Back to Journals » OncoTargets and Therapy » Volume 13
Circ_0001175 Promotes Hepatocellular Carcinoma Cell Proliferation and Metastasis by Regulating miR-130a-5p
Authors Li L, He K, Chen S, Wei W, Tian Z, Tang Y, Xiao C, Xiang G
Received 15 May 2020
Accepted for publication 12 November 2020
Published 31 December 2020 Volume 2020:13 Pages 13315—13327
DOI https://doi.org/10.2147/OTT.S262408
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Arseniy Yuzhalin
Liheng Li,1,* Ke He,2,* Siliang Chen,1 Wenjiang Wei,1 Zuofu Tian,1 Yinghong Tang,1 Chengjiang Xiao,1 Guoan Xiang2
1Department of Interventional Radiology, Guangdong Second Provincial General Hospital, Guangzhou, People’s Republic of China; 2Department of General Surgery, Guangdong Second Provincial General Hospital, Guangzhou, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Chengjiang Xiao; Guoan Xiang Email [email protected]; [email protected]
Objective: Many aberrantly expressed circular RNAs (circRNAs) play important roles in the development and progression of hepatocellular carcinoma (HCC). However, the exact function of circ_0001175 in HCC cells is unknown. Our study aimed to investigate the expression characteristics of circ_0001175 in HCC and its effects on the proliferation, migration and invasion of HCC cells, and to explore the potential mechanism.
Materials and Methods: Quantitative real-time polymerase chain reaction (qRT-PCR) and Western blot were carried out to detect circ_0001175, microRNA-130a-5p (miR-130a-5p) and sorting nexin 5 (SNX5) expressions in HCC tissues and cells; cell counting kit-8 (CCK-8), BrdU and Transwell assays were conducted to detect the proliferation, migration and invasion of HCC cells. A lung metastasis model in nude mice was used to examine the effect of circ_0001175 on the metastasis of HCC cells in vivo. Bioinformatics prediction, luciferase reporter gene experiment, Ago2-RIP experiment and RNA pull-down assay were adopted to identify the binding relationships among circ_0001175, miR-130a-5p and SNX5.
Results: Circ_0001175 and SNX5 expressions were up-regulated in HCC tissues and cell lines, while miR-130a-5p expression was down-regulated. Abnormal expressions of circ_0001175, miR-130a-5p and SNX5 were associated with poor clinicopathological features of HCC patients; circ_0001175 facilitated HCC cell proliferation, migration and invasion in vitro and promoted lung metastasis in vivo; miR-130a-5p inhibited the above malignant biological behaviors of HCC cells, and it could reverse the function of circ_0001175. SNX5 was identified as a target gene of miR-130a-5p, and circ_0001175 could sponge miR-130a-5p and up-regulate the expression of SNX5 in HCC cells.
Conclusion: Circ_0001175 is highly expressed in HCC and facilitates HCC progression through regulating miR-130a-5p/SNX5 axis.
Keywords: circ_0001175, miR-130a-5p, HCC, proliferation, metastasis
Introduction
According to the GLOBOCAN database, liver cancer is the sixth most common and the fourth most deadly cancer, causing about 840,000 new cases and 780,000 deaths per year worldwide.1 Hepatocellular carcinoma (HCC) accounts for 90% of primary liver cancer.2 Surgery can relatively effectively treat patients with HCC in early stage.3,4 However, the symptoms of HCC patients in early stage are not obvious, and the first diagnosis of most HCC patients is in advanced stage.5 Without effective therapy strategies, the prognosis of patients with advanced HCC is extremely poor, and the five-year survival rate is less than 10%.6,7 Therefore, finding new therapeutic targets is vital for improving the prognosis of HCC patients.
Circular RNAs (circRNAs) are closed-loop RNAs formed by reverse splicing of pre-mRNA.8 It is reported that the dysregulation of circRNAs is significantly associated with the carcinogenesis and progression of liver cancer.9–12 For example, circ_0000885 expression is up-regulated in HCC and circ_0000885 enhances HCC cell proliferation.11 Circ_0015756 is highly expressed in HCC and facilitates HCC cell proliferation and metastasis through regulating miR-7/FAK axis.12 Circ_0001175 is generated from the YTH N6-methyladenosine RNA-binding protein 1 (YTHDF1) transcript (Supplementary Figure 1A). YTHDF1 is reported to be up-regulated in HCC tissues and its high expression is associated with the unfavorable prognosis of HCC patients.13 However, the role of circ_0001175 in HCC is still obscure.
MicroRNAs (miRNAs) are endogenous small RNAs with a length of approximately 22 nucleotides, which directly bind to the 3ʹ-UTR of mRNA and repress translation.14 Many miRNAs are associated with the pathogenesis of HCC.15–18 For example, miR-144 is lowly expressed in HCC and it inhibits HCC progression by regulating CLK3 and Wnt/β-catenin signaling.17 MiR-223 reverses the chemoresistance of HCC cells by regulating autophagy.18 MiR-122, regulated by HNF1A, HNF3A and HNF3B, can inhibit the migration and invasion of HCC cells.19 MiR-130a-5p represses the growth and metastasis of glioma by down-regulating HMGB2 expression.20 Additionally, it is reported that low miR-130a-5p expression is correlated with poor prognosis of HCC patients.21 However, the function of miR-130a-5p in HCC and its regulatory mechanisms have not been fully elucidated.
The sorting nexin family (SNXs) are proteins that contain the SNX-PX domain.22 They regulate the structure and function of endoplasmic reticulum and Golgi apparatus, and modulate membrane cycling and endocytosis.23 Sorting nexin 5 (SNX5), belonging to SNX family, has key regulatory functions in cancer biology. Besides, SNX5 is highly expressed in thyroid cancer, and it promotes cancer progression.24 In HCC, SNX5 high expression is significantly related to vascular invasion, intrahepatic metastasis and the adverse prognosis of the patients; its overexpression also enhances HCC cell proliferation and metastasis.25 However, the mechanism that triggers the dysregulation of SNX5 in HCC is unclear.
As mentioned above, miR-130a-5p expression is down-regulated in HCC and miR-130a-5p functions as a tumor suppressor;21 meanwhile, our bioinformatics analysis indicated that miR-130a-5p was a potential target of circ_0001175. This relationship suggests that circ_0001175 can probably function as an oncogenic circRNA in HCC. This study verified that circ_0001175 expression was up-regulated in HCC. Moreover, circ_0001175 high expression was closely linked to increased TNM stage and low differentiation of tumor tissues. Additionally, it was demonstrated that circ_0001175 enhanced the proliferation and metastasis of HCC cells via regulating miR-130a-5p/SNX5 axis.
Materials and Methods
Samples Collection
HCC tissue samples and adjacent tissue samples were collected from the HCC patients who received surgery in Guangdong Second Provincial General Hospital (October 2017 to March 2018). None of the patients had received chemotherapy or interventional therapy before the surgery. All patients provided written informed consent, and the collection and use of human tissues were approved by the Research Ethics Committee of Guangdong Second Provincial General Hospital (2018-PWK-032). This study was performed based on the principles outlined in the Declaration of Helsinki.
Cell Culture
Normal liver cells (L02 cells) and HCC cell lines (SMMC-7721, Bel-7402, Huh-7, HepG2 and Hep3B cells) were available from the American Type Culture Collection (ATCC, Rockville, MD, USA) or China Center for Type Culture Collection (CCTCC, Wuhan, China). All cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM, HyClone, Logan, UT, USA) containing 10% fetal bovine serum (FBS, HyClone, Logan, UT, USA), 100 U/mL penicillin and 0.1 mg/mL streptomycin (Hyclone, Logan, UT, USA) at 37°C in 5% CO2. When the cell confluence reached 70% to 80%, they were trypsinized with 0.25% trypsin (Thermo Fisher Scientific, Shanghai, China) and subcultured.
Cell Transfection
The pCD5-ciR-circ_0001175 plasmid, pCD5-ciR-control plasmid, circ-0001175 siRNA, negative control siRNA (siNC), miR-130a-5p mimics, mimics control, miR-130a-5p inhibitors and inhibitors control were available from Ribobio (Guangzhou, China). Transfection was performed with FuGENE® HD Transfection Reagent (Roche, Shanghai, China) according to the manufacturer’s instruction. After 24 h of transfection, total RNA was extracted from cells and quantitative real-time polymerase chain reaction (qRT-PCR) was used to detect the transfection efficiency.
qRT-PCR
Total RNA was extracted from HCC tissues and cells by TRIzol regent (Invitrogen, Carlsbad, CA, USA) and then reversely transcribed into cDNA. The cDNA was then used as the template, and qRT-PCR was performed using SYBR Premix Ex Taq™ Kit (Takara, Dalian, China). MiRNA expression was determined using TaqMan MicroRNA Assay Kit (Applied Biosystems, Foster City, CA, USA). After that, the relative expressions of circ_0001175, miR-130a-5p and SNX5 mRNA were calculated using the 2−ΔΔCT method. The internal references for miRNA and mRNA/circRNA were U6 and GAPDH, respectively. All primer sequences were purchased from Genecopoeia (Guangzhou, China), and the details of primer sequences are shown in Table 1.
 |
Table 1 Sequences Used for RT-qPCR |
Cell Counting Kit-8 (CCK-8) Assay
Huh-7 and SMMC-7721 cells in the logarithmic growth phase were collected. Cell suspension was prepared and the cell density was modulated to 1×104 cells/mL. The cells were then inoculated into 96-well plates (100 μL of cell suspension/well), and 10 μL of CCK-8 solution (Beyotime, Shanghai, China) was supplemented into each well after 1, 2, and 3 d, respectively. On each day, after CCK-8 solution was added, the cells were incubated for 4 h. Subsequently, a microplate reader was employed to detect the optical density (OD value) of each well at a wavelength of 450 nm.
BrdU Experiment
Huh-7 and SMMC-7721 cells in the logarithmic growth phase were harvested to prepare the single-cell suspension, and the cells were then inoculated into a 24-well plate (1×105 cells/well) before BrdU staining reagent (Beyotime, Shanghai, China) was added. After the incubation for 12 h, the cells were fixed with 4% paraformaldehyde, incubated with anti-BrdU antibody (Beyotime, Shanghai, China) and then were stained with the DAPI staining solution (Beyotime, Shanghai, China) to label the nuclei. The BrdU positive cells and the DAPI positive cells in three visual fields were counted under a fluorescence microscope (Olympus, Tokyo, China). Cell proliferation rate=number of BrdU-positive cells/number of DAPI-positive cells.
Transwell Experiment
24-well plates supplemented with Transwell chambers (8 μm pore size, BD Biosciences, CA, USA) were used in Transwell assay. In migration assay, 600 μL of medium containing 20% FBS was added to each well of the 24-well plate, and 200 μL of cell suspension (containing 1×105 cells) was added to each Transwell chamber. Thirty-six hour later, the medium was discarded, and the Transwell chambers were taken out. A wet cotton swab was used to gently wipe off the cells that did not pass through the membrane. The migrated cells were fixed with methanol for 30 s, then stained with crystal violet solution for 1 min and washed with PBS. After the membrane was dried, the number of cells in three random visual fields was counted under a microscope (×200). In the invasion assay, the membranes of Transwell chambers were pre-coated with a layer of Matrigel (BD Biosciences, CA, USA), and the other procedures were the same as the migration assay.
Western Blot
Huh-7 and SMMC-7721 cells were harvested and washed three times with PBS. Then, RIPA lysis buffer (containing 1% PMSF) (Beyotime, Shanghai, China) was added, and the cells were lysed by sonication in ice water. The protein concentration was determined by Bradford method. After the proteins were denatured, an equal amount of protein was taken from each group for 10% SDS-PAGE, and the protein on the gel was then transferred onto the PVDF membrane (Millipore, Billerica, MA, USA). After that, the membrane was blocked with 5% skimmed milk at room temperature for 1 h. Next, the primary antibodies, including anti-SNX5 antibody (Abcam, ab241295, 1:500), anti-E-cadherin antibody (Abcam, ab40772, 1:1000) and anti-N-cadherin antibody (Abcam, ab76011, 1:1000), were added, and the PVDF membranes were incubated at 4°C overnight. After the membranes were washed with TBST, the secondary antibody (Beyotime, Shanghai, China, A0208, 1:2500) was added to incubate the membranes for 1 h at room temperature. After that, the membrane was washed with TBST. The development of the protein bands was performed using electrochemical luminescence (ECL) kit (Beyotime, Shanghai, China). GAPDH was used as the endogenous control.
Luciferase Reporter Assay
The luciferase reporter vectors of circ_0001175 wild type (WT)/mutant (MUT) or SNX5 WT/MUT were transfected into 293T cells together with miR-130a-5p mimic or control miRNA. The medium was removed after 48 h of culture. Subsequently, PBS was used to wash the cells, and then the washing solution was discarded. Next, lysis buffer was employed to lyse the cells. The cells were oscillated on a shaker at room temperature for 5 min and then centrifuged for 5 min, and the supernatant was harvested for luciferase activity measurement. According to the instruction of dual-luciferase reporter assay system (Promega, Madison, WI, USA), the luciferase activities of the samples were determined.
RNA Immunoprecipitation (RIP)
RIP assay was performed with Magna RIP RNA-binding protein immunoprecipitation kit (Millipore, Bedford, MA, USA). Approximately 1×107 cells were harvested and resuspended with RIP lysis buffer supplemented with protease and RNase inhibitors. Then, the cell lysate was incubated with anti-Ago2 antibody (Abcam, Cambridge, MA, USA) or negative control mouse immunoglobulin G (IgG; Millipore, Shanghai, China) at 4°C overnight. Next, proteinase K was used to remove the proteins, and immunoprecipitated RNA was extracted using the RNeasy MinElute Cleanup kit (Qiagen, Hilden, Germany) and then reversely transcribed using the GoScript™ Reverse Transcription System (Promega, Madison, WI, USA). Ultimately, the enrichment of circ_0001175 and miR-130a-5p was detected by qRT-PCR.
RNA Pull-Down Assay
Huh-7 and SMMC-7721 cells were lysed using RIPA lysis buffer (Biossci, Wuhan, China) and then incubated with biotinylated normal control circRNA (NC), biotinylated wild type circ_0001175 (WT) or biotinylated circ_0001175 mutant (MUT) (GenePharma, Shanghai, China) for 1 h at room temperature. Following that, Dynabeads® M-280 streptavidin (Invitrogen, Carlsbad, CA, USA) was used to isolate biotin-labeled RNA, and qRT-PCR was utilized for detecting the enrichment of miR-130a-5p.
Lung Metastasis Model in vivo
BALB/c nude mice (male, 4–5 weeks old) were purchased from the Guangzhou Experimental Animal Centre (Guangzhou, China). The mice were randomly divided into two groups (control group and circ_0001175 overexpression group, n=5 per group). Huh-7 cells (1×107 cells/per mouse) were injected into each mouse via tail vein. Three weeks later, the mice were euthanized, and the lungs were fixed in formalin and embedded in paraffin. Then, hematoxylin/eosin staining was performed for pathological examination of lung nodules of the mice. The number of pulmonary metastatic nodules in each section was counted in five randomly selected visual fields under the microscope (Olympus, Tokyo, China).
Immunohistochemical (IHC) Staining
HCC tissues were cut into sections (thickness: 3 μm). The sections were deparaffinized with xylene and then hydrated. The sections were then heated in citrate buffer at 100°C for 1 min for antigen retrieval, and endogenous peroxidases were inactivated in 3% hydrogen peroxide for 10 min at 37°C. The sections were then incubated with primary anti-SNX5 antibody (ab250218, 1:100, Abcam, Shanghai, China) at 4°C overnight, followed by the incubation with secondary antibody (A0277, 1:1000, Beyotime, Shanghai, China) for 60 min at 37°C. Finally, 3,3-diaminobenzidine tetrahydrochloride was used to develop the color, and each section was scored by two independent pathologists. IHC score was calculated based on staining intensity score (0, no staining; 1, weak staining; 2, moderate staining; and 3, intense staining) and the proportion score (0, no staining; 1, 1–25% of the tumor cells were stained; 2, 26–50%; 3, 51–75%; and 4, more than 75% of the tumor cells were stained). IHC score = staining intensity score × proportion score. IHC score was used to evaluate the expression of SNX5: 0 points, negative; 1–6 points, weakly positive; 8–12 points, strongly positive.
Statistical Analysis
All data were statistically analyzed using SPSS 22.0 (SPSS Inc., Chicago, IL, USA). The results were expressed as mean ± standard deviation. Differences between two or more groups were analyzed by Student’s t-test or one-way ANOVA. P < 0.05 signified statistical significance.
Results
Circ_0001175 Expression Was Up-Regulated in HCC Tissues and Cell Lines
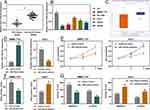
First of all, we tried to detect the circular structure of circ_0001175. RNase R assay indicated that circ_0001175 in Huh-7 cells was more resistant to RNAase R, compared with linear GAPDH mRNA and YTHDF1 mRNA (Supplementary Figure 1B). Next, we examined the specificity of siRNA and primers for circ_0001175 used in this work. As shown, siRNA targeting circ_0001175 specifically repressed the expression of circ_0001175, but did not impede the expressions of YTHDF1 and circ_0061146 (another circRNA generated from YTHDF1 transcript, Supplementary Figure 1C). Next, we used qRT-PCR to detect circ_0001175 expression in HCC tissues and cell lines. The results showed that compared with adjacent tissues, circ_0001175 expression was remarkably up-regulated in HCC tissues (Figure 1A). Circ_0001175 expression in HCC cell lines was also significantly up-regulated compared with that in normal liver cells (Figure 1B). Additionally, it was found that the high expression of circ_0001175 was associated with the high TNM stage of HCC patients and the low differentiation of tumor tissues (Figure 1C and D).
Circ_0001175 Significantly Enhanced the Proliferation, Migration, Invasion and Lung Metastasis of HCC Cells
Among the 5 HCC cell lines, circ_0001175 had the lowest expression in Huh-7 cells and the highest expression in SMMC-7721 cells. Therefore, we transfected the circ_0001175 overexpression plasmid into Huh-7 cells and the circ_0001175 siRNA into SMMC-7721 cells. qRT-PCR showed that the transfection was successful (Figure 2A). Through CCK-8 and BrdU assays, it was found that in comparison with the control group, the cell proliferation of the Huh-7 cells in circ_0001175 overexpression group was markedly increased while the cell proliferation of the SMMC-7721 cells in circ_0001175 knockdown group was markedly reduced (Figure 2B and C). Transwell experiment manifested that circ_0001175 overexpression enhanced the migration and invasion of Huh-7 cells while circ_0001175 knockdown inhibited the migration and invasion of SMMC-7721 cells (Figure 2D). Then, we detected the expressions of N-cadherin and E-cadherin by Western blot, the results of which unmasked that overexpression of circ_0001175 increased the expression of N-cadherin and inhibited the expression of E-cadherin in Huh-7 cells; knockdown of circ_0001175 functioned oppositely in SMMC-7721 cells (Supplementary Figure 1D). Additionally, a lung metastasis model in nude mice was used to evaluate the metastasis of HCC cells in vivo, and it showed that overexpression of circ_0001175 enhanced the pulmonary metastasis of Huh-7 cells in vivo (Supplementary Figure 1E). The above experiments implied that circ_0001175 facilitated the proliferation, migration, invasion, epithelial–mesenchymal transition (EMT) and metastasis of HCC cells.
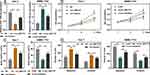
Circ_0001175 Specifically Regulated miR-130a-5p
StarBase database indicated that the sequence of circ_0001175 contained a possible binding site for miR-130a-5p (Figure 3A). Dual-luciferase reporter gene assay implied that compared to miR-NC, miR-130a-5p significantly suppressed the relative luciferase activity of wild-type circ_0001175 reporter while it showed no remarkable effect on the mutant circ_0001175 reporter (Figure 3B). Subsequently, Ago2-RIP experiments and RNA pull-down assay were used to further verify the interaction between circ_0001175 and miR-130a-5p, and the results indicated the direct interaction between them (Figure 3C and D). Additionally, we used qRT-PCR to detect the effect of circ_0001175 on miR-130a-5p expression. It was revealed that circ_0001175 overexpression inhibited miR-130a-5p expression in Huh-7 cells, and circ_0001175 knockdown promoted miR-130a-5p expression in SMMC-7721 cells (Figure 3E). Furthermore, after detecting circ_0001175 and miR-130a-5p expressions in the cancer tissues of the 45 HCC patients, we found that their expressions were negatively correlated (Figure 3F). The above data authenticated that circ_0001175 directly targeted and negatively regulated miR-130a-5p.
MiR-130a-5p Inhibited Proliferation, Migration and Invasion of HCC Cells
qRT-PCR showed that miR-130a-5p expression was markedly down-regulated in HCC tissues and cell lines (Figure 4A and B). Consistently, the data in StarBase database also indicated that miR-130a-5p expression in HCC samples was remarkably lower than in normal tissues (Figure 4C). Notably, the underexpression of miR-130a-5p was associated with a higher TNM stage and the poor differentiation of tumor tissues (Supplementary Figure 2A and B). Furthermore, miR-130a-5p mimics and miR-130a-5p inhibitors were transfected into SMMC-7721 cells and Huh-7 cells, respectively. qRT-PCR showed that the transfection was successful (Figure 4D). Through CCK-8, BrdU and Transwell assays, it was found that miR-130a-5p mimics significantly repressed the proliferation, migration and invasion of HCC cells; conversely, miR-130a-5p inhibitors promoted the above malignant biological behaviors (Figure 4E–G). The data revealed that miR-130a-5p was a tumor suppressor in HCC.
Circ_0001175 Facilitated the Proliferation, Migration and Invasion of HCC Cells by Adsorbing miR-130a-5p
To investigate whether circ_0001175 played a role in HCC cells through regulating miR-130a-5p, we co-transfected circ_0001175 overexpression plasmid+miR-130a-5p mimics into Huh-7 cells, and circ_0001175 siRNA+miR-130a-5p inhibitors were co-transfected into SMMC-7721 cells. qRT-PCR showed successful transfection (Figure 5A). Through CCK-8, BrdU and Transwell assays, it was found that miR-130a-5p mimics could partially weaken the promotion of HCC cell proliferation, migration and invasion induced by circ_0001175 overexpression; miR-130a-5p inhibitors could partly reverse the inhibitory effect of circ_0001175 knockdown on these malignant biological behaviors of HCC cell (Figure 5B–D). The above findings implied that circ_0001175 facilitated the proliferation, migration and invasion of HCC cells by adsorbing miR-130a-5p.
MiR-130a-5p Targeted SNX5
SNX5was predicted as one of the downstream targets of miR-130a-5p through the TargetScan database (Figure 6A). Subsequently, we verified the binding relationship through dual-luciferase reporter gene experiment: the data revealed that compared to miR-NC, miR-130a-5p significantly inhibited the relative luciferase activity of wild-type SNX5 reporter, but it had no remarkable effect on the relative luciferase activity of SNX5 MUT reporter (Figure 6B). Additionally, circ_0001175 overexpression or inhibiting miR-130a-5p expression promoted the expression of SNX5 in HCC cells while circ_0001175 knockdown or miR-130a-5p overexpression worked oppositely (Figure 6C and D). Furthermore, qRT-PCR was performed to detect SNX5 expression in HCC tissues, and the data revealed that SNX5 expression mRNA was remarkably higher than that in adjacent tissues (Figure 6E). Additionally, high expression of SNX5 mRNA was associated with higher TNM stage and poor differentiation of tumor tissues (Supplementary Figure 2C and D). In addition, the results of IHC indicated that SNX5 protein was overexpressed in HCC tumor tissues (Supplementary Figure 2E). Moreover, we used Pearson’s correlation analysis to analyze the correlation between the expressions of miR-130a-5p and SNX5 mRNA in HCC samples, the findings of which showed that miR-130a-5p expression was negatively correlated with SNX5 expression (Figure 6F). The above data indicated that miR-130a-5p directly targeted and negatively regulated SNX5 expression and SNX5 expression could be positively regulated by circ_0001175.
Discussion
Previous research demonstrates that circRNAs can regulate the progression of HCC through various mechanisms.26–28 For example, circ-DB expression is up-regulated in HCC patients with higher body fat ratios, and circ-DB inhibits miR-34a, activates the USP7/Ciclin A2 signaling pathway, accelerates tumor growth and reduces DNA damage;29 Twist1 promotes vimentin expression by increasing circ-10720 expression, thereby promoting EMT in HCC;30 circ-ADD3 promotes EZH2 degradation through CDK1-mediated ubiquitination, thereby inhibiting the metastasis of HCC cells.31 In this research, for the first time, we found that circ_0001175 was highly expressed in HCC tissues and cells, and its high expression was remarkably associated with increased TNM stage and low differentiation of tumor tissues. Functional experiments in vitro and in vivo demonstrated that circ_0001175 overexpression promoted the proliferation and metastasis of HCC cells. These data indicated that circ_0001175 played an oncogenic role in HCC.
MiRNAs participate in a variety of physiological and pathological processes.32–34 Reportedly, multiple miRNAs regulate the malignant biological behaviors of cancer cells, including proliferation, migration, apoptosis, autophagy, drug resistance and angiogenesis in diverse cancers, HCC included.16,35,36 MiR-130a-5p is identified as a tumor suppressor in several cancers, and it can inhibit the expressions of a variety of oncogenes. For example, miR-130a-5p down-regulates ZEB1 expression, thereby inhibiting proliferation and EMT of cancer cells in esophageal squamous cell carcinoma.37 In gastric cancer, miR-130a-5p targets CB1R and suppresses Wnt/β-catenin signaling pathway, thereby inhibiting the growth and metastasis of cancer cells.38 In the work, we found that miR-130a-5p expression was down-regulated in HCC samples and cell lines. As expected, it was demonstrated that miR-130a-5p repressed the proliferation, migration and invasion of HCC cells. Collectively, we proved that miR-130a-5p was a tumor suppressor of HCC.
Accumulating evidence shows that the circRNA-miRNA regulatory axis has important regulatory roles in HCC: in this mechanism, circRNA can function as a molecular sponge to decoy miRNAs and increase the expression level of mRNA, thus regulating gene expression.39,40 For example, circ_0001955 promotes the tumorigenesis of HCC by down-regulating miR-516a-5p expression and up-regulating TRAF6 and MAPK11 expressions;40 circ_0091579 facilitates the proliferation and metastasis of HCC cells by down-regulating miR-490-3p expression.41 In the present research, it was revealed that circ_0001175 directly adsorbed miR-130a-5p and repressed its expression; additionally, rescue experiments showed that the biological functions of circ_0001175 were dependent on miR-130a-5p. Our data suggested that circ_0001175 contributed to the dysregulation of miR-130a-5p and miR-130a-5p was an important downstream molecule of circ_0001175 in HCC pathogenesis.
SNX5expression is up-regulated in head and neck squamous cell carcinoma expression, and SNX5 inhibits FBW7-mediated oncoprotein ubiquitination, promoting the cancer progression.22 In HCC, SNX5 is also proved to be oncogenic. A recent study reports that SNX5 enhances HCC cell proliferation, migration and invasion by inhibiting EGF-mediated EGFR endocytosis and activating the EGFR-ERK1/2 pathway; importantly, its high expression implies poor prognosis of HCC patients.25 In the research, consistently, it was demonstrated that SNX5 expression was up-regulated in HCC samples, and SNX5 was validated as a target gene of miR-130a-5p and could be positively regulated by circ_0001175. Our work gave a possible explanation to clarify the mechanism of SNX5 dysregulation in HCC.
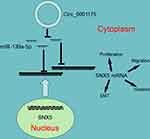
Taken together, this research reveals that circ_0001175 expression is up-regulated in HCC and circ_0001175 promotes HCC progression, and its high expression is significantly associated with adverse pathological parameters. Further mechanism investigation indicates that circ_0001175 regulates miR-130a-5p/SNX5 axis (Figure 7). Our work offers potential biomarkers and therapy targets for the diagnosis and treatment of HCC.
Ethics Statement
Our study was approved by the Ethics Review Board of Guangdong Second Provincial General Hospital.
Acknowledgment
We thank Hubei Yican Health Industry Co., Ltd for its linguistic assistance during the preparation of this manuscript.
Author Contributions
All authors made a significant contribution to the work (conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas); took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; agreed on the journal to which the article has been submitted; agreed to be accountable for all aspects of the work.
Funding
This work was supported by a grant from the National Natural Science Foundation of China (No. 81071990, No. 81641110).
Disclosure
The authors declare that they have no conflicts of interest for this work.
References
1. Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424.
2. Morse MA, Sun W, Kim R, et al. The role of angiogenesis in hepatocellular carcinoma. Clin Cancer Res. 2019;25(3):912–920. doi:10.1158/1078-0432.CCR-18-1254
3. Shiina S, Sato K, Tateishi R, et al. Percutaneous ablation for hepatocellular carcinoma: comparison of various ablation techniques and surgery. Can J Gastroenterol Hepatol. 2018;2018:4756147. doi:10.1155/2018/4756147
4. Yi PS, Li Y, Yan S, et al. Surgery combined with post-operative transcatheter arterial chemoembolization improves survival of intermediate hepatocellular carcinoma. Scand J Gastroenterol. 2019;54(2):240–245. doi:10.1080/00365521.2019.1577487
5. Gong XL, Qin SK. Progress in systemic therapy of advanced hepatocellular carcinoma. World J Gastroenterol. 2016;22(29):6582–6594. doi:10.3748/wjg.v22.i29.6582
6. Su CW, Chau GY, Hung HH, et al. Impact of steatosis on prognosis of patients with early-stage hepatocellular carcinoma after hepatic resection. Ann Surg Oncol. 2015;22(7):2253–2261. doi:10.1245/s10434-014-4221-5
7. Goldaracena N, Mehta N, Scalera I, et al. Multicenter validation of a score to predict prognosis after the development of HCC recurrence following liver transplantation. HPB. 2019;21(6):731–738. doi:10.1016/j.hpb.2018.10.005
8. Cai X, Nie J, Chen L, Yu F. Circ_0000267 promotes gastric cancer progression via sponging MiR-503-5p and regulating HMGA2 expression. Mol Genet Genomic Med. 2019;8:e1093.
9. Gao J, Dai C, Yu X, Yin XB, Zhou F. circ-TCF4.85 silencing inhibits cancer progression through microRNA-486-5p-targeted inhibition of ABCF2 in hepatocellular carcinoma. Mol Oncol. 2019.
10. Lin T, Dai Y, Guo X, et al. Silencing of hsa_circ_0008450 represses hepatocellular carcinoma progression through regulation of microRNA-214-3p/EZH2 Axis. Cancer Manag Res. 2019;11:9133–9143. doi:10.2147/CMAR.S222716
11. Li XQ, Song JY, Lv W, Zhang D, Wu JZ. Circular circ_0000885 promotes hepatocellular carcinoma proliferation by epigenetically upregulating Caprin1. Eur Rev Med Pharmacol Sci. 2019;23(18):7848–7854.
12. Liu L, Yang X, Li NF, Lin L, Luo H. Circ_0015756 promotes proliferation, invasion and migration by microRNA-7-dependent inhibition of FAK in hepato cellular carcinoma. Cell Cycle. 2019;18(21):2939–2953. doi:10.1080/15384101.2019.1664223
13. Zhao X, Chen Y, Mao Q, et al. Overexpression of YTHDF1 is associated with poor prognosis in patients with hepatocellular carcinoma. Cancer Biomark. 2018;21(4):859–868.
14. Guo S, Fesler A, Huang W, et al. Functional significance and therapeutic potential of miR-15a mimic in pancreatic ductal adenocarcinoma. Mol Ther Nucleic Acids. 2019;19:228–239. doi:10.1016/j.omtn.2019.11.010
15. Pratama MY, Pascut D, Massi MN, Tiribelli C. The role of microRNA in the resistance to treatment of hepato cellular carcinoma. Ann Transl Med. 2019;7(20):577. doi:10.21037/atm.2019.09.142
16. Baldari S, Di Rocco G, Magenta A, Picozza M, Toietta G. Extracellular vesicles-encapsulated MicroRNA-125b produced in genetically modified mesenchymal stromal cells inhibits hepatocellular carcinoma cell proliferation. Cells. 2019;8(12):1560. doi:10.3390/cells8121560
17. Li H, Cui X, Hu Q, Chen X, Zhou P. CLK3 is a direct target of miR-144 and contributes to aggressive progression in hepato cellular carcinoma. Onco Targets Ther. 2019;12:9201–9213. doi:10.2147/OTT.S224527
18. Zhou Y, Chen E, Tang Y, et al. MiR-223 overexpression inhibits doxorubicin-induced autophagy by targeting FOXO3a and reverses chemoresistance in hepato cellular carcinoma cells. Cell Death Dis. 2019;10(11):843. doi:10.1038/s41419-019-2053-8
19. Coulouarn C, Factor VM, Andersen JB, et al. Loss of miR-122 expression in liver cancer correlates with suppression of the hepatic phenotype and gain of metastatic properties. Oncogene. 2009;28(40):3526–3536. doi:10.1038/onc.2009.211
20. Xu CH, Xiao LM, Liu Y, et al. The lncRNA HOXA11-AS promotes glioma cell growth and metastasis by targeting miR-130a-5p/HMGB2. Eur Rev Med Pharmacol Sci. 2019;23(1):241–252.
21. Li B, Huang P, Qiu J, et al. MicroRNA-130a is down-regulated in hepatocellular carcinoma and associates with poor prognosis. Med Oncol. 2014;31(10):230. doi:10.1007/s12032-014-0230-2
22. Cai J, Sun M, Hu B, et al. Sorting Nexin 5 controls head and neck squamous cell carcinoma progression by modulating FBW7. J Cancer. 2019;10(13):2942–2952. doi:10.7150/jca.31055
23. Sheng J, Wang L, Han Y, et al. Dual roles of protein as a template and a sulfur provider: a general approach to metal sulfides for efficient photothermal therapy of cancer. Small. 2018;14(1):1702529. doi:10.1002/smll.201702529
24. Ara S, Kikuchi T, Matsumiya H, et al. Sorting nexin 5 of a new diagnostic marker of papillary thyroid carcinoma regulates Caspase-2. Cancer Sci. 2012;103(7):1356–1362. doi:10.1111/j.1349-7006.2012.02296.x
25. Zhou Q, Huang T, Jiang Z, et al. Upregulation of SNX5 predicts poor prognosis and promotes hepatocellular carcinoma progression by modulating the EGFR-ERK1/2 signaling pathway. Oncogene. 2019.
26. Xiong DD, Feng ZB, Lai ZF, et al. High throughput circRNA sequencing analysis reveals novel insights into the mechanism of nitidine chloride against hepatocellular carcinoma. Cell Death Dis. 2019;10(9):658. doi:10.1038/s41419-019-1890-9
27. Su Y, Xu C, Liu Y, Hu Y, Wu H. Circular RNA hsa_circ_0001649 inhibits hepatocellular carcinoma progression multiple miRNAs sponge. Aging. 2019;11(10):3362–3375. doi:10.18632/aging.101988
28. Zhang X, Xu Y, Qian Z, et al. CircRNA_104075 stimulates YAP-dependent tumorigenesis through the regulation of HNF4a and may serve as a diagnostic marker in hepatocellular carcinoma. Cell Death Dis. 2018;9(11):1091. doi:10.1038/s41419-018-1132-6
29. Zhang H, Deng T, Ge S, et al. Exosome circRNA secreted from adipocytes promotes the growth of hepatocellular carcinoma by targeting deubiquitination-related USP7. Oncogene. 2019;38(15):2844–2859. doi:10.1038/s41388-018-0619-z
30. Meng J, Chen S, Han J-X, et al. Twist1 regulates vimentin through Cul2 circular RNA to promote EMT in hepatocellular carcinoma. Cancer Res. 2018;78(15):4150–4162. doi:10.1158/0008-5472.CAN-17-3009
31. Sun S, Wang W, Luo X, et al. Circular RNA circ-ADD3 inhibits hepatocellular carcinoma metastasis through facilitating EZH2 degradation via CDK1-mediated ubiquitination. Am J Cancer Res. 2019;9(8):1695–1707.
32. Li X, Liu H, Lv Y, Yu W, Liu X, Liu C. MiR-130a-5p/Foxa2 axis modulates fetal lung development in congenital diaphragmatic hernia by activating the Shh/Gli1 signaling pathway. Life Sci. 2020;241:117166.
33. Peng J, Liu F, Zheng H, Wu Q, Liu S. Long noncoding RNA ZFAS1 promotes tumorigenesis and metastasis in nasopharyngeal carcinoma by sponging miR-892b to up-regulate LPAR1 expression. J Cell Mol Med. 2019;24(2):1437–1450. doi:10.1111/jcmm.14823
34. Tomasetti M, Gaetani S, Monaco F, Neuzil J, Santarelli L. Epigenetic regulation of miRNA expression in malignant mesothelioma: miRNAs as biomarkers of early diagnosis and therapy. Front Oncol. 2019;9:1293. doi:10.3389/fonc.2019.01293
35. Guo J, Zhao P, Liu Z, et al. MiR-204-3p inhibited the proliferation of bladder cancer cells via modulating lactate dehydrogenase-mediated glycolysis. Front Oncol. 2019;9:1242. doi:10.3389/fonc.2019.01242
36. Xia H, Zhao Y. miR-155 is high-expressed in polycystic ovarian syndrome and promotes cell proliferation and migration through targeting PDCD4 in KGN cells. Artif Cells Nanomed Biotechnol. 2020;48(1):197–205. doi:10.1080/21691401.2019.1699826
37. Wang W, Wu D, He X, et al. CCL18-induced HOTAIR upregulation promotes malignant progression in esophageal squamous cell carcinoma through the miR-130a-5p-ZEB1 axis. Cancer Lett. 2019;460:18–28. doi:10.1016/j.canlet.2019.06.009
38. Xian X, Tang L, Wu C, Huang L. miR-23b-3p and miR-130a-5p affect cell growth, migration and invasion by targeting CB1R via the Wnt/β-catenin signaling pathway in gastric carcinoma. Onco Targets Ther. 2018;11:7503–7512. doi:10.2147/OTT.S181706
39. Tian F, Yu C, Wu M, Wu X, Wan L, Zhu X. MicroRNA-191 promotes hepatocellular carcinoma cell proliferation by has_circ_0000204/miR-191/KLF6 axis. Cell Prolif. 2019;52(5):e12635. doi:10.1111/cpr.12635
40. Yao Z, Xu R, Yuan L, et al. Circ_0001955 facilitates hepatocellular carcinoma (HCC) tumorigenesis by sponging miR-516a-5p to release TRAF6 and MAPK11. Cell Death Dis. 2019;10(12):945. doi:10.1038/s41419-019-2176-y
41. Niu WY, Chen L, Zhang P, Zang H, Zhu B, Shao WB. Circ_0091579 promotes proliferative ability and metastasis of liver cancer cells by regulating microRNA-490-3p. Eur Rev Med Pharmacol Sci. 2019;23(23):10264–10273.
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.