Back to Journals » Cancer Management and Research » Volume 13
Circ_0001806 Promotes the Proliferation, Migration and Invasion of NSCLC Cells Through miR-1182/NOVA2 Axis
Authors Yi S, Li Z, Wang X, Du T , Chu X
Received 2 November 2020
Accepted for publication 19 March 2021
Published 8 April 2021 Volume 2021:13 Pages 3067—3077
DOI https://doi.org/10.2147/CMAR.S290059
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Eileen O'Reilly
Shenni Yi,1,* Zhan Li,1,* Xiuqin Wang,1,* Tiantian Du,1 Xiuhong Chu2
1Department of Respiratory and Critical Medicine, Yantai Yuhuangding Hospital, Yantai, Shandong Province, 264000, People’s Republic of China; 2Department of Nursing, Yantai YEDA Hospital, Yantai, Shandong Province, 264006, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Tiantian Du
Department of Respiratory and Critical Medicine, Yantai Yuhuangding Hospital, East Yuhuangding Road NO. 20, Yantai, Shandong Province, 264000, People’s Republic of China
Email [email protected]
Xiuhong Chu
Department of Nursing, Yantai YEDA Hospital, Taishan Road No. 11, Yantai Development Area, Yantai, Shandong Province, 264006, People’s Republic of China
Email [email protected]
Background: Circular RNAs (circRNAs) are non-coding RNAs with a covalently closed loop. circRNAs affect the progression of diverse cancers. Nonetheless, circ_0001806 expression and function in non-small cell lung cancer (NSCLC) are undefined.
Methods: qRT-PCR was executed to examine circ_0001806, miR-1182 and NOVA2 mRNA expression levels in NSCLC tissues and cells. CCK-8, EdU, cell scratch test and Transwell assay were conducted to examine the viability, multiplication, migration and invasion of NSCLC cell lines H1650 and HCC827. The binding sites between circ_0001806 and miR-1182, miR-1182 and NOVA2 mRNA were predicted by the circular RNA Interactome and TargetScan databases, and the dual-luciferase reporter gene experiment was employed for verification. Western blot was implemented to examine NOVA2 expression.
Results: Circ_0001806 expression in NSCLC tissues and cell lines was substantially augmented, while miR-1182 expression was markedly decreased. Circ_0001806 facilitated the multiplication, migration and invasion of H1650 and HCC827 cells, while miR-1182 exerted the opposite effect. Circ_0001806 indirectly enhanced NOVA2 expression by specifically down-modulating miR-1182.
Conclusion: Circ_0001806 augments NOVA2 expression by targeting miR-1182 to enhance the multiplication, migration and invasion of NSCLC cells.
Keywords: circ_0001806, miR-1182, NOVA2, NSCLC
Introduction
Non-small cell lung cancer (NSCLC) takes up 80% of all lung cancer cases and is the most common type of human lung cancer.1 Although surgery, radiotherapy, and chemotherapy have great progress, the 5-year survival rate of NSCLC patients is still less than 19% due to relapse and drug resistance, and the lack of sensitive and efficient early diagnostic markers.2–5 Therefore, it is imperative to conduct an in-depth research of the molecular mechanism of the occurrence and development of NSCLC, so as to provide a theoretical basis for diagnosis and therapy of NSCLC.
Circular RNAs (circRNAs) are circular non-coding RNAs formed by reverse splicing at the 3ʹ and 5ʹ ends of covalent junctions that modulate eukaryotic gene expression.6 Accumulating research has revealed that circRNA is closely related to the occurrence and development of NSCLC.7 For instance, circFGFR1 up-modulates CXCR4 expression by working as a sponge of miR-381-3p, thereby enhancing the multiplication, migration and invasion of NSCLC cells;8 circPTPRA specifically down-modulates miR-96-5p to impede the epithelial-mesenchymal transition (EMT), migration and invasion of NSCLC cells.9 Nevertheless, the role and mechanism of circ_0001806 in NSCLC are blurred.
MicroRNA (miRNA) is short-stranded RNA with a length of 21–25 nts. Many miRNAs are reported to participate in the occurrence and development of NSCLC. For instance, miR-140 is uncovered to be markedly reduced in NSCLC tissues. By targeting PD-L1, miR-140 restrains the multiplication of A549 and NCI-H1650 cells.10 miR-1182 is revealed to be remarkably under-expressed in NSCLC tissues relative to normal lung tissues. By targeting and modulating KLF8, miR-1182 markedly suppresses the multiplication, migration and invasion of H358 and H23 cells.11 Similarly, another research validates the inhibitory effect of miR-1182 on the multiplication and invasion of NSCLC cells through A549 and H1299 cell lines.12 Nevertheless, the upstream and downstream regulatory mechanism of miR-1182 in NSCLC is vague.
This work is aimed to probe the role of circ_0001806 and miR-1182 in NSCLC and its regulatory mechanism to clarify the molecular basis that affects NSCLC progression.
Materials and Methods
Tissue Specimens
This work analyzed specimens of cancerous tissues and adjacent tissues (at least 3 cm from the surgical margin) of 45 NSCLC patients recruited from Yantai Yuhuangding Hospital. All subjects had not undergone radiotherapy, chemotherapy or targeted therapy before surgery. This work was endorsed by the Ethics Committee of Yantai Yuhuangding Hospital, and all subjects signed an informed consent form before surgery. All specimens were immediately preserved in liquid nitrogen at −196°C for subsequent experiments.
Cell Culture
NSCLC cell lines (A549, H1975, H1650 and HCC827) and non-cancerous bronchial epithelial cell line (BEAS-2B) were procured from American Type Culture Collection (ATCC, Manassas, VA, USA). Cells were cultured in RPMI1640 (Thermo Fisher Scientific, Waltham, MA, USA) containing 100 U/mL penicillin, 100 U/mL streptomycin (Invitrogen, Carlsbad, CA, USA) and 10% fetal bovine serum (FBS) (Thermo Fisher Scientific, Waltham, MA, USA). The medium was maintained at 37°C with 5% CO2. 0.25% trypsin (Thermo Fisher Scientific, Waltham, MA, USA) was applied for trypsinization and subculture during logarithmic cell growth.
Cell Transfection
The H1650 and HCC827 cells were prepared into the cell suspension, the cell density was modulated to 1×106 cells/mL, and the cells were seeded in a 12-well plate. The cells were then cultured at 37°C with 5% CO2 until the confluence reached about 70%. Cell transfection was executed using lipofectamine 2000 reagent (Thermo Fisher Science, Waltham, MA, USA). Circ_0001806 overexpression plasmid (circ_0001806), empty plasmid (NC), shRNAs targeting circ_0001806 (sh-circ_0001806#1, #2 and #2), negative control for shRNA (sh-NC), miR-1182 mimic (miR-1182 mim), negative control for miR-1182 mimic (NC-mim), miR-1182 inhibitor (miR-1182 inh) or negative control for miR-1182 inhibitor (NC-inh) were mixed with the same volume of transfection reagent, and the cells were transfected at 37°C under the guidance of the protocols.
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
TRIzol reagent (Invitrogen, Carlsbad, CA, USA) was adopted to extract total RNA from NSCLC cells, BEAS-2B cells or NSCLC tissues, and the primers were designed by Primer6.0, and synthesized by TaKaRa (Dalian, China). The target and internal reference genes of each sample were amplified simultaneously. The 2−ΔΔCt method was utilized to calculate circ_0001806, miR-1182 and NOVA2 mRNA expression levels. U6 was the internal control of miR-1182, and GAPDH was the internal control of circ_0001806 and NOVA2 mRNA.
CCK-8 Assay
The cell density of H1650 and HCC827 was modulated to 2×103 cells/well, planted in a 96-well plate, and cultured under the conditions of 100% humidity, 37°C, and 5% CO2. Afterward, cells were cultured for 1, 2 or 3 d, respectively. On each day, 10μL of CCK-8 reagent (Beyotime Biotechnology, Shanghai, China) was supplemented to each well, and the cells were then cultured for another 1 h. The microplate reader was employed to measure the absorbance (450nm) of H1650 and HCC827 cells.
EdU Assay
H1650 and HCC827 cells were seeded in 24-well plates and cultured for 12 h. Subsequently, 200 μL of 50 μmol/L EdU medium (Beyotime Biotechnology, Shanghai, China) was added to each well, incubated for 2 hours, and rinsed with PBS. Thereafter, the cells were fixed with paraformaldehyde and incubated for 10 min. After supplementing 200 μL of glycine at a concentration of 2 mg/mL and incubating for 5 min, the cells were rinsed with PBS on a shaker for 5 min. 100 μL of PBS containing 0.5% TritonX-100 was supplemented to each well, and the cells were decolorized and incubated on a shaker for 10 min, and PBS was used to wash the cells twice for 5 min each time. The cells were stained using Apollo method in the dark for 30 min, and then incubated with DAPI staining solution in the dark for 20 min. Ultimately, the cells were rinsed with PBS, photographed and counted under a fluorescence microscope.
Wound Healing Assay
H1650 and HCC827 cells were planted in a 6-well plate. When the cells were cultured to 80–90% confluency, the center of the plate was scratched with a pipette to ensure that there were no cells in the scratch. Then, RPMI1640 containing 2.5% FBS (Thermo Fisher Scientific, Waltham, MA, USA) was supplemented. Then the cells were cultured at 37°C and 5% CO2 for 24h. The scratches at 0h and 24h were photographed and recorded under the microscope.
Transwell Invasion Assay
Matrigel (BD Biosciences, San Jose, CA, USA) was pre-laid in the Tranwell chamber (BD Biosciences, San Jose, CA, USA). 600 μL of RPMI1640 medium containing 10% FBS (Thermo Fisher Scientific, Waltham, MA, USA) was replenished to the lower chamber, and 200 μL of serum-free medium containing 1×105 cells was replenished to the upper chamber, and the cells were cultured for 24 h. After that, the fluid in the upper chamber was discarded, and cells that had not passed through the membrane was removed with a swab. Cells were fixed with methanol, stained with crystal violet, rinsed with PBS, and the membrane of the chamber was cut off and the cells were observed under a microscope. Five randomly selected fields of view under the microscope were applied to calculate the mean number of the cells.
Subcellular Fractionation
1×107 H1650 or HCC827 cells were resuspended, incubated on ice for 10 min. Then, RNA was extracted and isolated from cell nuclei and cytoplasm, respectively, using the PARIS™ Kit (Ambion, Austin, TX, USA) under the guidance of the protocols. The isolated RNA was subjected to qRT-PCR analysis, normalizing to U6 (nucleus control) and GAPDH (cytoplasm control).
Dual-Luciferase Reporter Gene Assay
Circ_0001806 and NOVA2 wild-type luciferase reporter vector (circ_0001806 wt and NOVA2 wt), circ_0001806 and NOVA2 mutant luciferase reporter vector (circ_0001806 mut and NOVA2 mut) were established by Promega (Madison, WI, USA). H1650 or HCC827 cells were planted in 48-well plates (4.5×104/well) and cultured to 70% confluence. Then, the above vectors were co-transfected with miR-1182 mim, miR-1182 inh, NC-mim or NC-inh into H1650 or HCC827 cells using lipofectamine 2000. After 48 h of transfection, the luciferase activity was measured.
Western Blot
H1650 and HCC827 cells were collected, rinsed with pre-cooled PBS, and the total protein was extracted with RIPA lysis buffer (Beyotime Biotechnology, Shanghai, China), and the protein concentration was determined by the Bradford method. The same amount of protein samples were separated by 12% SDS-PAGE, transferred to PVDF membrane (Millipore, Bedford, MA, USA), blocked with 5% skimmed milk for 2 h, and incubated with the primary antibody anti-NOVA2 (ab51004, 1:2000, Abcam, Shanghai, China) overnight at 4°C. After washing with PBST, the protein was then incubated with HRP-labeled specific secondary antibody (goat anti-mouse IgG, ab205719, 1:2500, Abcam, Shanghai, China) for 2 h at temperature, the membrane was rinsed with PBST 3 times. Finally, ECL reagent (Amersham Pharmacia Biotech, Little Chalfont, UK) was applied to show the protein bands on X-ray film. GAPDH was regarded as the endogenous control.
Statistical Analysis
SPSS 20.0 statistical software (SPSS Inc., Chicago, IL, USA) was applied for statistical analysis. All experiments were executed in triplicate. Measurement data were represented by means±standard deviation (x±s). The comparison among multiple groups was conducted by one-way ANOVA, and the comparison between the two groups was executed by Students’ t-test. P<0.05 signified statistical analysis.
Results
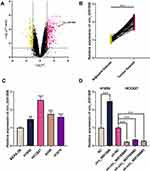
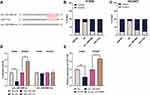
circ_0001806 Was Overexpressed in NSCLC Tissues and Cell Lines
To probe the circRNAs that were involved in NSCLC progression, GSE101684 was employed to analyze the abnormally expressed circRNAs in NSCLC tissues, and it was revealed that circ_0001806 expression was remarkably up-modulated in NSCLC tissues relative to adjacent tissues (Figure 1A). Consistently, through qRT-PCR analysis, a remarkable up-modulation of circ_0001806 expression was observed in NSCLC tissues and cell lines (Figure 1B and C). To explore the function of circ_0001806, we transfected circ_0001806 overexpression plasmid into H1650 cells with the lowest expression of circ_0001806, and circ_0001806 shRNAs into HCC827 cells with the highest expression of circ_0001806 (Figure 1D).
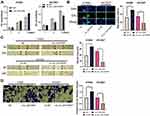
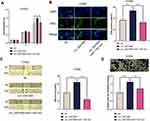
circ_0001806 Enhanced the Multiplication, Migration and Invasion of NSCLC Cells
Next, the biological role of circ_0001806 in NSCLC cells was investigated. Because sh-circ_0001806#1 had the highest knockdown efficiency, this shRNA was chosen for subsequent experiments. The data of CCK-8 and EdU experiments implied that circ_0001806 overexpression substantially enhanced the multiplication of H1650 cells, while knocking down circ_0001806 displayed the opposite phenomenon (Figure 2A and B). Additionally, through the cell scratch experiment and Transwell experiment, we observed that relative to the NC group, the cell migration and invasion of the circ_0001806 group was markedly improved, while relative to the sh-NC group, the cell migration and invasion of the sh-circ_0001806 group was markedly weakened (Figure 2C and D).
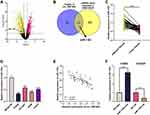
miR-1182 Was Under-Expressed in NSCLC Tissues and Cell Lines
To elaborate on the mechanism of circ_0001806, GSE135918 was employed to analyze the abnormally expressed miRNAs in NSCLC. MiR-1182 was unveiled to be remarkably down-modulated in NSCLC tissues relative to adjacent tissues, and it was also predicted by the Circular RNA Interactome database as one of the downstream targets of circ_0001806 (Figure 3A and B). We confirmed its expression by qRT-PCR analysis and a remarkable down-modulation of miR-1182 expression was observed in both NSCLC tissues and cell lines, and miR-1182 and circ_0001806 expression was negatively correlated in NSCLC tissues (Figure 3C–E). Therefore, we hypothesized that circ_0001806 may work through miR-1182. To explore the mechanism of miR-1182, we transfected miR-1182 mim into H1650 cells with the lowest miR-1182 expression and miR-1182 inh into HCC827 cells with the highest miR-1182 expression (Figure 3F).
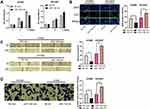
miR-1182 Impeded the Multiplication, Migration and Invasion of NSCLC Cells
The biological role of miR-1182 in NSCLC cells was then detected. In CCK-8 and EdU experiments, the transfection of miR-1182 mimics was observed to remarkably inhibit the multiplication of H1650 cells, but the multiplication of HCC827 was markedly enhanced after transfection with miR-1182 inhibitors (Figure 4A and B). Through cell scratch experiments and Transwell experiments, the migration and invasion of H1650 cells overexpressing miR-1182 was remarkably reduced, while the opposite effects were observed in HCC827 cells with low expression of miR-1182 (Figure 4C and D). The above data implied that miR-1182 was a tumor suppressor in NSCLC progression.
circ_0001806 Specifically Bound to miR-1182
To probe the potential downstream mechanism of circ_0001806, Circular RNA Interactome database was utilized to predict the targets of circ_0001806 and miR-1182 was found (Figure 5A). The subcellular distribution of circ_0001806 and miR-1182 was examined and unearthed that in H1650 and HCC827 cells, circ_0001806 and miR-1182 were mainly distributed in the cytoplasm (Figure 5B and C). The results of the dual-luciferase reporter gene experiment indicated that miR-1182 mim remarkably reduced the luciferase activity of circ_0001806 wt, while miR-1182 inh had an enhanced effect on it. No significant changes of luciferase activity were observed in circ_0001806 mut group (Figure 5D). Moreover, circ_0001806 overexpression remarkably suppressed miR-1182 expression in NSCLC cells, and miR-1182 expression was markedly augmented after repressing circ_0001806 (Figure 5E). The data revealed that miR-1182 was targetedly inhibited by circ_0001806.
miR-1182 Mim Counteracted the Effects of circ_0001806 on HCC827 Cells
Next, we co-transfected H1650 cells with circ_0001806 overexpression plasmid and miR-1182 mim to validate whether circ_0001806 worked through miR-1182. Through CCK-8, EdU, cell scratch and Transwell experiments, we observed that the multiplication, migration and invasion of H1650 cells were promoted after circ_0001806 overexpression but the phenomenon was reversed by co-transfected miR-1182 mim (Figure 6A–D).
circ_0001806 Indirectly Up-Modulated NOVA2 Expression by Down-Modulating miR-1182
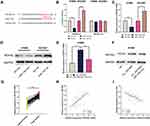
To probe the downstream mechanism of miR-1182, the TargetScan database was utilized to predict its downstream target genes and we observed that neuro-oncological ventral antigen 2 (NOVA2), which affected the NSCLC progression,13 was one of its downstream targets (Figure 7A). The dual-luciferase reporter gene experiment manifested that miR-1182 mim remarkably suppressed the luciferase activity of NOVA2 wt, while miR-1182 inh markedly enhanced the luciferase activity of NOVA2 wt; but no significant changes of luciferase activity in NOVA2 mut group was found (Figure 7B). The data of qRT-PCR and Western blot indicated that miR-1182 restrained NOVA2 expression at the mRNA and protein levels; NOVA2 expression was augmented after circ_0001806 overexpression, and this effect could be counteracted by co-transfected miR-1182 mim (Figure 7C–F). Therefore, we concluded that circ_0001806 indirectly up-modulated NOVA2 expression by targeting miR-1182. Furthermore, relative to adjacent tissues, NOVA2 mRNA expression was observed to be remarkably up-modulated in NSCLC tissues (Figure 7G), and its expression was positively correlated with circ_0001806 expression and negatively correlated with miR-1182 expression (Figure 7H and I).
Discussion
CircRNAs are vital regulators in cancer progression. The highly conserved and stable properties of circRNAs make them suitable as biomarkers for diverse diseases including cancer,14 and are therefore popular in the field of cancer research. For instance, circ_0005576 is validated to enhance the multiplication, migration and invasion of cervical cancer cells through the miR-153-3p/KIF20A axis.15 In NSCLC, circRNAs are also crucial. For instance, circPIP5K1A works as miR-600 sponge and facilitates the multiplication and metastasis of NSCLC cells by up-modulating HIF-1α expression.16 Circ_0102533 is reported to be remarkably increased in NSCLC tissues and patients’ serum, and it has the potential to diagnose NSCLC with relative high sensitivity and specificity, and knocking down circ_0102533 markedly impedes the multiplication of NSCLC cells and induces apoptosis.17 Reportedly, circMTDH.4 up-modulates AEG-1 expression through sponging miR-630, thereby promoting multiplication, migration and invasion of NCI-H1650 and A549 cells, and enhancing their chemoresistance and radioresistance.18 These studies indicate that circRNAs have great potential in the diagnosis and therapy of NSCLC. This work probed the expression characteristics and biological effects of circ_0001806 in NSCLC cells, and revealed that circ_0001806 was remarkably up-modulated in NSCLC tissues and cell lines. circ_0001806 markedly enhanced the multiplication, migration and invasion of NSCLC cells H1650 and HCC827. Our results suggested that circ_0001806 was an oncogenic circRNA in NSCLC.
The role of miR-1182 in cancer has been explored in the previous studies. MiR-1182 is reported to repress the viability of colorectal cancer cells HCT116 and restrains the multiplication, migration and invasion of prostate cancer cells.19,20 In NSCLC, miR-1182 exerts a tumor-suppressive effect, suppressing the multiplication, migration and invasion of NSCLC cells H358 and H23, and promoting their apoptosis.11 Similarly, miR-1182 is uncovered to suppress the multiplication, migration and invasion of NSCLC cells H522 and H1975.21 Accumulating evidence demonstrates that circRNA can act as ceRNA by targeting miRNA, thereby reducing the inhibitory effect of miRNA on its target.22 For instance, circWHSC1 modulates MUC1 and hTERT expression through sponging miR-1182, enhancing the multiplication, migration and invasion of ovarian cancer cells, and repressing the apoptosis of ovarian cancer cells.23 In this work, the tumor-suppressive effect of miR-1182 in NSCLC was validated through H1650 and HCC827 cells, and it was proved that miR-1182 was targeted by circ_0001806.
NOVA2 belongs to the NOVA protein family and participates in alternative splicing and transport of mRNA.24 Reportedly, NOVA2 is remarkably up-modulated in NSCLC tissues, and NOVA2 overexpression markedly facilitates the multiplication, migration and invasion of A549 and SPCA-1 cells.13,21 Mechanistically, the NOVA1/NOVA2 heterodimer enhances the stability of β-catenin mRNA, and therefore augments β-catenin accumulating, which activates the WNT/β-catenin pathway and subsequently enhances the growth, metastasis and EMT of cancer cells,24,25 which suggests that targeting NOVA2 may be one of the potential means to suppress NSCLC progression. In this work, miR-1182 was proved to target NOVA2 mRNA and down-modulate NOVA2 expression, while circ_0001806 indirectly up-modulated NOVA2 by down-modulating miR-1182. We clarified the cause of NOVA2 abnormal expression and its upstream regulatory mechanism.
In summary, this work probes the biological role of circ_0001806 and miR-1182 in NSCLC, and demonstrates that circ_0001806 in NSCLC cells indirectly up-modulates NOVA2 expression by down-modulating miR-1182 to enhance NSCLC cell multiplication, migration and invasion, which clarifies the molecular mechanism of NSCLC progression.
Data Sharing Statement
The data used to support the findings of this study are available from the corresponding author upon request.
Ethics Statement
This study was approved by the Ethics Committees of the Yantai Yuhuangding Hospital. All the participants provided written informed consents and all protocols were conducted in accordance with the principles of the Declaration of Helsinki.
Disclosure
The authors declare that they have no competing interests.
References
1. Shangguan W-J, Liu H-T, Que Z-J, Qian -F-F, Liu L-S, Tian J-H. TOB1-AS1 suppresses non-small cell lung cancer cell migration and invasion through a ceRNA network. Exp Ther Med. 2019;18(6):4249–4258. doi:10.3892/etm.2019.8103
2. Yu W, Sun Z, Yang L, et al. lncRNA PTAR promotes NSCLC cell proliferation, migration and invasion by sponging microRNA‑101. Mol Med Rep. 2019;20(5):4168–4174.
3. Wang G, Liu L, Zhang J, et al. LncRNA HCG11 suppresses cell proliferation and promotes apoptosis via sponging miR-224-3p in non-small- cell lung cancer cells. Onco Targets Ther. 2020;13:6553–6563. doi:10.2147/OTT.S244181
4. Tu Z, He D, Deng X, et al. An eight-long non-coding RNA signature as a candidate prognostic biomarker for lung cancer. Oncol Rep. 2016;36(1):215–222. doi:10.3892/or.2016.4817
5. Nagasaka M, Gadgeel SM. Role of chemotherapy and targeted therapy in early-stage non-small cell lung cancer. Expert Rev Anticancer Ther. 2018;18(1):63–70. doi:10.1080/14737140.2018.1409624
6. Zhao CH, Qu L, Zhang H, Qu R. Identification of breast cancer-related circRNAs by analysis of microarray and RNA-sequencing data: an observational study. Medicine (Baltimore). 2019;98(46):e18042. doi:10.1097/MD.0000000000018042
7. Zhang Q, Zhang C, Ma JX, Ren H, Sun Y, Xu JZ. Circular RNA PIP5K1A promotes colon cancer development through inhibiting miR-1273a. World J Gastroenterol. 2019;25(35):5300–5309. doi:10.3748/wjg.v25.i35.5300
8. Zhang PF, Pei X, Li KS, et al. Circular RNA circFGFR1 promotes progression and anti-PD-1 resistance by sponging miR-381-3p in non-small cell lung cancer cells. Mol Cancer. 2019;18(1):179. doi:10.1186/s12943-019-1111-2
9. Wei S, Zheng Y, Jiang Y, et al. The circRNA circPTPRA suppresses epithelial-mesenchymal transitioning and metastasis of NSCLC cells by sponging miR-96-5p. EBioMedicine. 2019;44:182–193. doi:10.1016/j.ebiom.2019.05.032
10. Xie WB, Liang LH, Wu KG, et al. MiR-140 expression regulates cell proliferation and targets PD-L1 in NSCLC. Cell Physiol Biochem. 2018;46(2):654–663. doi:10.1159/000488634
11. Zhao M, Ma W, Ma C. Circ_0067934 promotes non-small cell lung cancer development by regulating miR-1182/KLF8 axis and activating Wnt/β-catenin pathway. Biomed Pharmacother. 2020;129:110461. doi:10.1016/j.biopha.2020.110461
12. Li W, Jiang W, Liu T, Lv J, Guan J. Enhanced expression of circ_0000735 forecasts clinical severity in NSCLC and promotes cell progression via sponging miR-1179 and miR-1182. Biochem Biophys Res Commun. 2019;510(3):467–471. doi:10.1016/j.bbrc.2019.01.134
13. Xiao H. MiR-7-5p suppresses tumor metastasis of non-small cell lung cancer by targeting NOVA2. Cell Mol Biol Lett. 2019;24:60. doi:10.1186/s11658-019-0188-3
14. Chen Y, Li C, Tan C, Liu X. Circular RNAs: a new frontier in the study of human diseases. J. Med Genet. 2016;53(6):359–365. doi:10.1136/jmedgenet-2016-103758
15. Ma H, Tian T, Liu X, et al. Upregulated circ_0005576 facilitates cervical cancer progression via the miR-153/KIF20A axis. Biomed Pharmacother. 2019;118:109311. doi:10.1016/j.biopha.2019.109311
16. Chi Y, Luo Q, Song Y, et al. Circular RNA circPIP5K1A promotes non-small cell lung cancer proliferation and metastasis through miR-600/HIF-1α regulation. J Cell Biochem. 2019;120(11):19019–19030. doi:10.1002/jcb.29225
17. Zhou X, Liu HY, Wang WY, Zhao H, Wang T. Hsa_circ_0102533 serves as a blood-based biomarker for non-small-cell lung cancer diagnosis and regulates apoptosis in vitro. Int J Clin Exp Pathol. 2018;11(9):4395–4404.
18. Li YH, Xu CL, He CJ, Pu HH, Liu JL, Wang Y. circMTDH.4/miR-630/AEG-1 axis participates in the regulation of proliferation, migration, invasion, chemoresistance, and radioresistance of NSCLC. Mol Carcinog. 2020;59(2):141–153. doi:10.1002/mc.23135
19. Li Y, Li C, Xu R, Wang Y, Li D, Zhang B. A novel circFMN2 promotes tumor proliferation in CRC by regulating the miR-1182/hTERT signaling pathways. Clin Sci (Lond). 2019;133(24):2463–2479. doi:10.1042/CS20190715
20. Huang C, Deng H, Wang Y, et al. Circular RNA circABCC4 as the ceRNA of miR-1182 facilitates prostate cancer progression by promoting FOXP4 expression. J Cell Mol Med. 2019;23(9):6112–6119. doi:10.1111/jcmm.14477
21. Li C, Liu H, Niu Q, Gao J. Circ_0000376, a novel circRNA, promotes the progression of non-small cell lung cancer through regulating the miR-1182/NOVA2 network. Cancer Manag Res. 2020;12:7635–7647. doi:10.2147/CMAR.S258340
22. Wang J, Zhao X, Wang Y, et al. circRNA-002178 act as a ceRNA to promote PDL1/PD1 expression in lung adenocarcinoma. Cell Death Dis. 2020;11(1):32. doi:10.1038/s41419-020-2230-9
23. Zong ZH, Du YP, Guan X, Chen S, Zhao Y. CircWHSC1 promotes ovarian cancer progression by regulating MUC1 and hTERT through sponging miR-145 and miR-1182. J Exp Clin Cancer Res. 2019;38(1):437. doi:10.1186/s13046-019-1437-z
24. Tang S, Zhao Y, He X, et al. Identification of NOVA family proteins as novel β-catenin RNA-binding proteins that promote epithelial-mesenchymal transition. RNA Biol. 2020;17(6):881–891. doi:10.1080/15476286.2020.1734372
25. Yang S, Liu Y, Li MY, et al. FOXP3 promotes tumor growth and metastasis by activating Wnt/β-catenin signaling pathway and EMT in non-small cell lung cancer. Mol Cancer. 2017;16(1):124. doi:10.1186/s12943-017-0700-1
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.