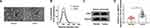Back to Journals » Cancer Management and Research » Volume 13
Circ_0004771 Accelerates Cell Carcinogenic Phenotypes via Suppressing miR-1253-Mediated DDAH1 Inhibition in Breast Cancer
Authors Ding X, Zheng J, Cao M
Received 27 July 2020
Accepted for publication 4 December 2020
Published 6 January 2021 Volume 2021:13 Pages 1—11
DOI https://doi.org/10.2147/CMAR.S273783
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Sanjeev K. Srivastava
Xubei Ding,1,* Junjun Zheng,2,* Mingxiang Cao3
1Thyroid and Breast Surgery, Jingmen No.1 People’s Hospital, Jingmen, Hubei, People’s Republic of China; 2Pharmacy Intravenous Admixture Services, Jingmen No.2 People’s Hospital, Jingmen, Hubei, People’s Republic of China; 3Department of Anesthesiology, Jingmen No.1 People’s Hospital, Jingmen, Hubei, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Mingxiang Cao
Department of Anesthesiology, Jingmen No.1 People’s Hospital, No. 168, Xiangshan Street, Jingmen, Hubei 448000, People’s Republic of China
Email [email protected]
Background: Circ_0004771 was demonstrated to mediate cell growth promotion and apoptosis suppression in breast cancer (BC). Herein, the precise functions and mechanism of circ_0004771 in the biological property of BC cells were investigated.
Methods: The expression of circ_0004771, microRNA (miR)-1253 and dimethylarginine dimethylaminohydrolase 1 (DDAH1) mRNA was analyzed using quantitative real-time polymerase chain reaction. The proliferation, apoptosis, migration, invasion, adhesion, Western blot and in vivo tumorigenesis assays were employed to evaluate the roles of circ_0004771 and DDAH1 in BC tumorigenesis. The interaction between miR-1253 and circ_0004771 or DDAH1 was validated by dual-luciferase reporter, pull-down and RNA immunoprecipitation (RIP) assay. Exosomes were isolated by Exoquick-TC® methods, and qualified using Nanosight™ technology and Western blot.
Results: Circ_0004771 or DDAH1 expression was elevated in BC, and silencing either of them suppressed cell malignant phenotypes, thus impeding BC progression. Importantly, circ_0004771 up-regulation attenuated the anticancer action of DDAH1-knockdown in BC. Additionally, we confirmed that circ_0004771 functioned as a sponge of miR-1253 to up-regulate DDAH1 expression. Moreover, xenograft assay exhibited that circ_0004771 knockdown also hindered tumor growth in vivo via regulating DDAH1 and miR-1253. Besides that, it was found that circ_0004771 was packaged into exosomes isolated from the serum of BC.
Conclusion: Circ_0004771 accelerated cell carcinogenic phenotypes via up-regulating DDAH1 expression through absorbing miR-1253 in BC. Besides, circ_0004771 was packaged into exosomes isolated from the serum of BC. All these findings suggested a promising molecular target for BC treatment.
Keywords: circ_0004771, miR-1253, DDAH1, breast cancer, exosome
Introduction
On a global scale, breast cancer (BC) is the most common malignancy in women, and its incidence and mortality rates are increased significantly.1 Although great improvements in the cancer research setting, breast cancer is still a major health threat for women.2,3 Hence, an investigation of the molecular mechanisms of this cancer is necessary to develop novel treatment strategies for patients with breast cancer.
Circular RNAs (circRNAs) are a subgroup of noncoding RNA transcripts highlighted by a ring structure that render them resistant to RNase R decay, so they can be more stably present in tissues and cells.4 CircRNAs are largely generated from exons, introns, or intergenic regions, and abundantly represented in eukaryotes with cell/tissue-specific expression patterns.5 Additionally, increasing evidence indicates that circRNAs are involved in the occurrence and development of malignant cancers via the modulation of cell crucial biological and pathological processes, like cell growth, metastasis, differentiation, chemoresistance and metabolism.6–8 Thus, circRNAs may be ideal candidates for the early detection, treatment and prognosis of cancers. Recently, the aberrant expression of some circRNAs has been found to be associated with the development and progression of breast cancer.9 Circ_0004771 is a functional circRNA, which was found to function as a sponge of microRNA (miRNA/miR)-339-5p to modulate CDC25A expression, and subsequently facilitated the proliferation and growth of esophageal squamous cell cancer.10 Importantly, Xie et al uncovered an elevation of circ_0004771 expression in breast cancer, which stimulated cell growth and suppressed cell apoptosis in breast cancer.11 Thus, circ_0004771 might play vital roles in the progression of breast cancer. Nevertheless, the molecular mechanisms underlying circ_0004771 action in the tumorigenesis of breast cancer remain vague.
Dimethylarginine dimethylaminohydrolase 1 (DDAH1) is the critical enzyme that metabolizes asymmetric dimethylarginine (ADMA) and N-monomethyl L-arginine (L-NMMA), inhibitors of endogenous nitric oxide synthase (NOS), and DDAH1 overexpression induces ADMA decrease, NO production increase and NOS activity,12,13 which has been exhibited to enhance angiogenesis of cancers accompanying by an increase in tumor growth and metastatic potential.14,15 DDAH1 was found to be elevated in many of types of cancers, such as gastric cancers,16 glioma17 and prostate cancer,18 and acted as an oncogene to promote cancer progression through enhancing cancer cell carcinogenic biological behaviors. Besides that, recent evidence reported that DDAH1 was also increased in breast cancer, and conversed ADMA to citrulline and promoted cell vasculogenic mimicry and migration.19,20 Thus, DDAH1 is a potential therapeutic target for breast cancer.
Herein, this study focused on investigating the role of circ_0004771 and DDAH1 in the malignant phenotypic changes of breast cancer cells, and explored the regulatory network between circ_0004771 and DDAH1 in breast cancer progression.
Materials and Methods
Clinical Samples
BC tissues and adjacent non-tumor tissues of 70 BC patients were collected from the surgical specimen archives of Jingmen No.1 People’s Hospital, and then stored in liquid nitrogen at −80°C. In the meanwhile, blood samples were collected from 29 BC patients and 30 healthy controls. Following the centrifugation at 3000 g for 10 min, the supernatant serum was obtained and then stored at −80°C until used. This study was conducted in accordance with the Declaration of Helsinki. All the procedures were permitted by the Ethics Committee of Jingmen No.1 People’s Hospital, and all enrolled individuals had provided written informed consent.
Cell Culture and Transfection
Human breast cancer cell lines (T47D and MB231) and MCF-10A cell lines purchased from Beijing Institute for Cancer Research Collection (Beijing, China) were grown in the Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 100 U/mL penicillin, and 100 U/mL streptomycin (Gibco, Carlsbad, CA, USA) at 37°C in a 5% CO2 humidified atmosphere.
When the confluence reached to 60–70%, 50 nM of siRNA against circ_0004771 (si-circ#1, si-circ#2, si-circ#3), siRNA against DDAH1 (si-DDAH1#1, si-DDAH1#2, si-DDAH1#3) or a scramble siRNA (si-NC) (GenePharma, Shanghai, China), 100 ng of pcDNA3.1 circ_0004771 overexpression vector (circ_0004771) or nontarget plasmid (vector) (Promega, Madison, WI, USA), or 50 nM of miR-1253 mimic or miR-1253 inhibitor (miR-1253, anti-miR-1253) or their negative control (miR-NC, anti-NC) (GenePharma) were transfected into T47D and MB231 cells using Lipofectamine 3000 reagent obtained from Invitrogen (Carlsbad, CA, USA).
RNA Extraction and qRT-PCR
Whole-RNA extracts were conducted in certain cells and tissues using TRIzol reagent (Invitrogen). Then complementary DNA (cDNA) was generated from extracted RNA through a reverse transcription kit (TaKaRa, Tokyo, Japan), and the synthesized cDNA template was determined by SYBR-Green PCR kit (TaKaRa) on an ABI 7500 Real-Time PCR system. The relative fold changes were normalized to glyceraldehyde 3-phosphate dehydrogenase (GADPH) or U6 and detected using a 2−ΔΔCt method. The primer in the present work was listed as follows: circ_0004771: F, 5ʹ-AGTTGCTCCAATGACAGAGTTACC-3ʹ and R, 5ʹ-CCTCCTTCAGTCAAGTGTGCATC-3ʹ; DDAH1: F, 5ʹ-ACTCACTGTGCCTGATGACA-3ʹ and R, 5ʹ-TCCAGTTCAGACATGCTCA-3ʹ; GADPH: F 5ʹ-GTCAACGGATTTGGTCTGTATT-3ʹ and R 5ʹ- AGTCTTCTGGGTGGCAGTGAT-3ʹ; miR-1253: F, 5ʹ- GCTGTAACAGCGGCGGAACT-3ʹ and R, 5ʹ-ATCCGCAGGAGTGTCCGAGG-3ʹ; U6: F, 5ʹ-CTCGCTTCGGCAGCACA −3ʹ and R, 5ʹ-AACGCTTCACGAATTTGCGT-3ʹ.
Western Blot
Whole protein was isolated from tissues and cells using RIPA buffer (Beyotime, Shanghai, China). Then approximately 30 μg of extracted protein was resolved on the 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), and blotted onto the polyvinylidene difluoride membranes (Millipore, Billerica, MA, USA). Afterwards, immunoblotting was conducted with the primary antibodies against DDAH1 (1:1000, ab2231), vimentin (1:1000, ab92547,), E-cadherin (1:3000, ab15148), CD81 (1:5000, ab109201), TSG101 (1:5000, ab125011) and HRP-conjugated secondary antibody (1:300, ab9482), which were all obtained from Abcam (Cambridge, MA, USA). Protein bands were visualized with ECL chromogenic reagent (Millipore) and β-actin (1:1000, Cat # 4967, Cell Signaling Technology, Beverly, MA, USA) was used as an internal reference.
Cell Proliferation Assay
Transfected T47D and MB231 cells were seeded in 96-well plates and co-incubated with 10 μL cell counting kit-8 (CCK-8) solution (Sigma-Aldrich, St. Louis, MO, USA) in a humidified incubator for 4 h at 37°C. Then cell proliferation was analyzed via reading the optical density (OD) 450 nm value.
Cell Apoptosis Analysis
After transfection, T47D and MB231 cells resuspended in binding buffer were doubled-stained with 10 μL of Annexin V-FITC and propidium iodide (Sigma-Aldrich). The apoptotic cells were determined by a FACScan flow cytometer.
Caspase3 Activity
A colorimetric assay kit (Sigma-Aldrich) was applied to detect caspase3 activity in T47D and MB231 cells referring to the recommendations of the manufacturer. Finally, a microplate reader was used to detect the absorbance at 450 nm.
Cell Adhesion Assay
Following assigned transfection, T47D and MB231 cells (1×105) were collected and seeded into 96-well plates which were pre-coated with 1:3 diluted Matrigel (Corning Incorporated, Corning, NY, USA). 30 min later, the corresponding medium containing non-adhered cells was discarded, and the loosely adhered cells were removed through washing with PBS. Lastly, adherent cells were counted by a microscope (100×)
Transwell Assay
For the migration assay, 5×104 T47D and MB231 cells in serum-free medium were plated onto the upper chamber of Transwell insert (Corning Incorporated). For the invasion assay, the Transwell insert was pre-coated with Matrigel and 1×105 cells were seeded onto the top side of the coated insert. Then medium with FBS was used in the bottom chamber. Following incubation for 24 h, the lower surface of the filter was fixed and stained, and migratory and invasive cells were counted using a microscope in five random fields (100×).
Dual-Luciferase Reporter Assay
T47D and MB231 cells were placed on 6-well plates and then transfected with constructed luciferase reporters, pRL-TK-WT-circ_0004771/DDAH1 3ʹUTR, or pRL-TK-MUT-circ_0004771/DDAH1 3ʹUTR (Promega), and miR-1253 or miR-NC using Lipofectamine 3000 (Invitrogen). Luciferase activities were determined with the help of a dual-luciferase reporter assay kit (Promega).
Pull-Down Assay
Biotinylated-miR-1253 (Bio-miR-1253) and biotinylated-miR-NC (Bio-NC) (GenePharma) were transfected into sonicated T47D and MB231 cells. 48 h later, cells were collected and lysed, followed by mixing with streptavidin-coated magnetic beads. Finally, the biotin-coupled RNA complex was eluted, isolated and analyzed using qRT-PCR.
RNA Immunoprecipitation (RIP) Assay
T47D and MB231 cells were lysed using RIP buffer (Millipore), and then incubated with magnetic beads coated with human Anti-Ago2 (Millipore) or normal mouse Anti-IgG (Millipore). After incubation with Proteinase K, the immunoprecipitated RNA was isolated and subjected to qRT-PCR analysis as described above.
Xenograft Experiments in vivo
The lentiviral particles expressing shRNA targeting circ_0004771 (sh-circ_0004771) or sh-NC were constructed by Invitrogen. Then M231 cells infected with sh-circ_0004771 or sh-NC were subcutaneously inoculated into each female BALB/c nude mouse (5-week-old, N=12) to establish tumor xenografts. Tumor size was measured and calculated every 4 days. At day 28, mice were euthanatized and tumors were excised for weight and subsequent molecular analyses. This animal study was approved by the Animal Ethics Committee of Jingmen No.1 People’s Hospital and undertaken according to NIH Guidelines for the Care and Use of Laboratory Animals.
Isolation and Quantification of Exosomes
Exosomes were exacted from serum samples with the help of the ExoQuick serum exosome precipitation solution (SBI, Mountain View, CA, USA) referring to the instructions of the manufacture. For the morphology of exosomes, purified exosomes were deposited onto the carbon-coated copper grid, labeled with 2% phosphotungstic acid solution for 1 min, and then captured using a transmission electron microscope (TEM). For the size and quantity of exosomes, exosomes were injected into the ZetaView PMX 110 (Particle Metrix, Meerbusch, Germany), and capture and analysis settings were manually set in accordance with the protocol of the manufacturer; then, data were analyzed using nanoparticle tracking analysis (NTA) software.
Statistical Analyses
GraphPad Prism 7 was used for statistical analysis and statistical significance was calculated by Student’s t-test (two groups) or one-way ANOVA (two groups above). The data were presented as the mean ± standard deviation (SD). Pearson correlation analysis was used to evaluate the correlation between two variables. P-value <0.05 suggested statistically significant.
Results
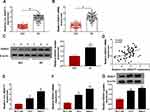
The Expression of Circ_0004771 and DDAH1 is Elevated in BC
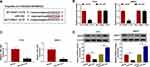
To analyze the functions of circ_0004771 and DDAH1 in BC oncogenic phenotypes, first, the expression of them in clinical samples was detected. Results showed circ_0004771 (Figure 1A) or DDAH1 (Figure 1B and C) expression was significantly elevated in BC tissues relative to the normal tissues, importantly, a positive correlation between their expression was observed (Figure 1D). Also, the level of circ_0004771 (Figure 1E) or DDAH1 (Figure 1F and G) in BC cells (T47D and MB231) was increased compared with MCF-10A nonmalignant breast epithelial cells. These results implied that abnormal circ_0004771 or DDAH1 expression might be associated with the development of BC.
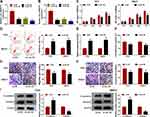
Knockdown of Circ_0004771 Suppresses Cell Oncogenic Phenotypes in BC
Next, the loss-of-function assay was performed using si-circ_0004771 plasmid in T47D and MB231 cells in vitro. As shown in Figure 2A and B, the expression of circ_0004771 was significantly reduced by the transfection of si-circ_0004771 (si-circ#1, si-circ#2, and si-circ#3) compared with si-NC in T47D and MB231 cells. si-circ#3 was selected for subsequent functional analyses due to the best interference efficiency. After that, functional experiments were performed. Results of CCK-8 assay and flow cytometry indicated that circ_0004771 knockdown suppressed cell proliferation (Figure 2C), but induced cell apoptosis in T47D and MB231 cells (Figure 2D). Moreover, the relative activity of caspase3 was enhanced in circ_0004771-down-regulated T47D and MB231 cells (Figure 2E), further suggesting circ_0004771 knockdown promoted apoptosis in BC. Besides that, cell adhesion assay showed that circ_0004771 knockdown attenuated cell adhesion ability (Figure 2F), and results of transwell assay exhibited that circ_0004771 decrease suppressed the migration and invasion abilities of T47D and MB231 cells (Figure 2G and H). Meanwhile, Western blot analysis showed circ_0004771 knockdown increased the level of E-cadherin and decreased the level of vimentin in T47D and MB231 cells (Figure 2I), suggesting down-regulation of circ_0004771 repressed cell epithelial-to-mesenchymal transition (EMT) in BC. Taken together, silencing of circ_0004771 suppressed BC progression by inhibiting cell malignant phenotypes.
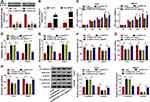
Circ_0004771 Regulates Cell Oncogenic Phenotypes in BC via DDAH1
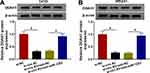
The detailed functions of DDAH1 on BC progression were then investigated. The si-DDAH1 were synthesized and transfected into T47D and MB231 cells, as expected, si-DDAH1, especially si-DDAH1#1, significantly reduced the expression of DDAH1 relative to the si-NC (Figure 3A). Thus, si-DDAH1#1 was selected for subsequent functional analyses. By contrast with si-NC group, DDAH1 down-regulation markedly suppressed cell proliferation (Figure 3C), adhesion (Figure 3F), migration (Figure 3G), and invasion (Figure 3H) abilities, but induced apoptosis (Figure 3D and E) in T47D and MB231 cells. Besides, DDAH1 knockdown also suppressed cell EMT, evidenced by the increase of E-cadherin and decrease of vimentin expression in T47D and MB231 cells (Figure 3I). Thus, DDAH1 knockdown suppressed BC progression.
Considering the positive correlation between DDAH1 and circ_0004771 expression in BC, whether circ_0004771 regulated cell oncogenic phenotypes via DDAH1 was explored. First, T47D and MB231 cells were transfected with vector or circ_0004771, and circ_0004771 expression was found to be significantly elevated after circ_0004771 transfection (Figure 3B). Next, si-DDAH1#1 was transfected into circ_0004771-increased T47D and MB231 cells to perform rescue assay. We demonstrated that the regulatory effects of si-DDAH1#1 on T47D and MB231 cell proliferation, apoptosis, adhesion, migration, invasion and EMT were partially overturned by circ_0004771 overexpression (Figure 3C–I). Collectively, circ_0004771 regulated BC progression via DDAH1.
Circ_0004771 is a Sponge of miR-1253
It has been documented that circRNAs can serve as miRNA sponges for miRNAs binding to modulate the depression of miRNA targets.21 Thus, we suspected that circ_0004771-mediated regulatory functions might operate through a sponge mechanism. Herein, through searching online databases (circinteractome), the potential target miRNA binding partners of circ_0004771 were identified, and we found that circ_0004771 harbored binding sites of miR-1253 (Figure 4A). Afterwards, results of dual-luciferase reporter assay showed that the luciferase activity was significantly declined in T47D and MB231 cells co-transfected with WT-circ_0004771 and miR-1253 (Figure 4B). Pull-down assay suggested circ_0004771 was highly enriched by Bio-miR-1253 probe compared to negative control (Figure 4C). Moreover, RIP assay displayed that circ_0004771 and miR-1253 were highly enriched in the complex precipitated by Anti-Ago2 relative to nonspecific Anti-IgG (Figure 4D). These results confirmed that circ_0004771 acted as a sponge for miR-1253. Importantly, it was also proved that miR-1253 expression in T47D and MB231 cells was elevated by circ_0004771 decrease, but was down-regulated by circ_0004771 increase (Figure 4E). Together, we verified that circ_0004771 bound to miR-1253 and negatively regulated its expression in BC.
DDAH1 is a Target of miR-1253
Through searching the online databases (Targetscan), the putative binding sites of miR-1253 on DDAH1 were also identified (Figure 5A). Then the reduction of luciferase activity in T47D and MB231 cells co-transfected with WT-DDAH1 3ʹ UTR and miR-1253 validated the direct interaction between miR-1253 and DDAH1 (Figure 5B). Subsequently, T47D and MB231 cells were transfected with anti-NC or anti-miR-1253, qRT-PCR analysis showed miR-1253 expression was significantly decreased by anti-miR-1253 transfection as expected (Figure 5C). Afterwards, Western blot analysis suggested that DDAH1 expression was reduced by miR-1253 overexpression, but was elevated by miR-1253 down-regulation (Figure 5D). Collectively, miR-1253 targetedly suppressed DDAH1 expression in BC cells.
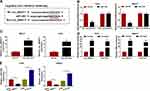
Circ_0004771 Regulates DDAH1 Expression via miR-1253
Given the association between miR-1253 and circ_0004771 or DDAH1, we then studies whether circ_0004771 could regulate DDAH1 via miR-1253. As shown in Figure 6A and B, circ_0004771 knockdown decreased the level of DDAH1 in T47D and MB231 cells, while this reduction was rescued by the inhibition of miR-1253. Thus, we concluded that circ_0004771 indirectly regulated DDAH1 expression by acting as miR-1253 sponge.
Circ_0004771 Hinders BC Tumor Growth and EMT in vivo
The role of circ_0004771 in vivo was further analyzed. As shown in Figure 7A and B, circ_0004771 knockdown impeded tumor growth in vivo, evidenced by the smaller in size and weight of xenograft tumors in sh-circ_0004771 groups. Besides, molecular analyses indicated that the levels of circ_0004771 and DDAH1 were decreased while miR-1253 level was increased in the tumors from sh-circ_0004771 groups (Figure 7C–E). Furthermore, compared with sh-NC groups, E-cadherin expression was increased and vimentin expression was decreased in sh-circ_0004771 groups (Figure 7E). Altogether, circ_0004771 knockdown hindered tumor growth and EMT in vivo via miR-1253/DDAH1 axis.
Circ_0004771 is Packaged into Exosomes Derived from the Serum of BC
Previous study has reported that circ_0004771 was up-regulated in exosomes derived from the serum of colorectal cancer (CRC) patients, and circulating exosomal circ-0004771 might act as a potential biomarker for early diagnosis of CRC.22 Thus, whether exosomal circ_0004771 was detectable in the serum of BC was investigated. Exosomes from the serum of BC patients and healthy controls were extracted and characterized. The vesicles, where the arrows pointed, showed a round shape with double-layer membrane structure under a TEM (Figure 8A). The result of NTA suggested that the size distribution of exosomes was approximately 100 nm in diameter (Figure 8B); besides, Western blot analysis displayed that the exosomal markers CD81 and TSG101 were detectable in the exosomes-enriched fractions (Figure 8C). These results demonstrated that exosomes were isolated from serum successfully. Then the relative expression of circ_0004771 in serum exosomes was analyzed. Results showed that circ_0004771 expression was higher in serum exosomes from BC patients than that from healthy controls (Figure 8D). Overall, exosomal circ_0004771 was packaged into exosomes derived from BC serum and might be closely associated with the development of BC.
Discussion
With the improvement of RNA sequencing technologies and bioinformatics, thousands of endogenous circRNAs have been identified in mammalian cells, emerging evidence reveals that circRNAs have significant roles in the development of diverse diseases, including atherosclerotic vascular disease risk,23 neurological disorders,24 and cancers.25 In BC, multiple circRNAs were discovered to show aberrant expression, and functioned as diagnostic or therapeutic biomarkers. For example, circANKS1B was significantly elevated in BC, acted as an independent risk factor for the overall survival of BC patients, and facilitated BC metastasis and invasion.26 CircTADA2A-E5/E6 was decreased in BC, whose down-regulation was related to poor prognosis, and up-regulation of them suppressed cell malignant biological behaviors in BC.27 Thus, circRNAs may be useful targets for regulating BC progression by serving as oncogenic or tumor-suppressive genes. In this study, we found circ_0004771 was up-regulated in BC, and knockdown of circ_0004771 induced cell proliferation, adhesion, invasion, migration, EMT suppression and apoptosis promotion in vitro, and blocked tumor growth and EMT in vivo. Taken together, circ_0004771 worked as an oncogene to regulate BC progression.
Recently, DDAH1 exhibits aberrant expression in multiform types of cancer, including BC and functioned as an oncogenic gene to regulate the malignancies of cancers,16–18,20 suggesting the carcinogenic role of DDAH1 in cancer progression. In the present work, an elevation of DDAH1 expression was also detected in BC, and then, functional experiments indicated that silencing of DDAH1 inhibited cell malignant phenotypes in BC, thus impeding BC progression. What’s more, we found circ_0004771 expression was positively correlated with DDAH1 in BC, and circ_0004771 overexpression mediated the anticancer effects of DDAH1-decrease in BC cells. However, the regulatory network of them remains unclear.
CircRNAs have been reported to mediate gene expression at the transcriptional or post-transcriptional level by as miRNA sponges or binding to other molecules.21,28 Therefore, whether circ_0004771-mediated regulatory functions were handled via a sponge mechanism was investigated. In this study, miR-1253 was validated to directly interact with circ_0004771 or DDAH1. Besides, circ_0004771 could indirectly regulate DDAH1 expression via serving as a miR-1253 sponge in vivo and in vitro. Thus, a circ_0004771/miR-1253/DDAH1 network in regulating BC progression was identified.
A recent study has discovered that circ_0004771 was highly expressed in exosomes derived from the serum of CRC patients, which have some diagnostic value for CRC.22 Exosomes are a type of extracellular vesicles, which can be secreted by a variety of types of cells, including tumor cells.29 Exosomes have been found to mediate tumor communication in cancer cells and extracellular microenvironment through transferring and exchanging their cargo, such as lipids, protein, noncoding or coding RNAs, thus involving in regulating cancer tumorigenesis.29,30 In this study, we also discovered that circ_0004771 was packaged into exosomes derived from BC serum, while whether exosomal circ_0004771 was implicated in the modulation of BC progression is needed further explored.
In conclusion, these findings uncovered that circ_0004771 contributed to the induction of cell malignant phenotypes via elevating DDAH1 expression through absorbing miR-1253 in BC (Figure S1), besides that, it was also found circ_0004771 was packaged into exosomes isolated from the serum of patients with BC. All these findings suggested a potential molecular target for BC therapy.
Funding
This work was supported by Jingmen guiding scientific research project (NO.2018YDKY048).
Disclosure
The authors declare that they have no conflicts of interest.
References
1. Seigel R, Naishadham D, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64(1):9–29. doi:10.3322/caac.21208
2. Anastasiadi Z, Lianos GD, Ignatiadou E, Harissis HV, Mitsis M. Breast cancer in young women: an overview. Updates Surg. 2017;69(3):313–317. doi:10.1007/s13304-017-0424-1
3. Goldvaser H, Amir E. Role of bisphosphonates in breast cancer therapy. Curr Treat Options Oncol. 2019;20(4):26. doi:10.1007/s11864-019-0623-8
4. Ebbesen KK, Kjems J, Hansen TB. Circular RNAs: identification, biogenesis and function. Biochim Biophys Acta. 2016;1859(1):163–168. doi:10.1016/j.bbagrm.2015.07.007
5. Qu S, Yang X, Li X, et al. Circular RNA: a new star of noncoding RNAs. Cancer Lett. 2015;365(2):141–148. doi:10.1016/j.canlet.2015.06.003
6. Kristensen LS, Andersen MS, Stagsted LVW, Ebbesen KK, Hansen TB, Kjems J. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20(11):675–691. doi:10.1038/s41576-019-0158-7
7. Yu T, Wang Y, Fan Y, et al. CircRNAs in cancer metabolism: a review. J Hematol Oncol. 2019;12(1):90. doi:10.1186/s13045-019-0776-8
8. Hua X, Sun Y, Chen J, et al. Circular RNAs in drug resistant tumors. Biomed Pharmacother. 2019;118:109233. doi:10.1016/j.biopha.2019.109233
9. Li Z, Chen Z, Hu G, Jiang Y. Roles of circular RNA in breast cancer: present and future. Am J Transl Res. 2019;11(7):3945–3954.
10. Huang E, Fu J, Yu Q, et al. CircRNA hsa_circ_0004771 promotes esophageal squamous cell cancer progression via miR-339-5p/CDC25A axis. Epigenomics. 2020;12(7):587–603. doi:10.2217/epi-2019-0404
11. Xie R, Tang J, Zhu X, Jiang H. Silencing of hsa_circ_0004771 inhibits proliferation and induces apoptosis in breast cancer through activation of miR-653 by targeting ZEB2 signaling pathway. Biosci Rep. 2019;39(5):BSR20181919. doi:10.1042/BSR20181919
12. Leiper J, Nandi M, Torondel B, et al. Disruption of methylarginine metabolism impairs vascular homeostasis. Nat Med. 2007;13(2):198–203. doi:10.1038/nm1543
13. Xu X, Zhang P, Kwak D, et al. Cardiomyocyte dimethylarginine dimethylaminohydrolase-1 (DDAH1) plays an important role in attenuating ventricular hypertrophy and dysfunction. Basic Res Cardiol. 2017;112(5):55. doi:10.1007/s00395-017-0644-z
14. Jacobi J, Sydow K, von Degenfeld G, et al. Overexpression of dimethylarginine dimethylaminohydrolase reduces tissue asymmetric dimethylarginine levels and enhances angiogenesis. Circulation. 2005;111(11):1431–1438. doi:10.1161/01.CIR.0000158487.80483.09
15. Yung BC, Li J, Zhang M, et al. Lipid nanoparticles composed of quaternary amine-tertiary amine cationic lipid combination (QTsome) for therapeutic delivery of AntimiR-21 for lung cancer. Mol Pharm. 2016;13(2):653–662. doi:10.1021/acs.molpharmaceut.5b00878
16. Ye J, Xu J, Li Y, et al. DDAH1 mediates gastric cancer cell invasion and metastasis via Wnt/β-catenin signaling pathway. Mol Oncol. 2017;11(9):1208–1224. doi:10.1002/1878-0261.12089
17. Gao F, Du Y, Zhang Y, Ren D, Xu J, Chen D. Circ-EZH2 knockdown reverses DDAH1 and CBX3-mediated cell growth and invasion in glioma through miR-1265 sponge activity. Gene. 2020;726:144196. doi:10.1016/j.gene.2019.144196
18. Reddy KRK, Dasari C, Duscharla D, et al. Dimethylarginine dimethylaminohydrolase-1 (DDAH1) is frequently upregulated in prostate cancer, and its overexpression conveys tumor growth and angiogenesis by metabolizing asymmetric dimethylarginine (ADMA). Angiogenesis. 2018;21(1):79–94. doi:10.1007/s10456-017-9587-0
19. Hulin JA, Tommasi S, Elliot D, Hu DG, Lewis BC, Mangoni AA. MiR-193b regulates breast cancer cell migration and vasculogenic mimicry by targeting dimethylarginine dimethylaminohydrolase 1. Sci Rep. 2017;7(1):13996. doi:10.1038/s41598-017-14454-1
20. Hulin JA, Tommasi S, Elliot D, Mangoni AA. Small molecule inhibition of DDAH1 significantly attenuates triple negative breast cancer cell vasculogenic mimicry in vitro. Biomed Pharmacother. 2019;111:602–612. doi:10.1016/j.biopha.2018.12.117
21. John-Aryankalayil M, Palayoor ST, Makinde AY, et al. Fractionated radiation alters oncomir and tumor suppressor miRNAs in human prostate cancer cells. Radiat Res. 2012;178(3):105–117. doi:10.1667/RR2703.1
22. Pan B, Qin J, Liu X, et al. Identification of serum exosomal hsa-circ-0004771 as a novel diagnostic biomarker of colorectal cancer. Front Genet. 2019;10:1096. doi:10.3389/fgene.2019.01096
23. Altesha MA, Ni T, Khan A, Liu K, Zheng X. Circular RNA in cardiovascular disease. J Cell Physiol. 2019;234(5):5588–5600. doi:10.1002/jcp.27384
24. Kumar L, Shamsuzzama HR, Baghel T, Nazir A. Circular RNAs: the emerging class of non-coding RNAs and their potential role in human neurodegenerative diseases. Mol Neurobiol. 2017;54(9):7224–7234. doi:10.1007/s12035-016-0213-8
25. Kristensen LS, Hansen TB, Venø MT, Kjems J. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37(5):555–565. doi:10.1038/onc.2017.361
26. Zeng K, He B, Yang BB, et al. The pro-metastasis effect of circANKS1B in breast cancer. Mol Cancer. 2018;17(1):160. doi:10.1186/s12943-018-0914-x
27. Xu JZ, Shao CC, Wang XJ, et al. circTADA2As suppress breast cancer progression and metastasis via targeting miR-203a-3p/SOCS3 axis. Cell Death Dis. 2019;10(3):175. doi:10.1038/s41419-019-1382-y
28. Panda AC. Circular RNAs Act as miRNA sponges. Adv Exp Med Biol. 2018;1087:67–79.
29. Cocucci E, Meldolesi J. Ectosomes and exosomes: shedding the confusion between extracellular vesicles. Trends Cell Biol. 2015;25(6):364–372. doi:10.1016/j.tcb.2015.01.004
30. Valencia K, Luis-Ravelo D, Bovy N, et al. miRNA cargo within exosome-like vesicle transfer influences metastatic bone colonization. Mol Oncol. 2014;8(3):689–703. doi:10.1016/j.molonc.2014.01.012
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.