transwell Assay
The migratory and invasive abilities of LoVo and SW620 cells were detected by transwell chamber without or with Matrigel (Corning, New York, Madison, USA). Shortly, cells were suspended in FBS-free RPMI-1640 media and added into the upper chamber. And RPMI-1640 media with 20% FBS was placed into the lower chamber. After 24 h, cells were washed using PBS (Thermo Fisher). Cells were immobilized using paraformaldehyde (Beyotime) and stained with crystal violet (Beyotime). Results were assessed with microscope (Olympus) with a 100 magnification.
Dual-Luciferase Reporter Assay
The binding sites between miR-942-5p and circ_0005927 or BATF2 were predicted by Circinteractome or TargetScan online database. The wide-type (WT) sequences of circ_0005927 and BATF2 3ʹ-untranslated regions (3ʹUTR) containing the binding sites of miR-942-5p were fused into pmirGLO vector (Promega, Madison, WI, USA), and named as circ_0005927 WT and BATF2 3ʹUTR WT, respectively. The mutant (MUT) binding sites of circ_0005927 (circ_0005927 MUT) and BATF2 3ʹUTR (BATF2 3ʹUTR MUT) in miR-942-5p were sub-cloned into pmirGLO vector (Promega). Plasmids were transduced into cells with miR-942-5p mimic or miR-NC using Lipofectamine 2000 (Thermo Fisher) and cells were cultured for 48 h. Luciferase activities were detected via dual luciferase reporter assay kit (Promega) with the normalization to Ranilla luciferase activity.
RNA Immunoprecipitation (RIP) Assay
The associated relationship between miR-942-5p and circ_0005927 was identified with RIP assay. Magna RIP Kit (Millipore, Bradford, MA, USA) was employed. Shortly, LoVo and SW620 cells were harvested and lysed with RIP lysis buffer (Millipore). The lysate was incubated with magnetic beads labeled with anti-argonaute2 (anti-AGO2) (1:50; Abcam, Cambridge, UK) or anti-immunoglobulin G (anti-IgG) (1:100; Abcam). After 24 h, beads were washed and protease K (Millipore) was used to digest proteins. RNA was purified and the contents of miR-942-5p and circ_0005927 were revealed by qRT-PCR.
Western Blot Analysis
CRC tissues and cells were lysed with RIPA buffer (Beyotime). Lysate was loaded by 12% sodium dodecyl SDS polyacrylamide gel (SDS-PAGE) (Beyotime). Protein bands were transfected into the nitrocellulose membranes (Membrane Solutions, Shanghai, China). After blocked in 5% fat-free milk (Beyotime), membranes were incubated with anti-BATF2 (1:2000; Affinity, Nanjing, china) and anti-GAPDH (1:5000; Affinity). After that, bands were incubated with the secondary antibody coated with horseradish peroxidase (1:5000; Affinity). Protein bands were visualized under enhanced chemiluminescence (KeyGen, Nanjing, China). GAPDH was employed as a control.
In vivo Tumor Formation Assay
BALB/c nude mice (6-week old) were obtained from Charles River (Beijing, China) and grown in gnotobasis. Nude mice were divided into the two groups (Lv-circ_0005927 group and Lv-NC group, N=6 per group). 5×106 cells transfected with Lv-circ_0005927 or Lv-NC were hypodermically injected into the mice. After 5 days, tumor volume was detected each 5 days. After 30 days, mice were euthanized and tumors were excised. And the size and weight of each tumor were measured. The effects of circ_0005927 overexpression on the expression of miR-942-5p and BATF2 were determined by qRT-PCR and Western blot, respectively. The Animal Care and Use Committee of the First Affiliated Hospital of Anhui Medical University agreed with this experiment. This study complied with the National Institutes of Health guide for the care and use of Laboratory animals (NIH Publications No. 8023, revised 1978).
Statistical Analysis
Data were analyzed with SPSS 21.0 software (IBM, Somers, NY, USA) based on at least 3 experiments. Data were shown as means ± standard deviations. Significant differences were assessed with two-tailed Student’s t-tests between the two groups or compared with one-way analysis of variance among three or more groups. Spearman correlation test was performed to analyze the significant differences in Spearman correlation analysis. A chi-square test was conducted to compare the groups between low and high circ_0005927 expression. P value <0.05 was considered to be statistically significant.
Results
Circ_0005927 Expression Was Downregulated in CRC Tissues and Cells with Poor Clinical Outcome
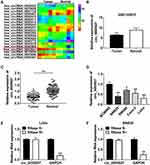
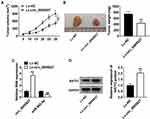
The differently expressed circRNAs in CRC tissues were firstly analyzed by GSE142837 database. Results showed that circ_0005927 expression was lower in CRC tissues than normal intestinal tissues (Figure 1A and B). QRT-PCR results showed that circ_0005927 was lowly expressed in CRC tissues (N=87) and SW620, SW480, Caco-2 and LoVo cells as compared to normal intestinal tissues (N=87) and NCM460 cells, respectively (Figure 1C and D). LoVo and SW620 cells were employed for further study based on the lower circ_0005927 expression in the two types of cells. Subsequently, the stability of circ_0005927 was explored by RNase R treatment assay. Data presented circ_0005927 expression had no obvious change after RNase R treatment, whereas GAPDH expression was significantly downregulated in LoVo and SW620 cells (Figure 1E and F). Furthermore, decreased expression of circ_0005927 was closely correlated with tumor size (≥5 cm), tumor (T) stage (T1+T2) and lymphatic node (N) metastasis (N0), but there was no relevant relation between circ_0005927 expression and the age and gender of patients or tumor differentiation (Table 1). These findings showed that circ_0005927 was lowly expressed in CRC tissues and cells with poor clinical characteristic.
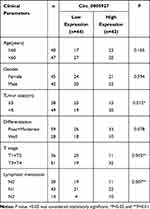 |
Table 1 Correlation Between Circ_0005927 Expression and Clinicopathological Characteristics of Patients with Colorectal Cancer |
Circ_0005927 Inhibited Cell Colony-Forming Ability, Migration and Invasion, Whereas Induced Cell Apoptosis in CRC
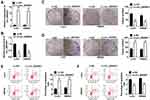
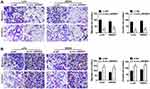
The function of circ_0005927 in CRC progression was further revealed. The overexpression and interfering plasmids of circ_0005927 were primarily constructed. Results showed circ_0005927 expression was dramatically upregulated and downregulated after Lv-circ_0005927 and si-circ_0005927 transfection, respectively (Figure 2A and B). Subsequently, cell colony formation assay unveiled that the colony numbers of LoVo and SW620 cells were significantly decreased after transfection of Lv-circ_0005927 and were evidently increased by si-circ_0005927 when compared with control groups (Figure 2C and D), suggesting that circ_0005927 suppressed cell colony-forming ability. Meanwhile, cell apoptosis was promoted after Lv-circ_0005927 transfection and was repressed by si-circ_0005927 in LoVo and SW620 cells (Figure 2E and F). Transwell assay disclosed that circ_0005927 overexpression repressed cell migration and invasion, and circ_0005927 knockdown enhanced cell migratory and invasive abilities in LoVo and SW620 cells (Figure 3A and B). All data revealed that circ_0005927 functioned as a cancer suppressor in CRC progression.
Circ_0005927 Acted as a Sponge of miR-942-5p
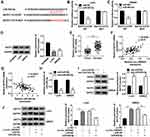
In order to explain how circ_0005927 affected CRC process, the associated miRNA with circ_0005927 was predicted by Circinteractome online database. Result showed that circ_0005927 contained the binding sites of miR-942-5p (Figure 4A). Subsequently, miR-942-5p mimic was synthesized and its overexpression efficiency was detected. Results displayed that miR-942-5p expression was dramatically upregulated after miR-942-5p mimic transfection (Figure 4B). And dual-luciferase reporter assay demonstrated that the luciferase activity of circ_0005927 WT+miR-942-5p group was markedly repressed, whereas the luciferase activity of circ_0005927 MUT+miR-942-5p group had no obvious change (Figure 4C and D). RIP assay revealed that circ_0005927 and miR-942-5p could be dramatically enriched by AGO2 compared with IgG in LoVo and SW620 cells (Figure 4E and F). Additionally, qRT-PCR results manifested miR-942-5p expression was upregulated in LoVo and SW620 cells and CRC tissues in contrast to control groups (Figure 4G and H). Spearman correlation analysis disclosed that circ_0005927 expression was negatively correlated with miR-942-5p expression (Figure 4I). Overall, our findings demonstrated that circ_0005927 was associated with miR-942-5p in LoVo and SW620 cells.
Circ_0005927 Sponged miR-942-5p to Induce BATF2
Given the effects of circ_0005927 on CRC development and the relationship between circ_0005927 and miR-942-5p, the target gene of miR-942-5p was continued to be explored. TargetScan online database showed that BATF2 3ʹUTR contained the binding sites of miR-942-5p (Figure 5A). Dual-luciferase reporter assay presented that luciferase activity was dramatically repressed after BATF2 3ʹUTR WT and miR-942-5p mimic co-transfection, but there was no significant change in BATF2 3ʹUTR MUT+miR-942-5p mimic group in LoVo and SW620 cells (Figure 5B and C). Subsequently, BATF2 expression was determined in LoVo and SW620 cells and CRC tissues. Results showed the protein expression of BATF2 was dramatically repressed in LoVo and SW620 cells in contrast to NCM460 cells (Figure 5D), and BATF2 mRNA expression was apparently downregulated in CRC tissues in contrast to normal intestinal tissues (Figure 5E). Spearman correlation analysis manifested that BATF2 expression was positively related to circ_0005927 expression and was negatively correlated with miR-942-5p expression (Figure 5F and G). Furthermore, the impact of miR-942-5p inhibitor on BATF2 protein expression was investigated. The interfering efficiency of miR-942-5p inhibitor was primarily determined, and result exhibited miR-942-5p expression was aberrantly downregulated by anti-miR-942-5p (Figure 5H). Western blot analysis exhibited that the protein expression of BATF2 was notably upregulated by miR-942-5p inhibitor in LoVo and SW620 cells (Figure 5I). In addition, BATF2 protein expression was dramatically increased by circ_0005927 overexpression, whereas this promotion effect was attenuated by miR-942-5p mimic in LoVo and SW620 cells (Figure 5J). All data revealed that circ_0005927 could upregulate BATF2 expression by sponging miR-942-5p in LoVo and SW620 cells.
Circ_0005927 Overexpression Inhibited CRC Progression by Regulating miR-942-5p and BATF2
The relation among circ_0005927, miR-942-5p and BATF2 on CRC progression was disclosed in this part. The knockdown efficiency of si-BATF2 was firstly determined. And results suggested that the protein expression of BATF2 was remarkably repressed by si-BATF2 (Figure 6A). Subsequently, cell colony formation assay revealed that circ_0005927 overexpression inhibited cell colony-forming ability, whereas this effect was restored by miR-942-5p mimic or BATF2 knockdown in LoVo and SW620 cells (Figure 6B). Flow cytometry analysis manifested that hypernormal circ_0005927 expression accelerated the apoptosis of LoVo and SW620 cells, but this impact was abolished by miR-942-5p overexpression or BATF2 depletion (Figure 6C). Transwell assay showed that circ_0005927 overexpression restrained cell migration and invasion; however, miR-942-5p mimic or BATF2 silencing hindered these influences (Figure 6D and E). Overall, these findings demonstrated that circ_0005927 suppressed CRC development by regulating miR-942-5p and BATF2.
Circ_0005927 Overexpression Inhibited Tumor Growth in vivo by miR-942-5p/BATF2 Pathway
The effects of circ_0005927 overexpression on tumor growth were further unveiled in vivo. Results displayed that circ_0005927 overexpression suppressed tumor volume and decreased tumor weight (Figure 7A and B). Subsequently, the impacts of circ_0005927 on miR-942-5p and BATF2 expression in vivo were investigated. The overexpression efficiency of Lv-circ_0005927 was firstly detected. Result demonstrated circ_0005927 expression was markedly upregulated after Lv-circ_0005927 transfection in vivo (Figure 7C), suggesting the overexpression plasmid of circ_0005927 was successfully built. QRT-PCR showed that circ_0005927 overexpression evidently repressed miR-942-5p expression in vivo (Figure 7C). In addition, BATF2 protein expression was obviously upregulated by circ_0005927 overexpression (Figure 7D). These data meant that circ_0005927 overexpression hindered tumor growth in vivo by upregulating BATF2 expression and downregulating miR-942-5p expression.
Discussion
CRC is a malignant tumor in colon and rectum worldwide and its incidence is increasing.2 And CRC poses a heavy burden to human’s health and seriously declines the quality of people’s life.28 However, the effective drug is lacking in treating CRC. CircRNA has been reported to regulate CRC process.29,30 Therefore, exploring the regulatory mechanism of CRC by circRNAs is necessary to search drug in treating CRC.
CircRNAs were unveiled to mainly act as a carcinogen in CRC progression. Yin et al indicated that circ_0007142 contributed to CRC development via sponging miR-122-5p.31 Chen et al revealed that circ_001971 silencing repressed CRC process by inhibiting cell proliferative and invasive abilities through binding to miR-29c-3p.32 In addition, circ_0001946 was found to enhance the proliferative and migratory abilities of CRC cells.33 In our study, circ_0005927 was disclosed to repress CRC progression for the first time. We found circ_0005927 expression was downregulated in CRC tissues and cells. Subsequently, to unveil the effects of circ_0005927 on CRC process, gain- and loss-of-function experiments were employed. Results confirmed that enforced circ_0005927 expression suppressed cell colony-forming ability, cell migration and cell invasion, whereas accelerated cell apoptosis in CRC. And circ_0005927 overexpression restrained tumor growth in vivo. To determine how circ_0001946 affected CRC progression, the associated miRNA with circ_0001946 was predicted and identified. We found circ_0005927 was associated with miR-942-5p. Additionally, miR-942-5p is bound to BATF2.
MiR-942-5p expression was found to be upregulated in CRC tissues and cells in this study. MiR-942-5p attenuated the inhibition effects of circ_0001946 overexpression on cell colony-forming, migration and invasion, and the promotion effect of enforced expression of circ_0001946 on cell apoptosis, which meant that miR-942-5p promoted CRC process. Wang et al unveiled that miR-942-5p mimic restored the inhibition of LIFR antisense RNA 1 on the number of migrated and invaded cells in lung cancer;24 Lu et al manifested circ-centrosomal protein 85 kDa-like (circ-CEP85L) inhibited cell proliferation and invasion by binding to miR-942-5p in gastric cancer.34 These findings also suggested that miR-942-5p served as a promoter in cancer progression.
BATF2 has been illustrated to participate in the process of gastric cancer.35–37 To reveal the influences of BATF2 on CRC development, Lv-circ_0001946 and si-BATF2 were transfected into LoVo and SW620 cells with control groups. Results displayed that circ_0001946 overexpression hindered cell colony-forming ability, migration and invasion, whereas promoted cell apoptosis in CRC; however, these impacts were abolished by BATF2 silencing. Videlicet, BATF2 suppressed CRC process. Liu et al demonstrated that enforced BATF2 expression facilitated cell apoptosis and hindered the proliferative, migratory and invasive abilities of CRC cells.26,27 Our findings were consistent with Liu’s results.
Collectively, the expression of circ_0001946 and BATF2 was distinctly downregulated, miR-942-5p expression was dramatically upregulated in CRC tissues and cells. Circ_0001946 overexpression hindered cell colony-forming ability, migration and invasion, but accelerated cell apoptosis via sponging miR-942-5p to upregulate BATF2. Additionally, enforced circ_0001946 expression inhibited tumor growth in vivo. Our results reveal a new mechanism in circRNA-mediated CRC development and provide a theoretical basis for the study of CRC treatment.
Funding
There is no funding to report.
Disclosure
The authors declare that they have no financial conflicts of interest. The authors report no conflicts of interest in this work.
References
1. Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. doi:10.3322/caac.20107
2. Brody H. Colorectal cancer. Nature. 2015;521(7551):S1. doi:10.1038/521S1a
3. Dekker E, Tanis PJ, Vleugels JLA, Kasi PM, Wallace MB. Colorectal cancer. Lancet. 2019;394(10207):1467–1480. doi:10.1016/S0140-6736(19)32319-0
4. Zhang W, Yang S, Liu Y, et al. Hsa_circ_0007534 as a blood-based marker for the diagnosis of colorectal cancer and its prognostic value. Int J Clin Exp Pathol. 2018;11(3):1399–1406.
5. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
6. Mármol I, Sánchez-de-Diego C, Pradilla Dieste A, Cerrada E, Rodriguez Yoldi MJ. Colorectal Carcinoma: A General Overview and Future Perspectives in Colorectal Cancer. Int J Mol Sci. 2017;18(1):197. doi:10.3390/ijms18010197
7. Qu S, Yang X, Li X, et al. Circular RNA: a new star of noncoding RNAs. Cancer Lett. 2015;365(2):141–148. doi:10.1016/j.canlet.2015.06.003
8. Ebbesen KK, Hansen TB, Kjems J. Insights into circular RNA biology. RNA Biol. 2017;14(8):1035–1045. doi:10.1080/15476286.2016.1271524
9. Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi:10.1038/nature11993
10. Geng Z, Wang W, Chen H, Mao J, Li Z, Zhou J. Circ_0001667 promotes breast cancer cell proliferation and survival via Hippo signal pathway by regulating TAZ. Cell Biosci. 2019;9:104. doi:10.1186/s13578-019-0359-y
11. Huang MS, Liu JY, Xia XB, et al. Hsa_circ_0001946 inhibits lung cancer progression and mediates cisplatin sensitivity in non-small cell lung cancer via the nucleotide excision repair signaling pathway. Front Oncol. 2019;9:508. doi:10.3389/fonc.2019.00508
12. Li H, Jin X, Liu B, Zhang P, Chen W, Li Q. CircRNA CBL.11 suppresses cell proliferation by sponging miR-6778-5p in colorectal cancer. BMC Cancer. 2019;19(1):826. doi:10.1186/s12885-019-6017-2
13. Xu H, Wang C, Song H, Xu Y, Ji G. RNA-Seq profiling of circular RNAs in human colorectal cancer liver metastasis and the potential biomarkers. Mol Cancer. 2019;18(1):8. doi:10.1186/s12943-018-0932-8
14. Li M, Liu Y, Liu J, et al. Circ_0006332 promotes growth and progression of bladder cancer by modulating MYBL2 expression via miR-143. Aging. 2019;11(22):10626–10643. doi:10.18632/aging.102481
15. Liu Z, Zhou Y, Liang G, et al. Circular RNA hsa_circ_001783 regulates breast cancer progression via sponging miR-200c-3p. Cell Death Dis. 2019;10(2):55. doi:10.1038/s41419-018-1287-1
16. Yang Z, Zhang J, Lu D, et al. Hsa_circ_0137008 suppresses the malignant phenotype in colorectal cancer by acting as a microRNA-338-5p sponge. Cancer Cell Int. 2020;20:67. doi:10.1186/s12935-020-1150-1
17. Du H, He Z, Feng F, et al. Hsa_circ_0038646 promotes cell proliferation and migration in colorectal cancer via miR-331-3p/GRIK3. Oncol Lett. 2020;20(1):266–274. doi:10.3892/ol.2020.11547
18. Moya L, Meijer J, Schubert S, Matin F, Batra J. Assessment of miR-98-5p, miR-152-3p, miR-326 and miR-4289 expression as biomarker for prostate cancer diagnosis. Int J Mol Sci. 2019;20(5):1154. doi:10.3390/ijms20051154
19. Lu TX, Rothenberg ME. MicroRNA. J Allergy Clin Immunol. 2018;141(4):1202–1207. doi:10.1016/j.jaci.2017.08.034
20. Lv F, Zheng K, Yu J, Huang Z. MicroRNA-661 expression is upregulated in pancreatic ductal adenocarcinoma and promotes cell proliferation. Oncol Lett. 2018;16(5):6293–6298. doi:10.3892/ol.2018.9454
21. Zhou Z, Wu L, Liu Z, et al. MicroRNA-214-3p targets the PLAGL2-MYH9 axis to suppress tumor proliferation and metastasis in human colorectal cancer. Aging (Albany NY). 2020;12(10):9633–9657. doi:10.18632/aging.103233
22. To KK, Tong CW, Wu M, Cho WC. MicroRNAs in the prognosis and therapy of colorectal cancer: From bench to bedside. World J Gastroenterol. 2018;24(27):2949-2973. doi:10.3748/wjg.v24.i27.2949
23. Ou R, Mo L, Tang H, et al. circRNA-AKT1 Sequesters miR-942-5p to Upregulate AKT1 and Promote Cervical Cancer Progression. Mol Ther Nucleic Acids. 2020;20:308–322. doi:10.1016/j.omtn.2020.01.003
24. Wang Q, Wu J, Huang H, et al. lncRNA LIFR-AS1 suppresses invasion and metastasis of non-small cell lung cancer via the miR-942-5p/ZNF471 axis. Cancer Cell Int. 2020;20:180. doi:10.1186/s12935-020-01228-5
25. Su ZZ, Lee SG, Emdad L, et al. Cloning and characterization of SARI (suppressor of AP-1, regulated by IFN). Proc Natl Acad Sci U S A. 2008;105(52):20906–20911. doi:10.1073/pnas.0807975106
26. Liu Z, Wei P, Yang Y, et al. BATF2 deficiency promotes progression in human colorectal cancer via activation of HGF/MET signaling: a potential rationale for combining MET inhibitors with IFNs. Clin Cancer Res. 2015;21(7):1752–1763. doi:10.1158/1078-0432.CCR-14-1564
27. Liu Z, Yang Y, Zhou X. BATF2 in human colorectal cancer. Aging (Albany NY). 2015;7(5):284–285. doi:10.18632/aging.100749
28. Jian X, He H, Zhu J, et al. Hsa_circ_001680 affects the proliferation and migration of CRC and mediates its chemoresistance by regulating BMI1 through miR-340. Mol Cancer. 2020;19(1):20. doi:10.1186/s12943-020-1134-8
29. Lu X, Yu Y, Liao F, Tan S. Homo sapiens circular RNA 0079993 (hsa_circ_0079993) of the POLR2J4 gene acts as an oncogene in colorectal cancer through the microRNA-203a-3p.1 and CREB1 axis. Med Sci Monit. 2019;25:6872–6883. doi:10.12659/MSM.916064
30. Xu H, Liu Y, Cheng P, et al. CircRNA_0000392 promotes colorectal cancer progression through the miR-193a-5p/PIK3R3/AKT axis. J Exp Clin Cancer Res. 2020;39(1):283. doi:10.1186/s13046-020-01799-1
31. Yin W, Xu J, Li C, Dai X, Wu T, Wen J. Circular RNA circ_0007142 facilitates colorectal cancer progression by modulating CDC25A expression via miR-122-5p. Onco Targets Ther. 2020;13:3689–3701. doi:10.2147/OTT.S238338
32. Chen C, Huang Z, Mo X, et al. The circular RNA 001971/miR-29c-3p axis modulates colorectal cancer growth, metastasis, and angiogenesis through VEGFA. J Exp Clin Cancer Res. 2020;39(1):91. doi:10.1186/s13046-020-01594-y
33. Deng Z, Li X, Wang H, et al. Dysregulation of CircRNA_0001946 contributes to the proliferation and metastasis of colorectal cancer cells by targeting microRNA-135a-5p. Front Genet. 2020;11:357. doi:10.3389/fgene.2020.00357
34. Lu J, Wang YH, Huang XY, et al. circ-CEP85L suppresses the proliferation and invasion of gastric cancer by regulating NFKBIA expression via miR-942-5p. J Cell Physiol. 2020;235(9):6287–6299. doi:10.1002/jcp.29556
35. Yang W, Wu B, Ma N, et al. BATF2 reverses multidrug resistance of human gastric cancer cells by suppressing Wnt/β-catenin signaling. In Vitro Cell Dev Biol Anim. 2019;55(6):445–452. doi:10.1007/s11626-019-00360-5
36. Yang W, Zhao S, Wu B, et al. BATF2 inhibits chemotherapy resistance by suppressing AP-1 in vincristine-resistant gastric cancer cells. Cancer Chemother Pharmacol. 2019;84(6):1279–1288. doi:10.1007/s00280-019-03958-4
37. Xie JW, Huang XB, Chen QY, et al. m6A modification-mediated BATF2 acts as a tumor suppressor in gastric cancer through inhibition of ERK signaling. Mol Cancer. 2020;19(1):114. doi:10.1186/s12943-020-01223-4
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.