Back to Journals » OncoTargets and Therapy » Volume 13
Circ_0001023 Promotes Proliferation and Metastasis of Gastric Cancer Cells Through miR-409-3p/PHF10 Axis
Authors Wang Y, Zhang J, Chen X , Gao L
Received 31 December 2019
Accepted for publication 2 April 2020
Published 22 May 2020 Volume 2020:13 Pages 4533—4544
DOI https://doi.org/10.2147/OTT.S244358
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Leo Jen-Liang Su
Yongxiang Wang,1 Jianbin Zhang,2,3 Xiaochen Chen,2,3 Liang Gao2,3
1Department of Abdominal Surgery, Cancer Hospital of the University of Chinese Academy of Sciences (Zhejiang Cancer Hospital), Hangzhou 310022, Zhejiang Province, People’s Republic of China; 2Department of Oncology, Zhejiang Provincial People’s Hospital, Hangzhou 310022, Zhejiang Province, People’s Republic of China; 3Department of Oncology, People’s Hospital of Hangzhou Medical College, Hangzhou 310014, Zhejiang Province, People’s Republic of China
Correspondence: Xiaochen Chen
Department of Oncology, Zhejiang Provincial People’s Hospital, Hangzhou 310022, Zhejiang Province, People’s Republic of China
Tel +86-571-85893183
Email [email protected]
Liang Gao
Department of Oncology, People’s Hospital of Hangzhou Medical College, Hangzhou 310014, Zhejiang Province, People’s Republic of China
Email [email protected]
Background: Circular RNAs (circRNAs) have been well documented to regulate the gene expression via sponging microRNA (miRNA) in diverse neoplasms including gastric cancer (GC).
Methods: In the present study, the expressions of circ_0001023, miR-409-3p, and plant homeodomain finger 10 (PHF10) in GC tissues were detected by qRT-PCR. Chi-square test was performed to analyze the associations between circ_0001023 and pathological parameters. Cell Counting Kit-8 assay, colony formation assay, flow cytometry, and transwell assay were adopted to detect the role of circ_0001023/miR-409-3p axis in the proliferation, apoptosis, and migration of GC cells, respectively. The targeting relationship between circ_0001023 and miR-409-3p was investigated by dual-luciferase gene reporter gene assay. Additionally, subcutaneous xenotransplanted tumor model in nude mice was established to detect the function of circ_0001023 on GC growth in vivo.
Results: Compared with adjacent tissues, the expression of circ_0001023 was significantly upregulated and correlated with lymph node invasion and higher T stage of GC patients. It has also been proved that circ_0001023 could target miR-409-3p. Silencing circ_0001023 can impede the proliferation of GC cells and promote apoptosis, while miR-409-3p inhibitors can partially reverse the biological behavior of GC cells mentioned above. Moreover, the expression of circ_0001023 was reversely associated with miR-409-3p expression but positively correlated with PHF10, a downstream oncogene of miR-409-3p.
Conclusion: Collectively, it is concluded that circ_0001023 promotes the progression of GC via regulating miR-409-3p/PHF10 axis.
Keywords: gastric cancer, circ_0001023, miR-409-3p, PHF10
Introduction
Gastric cancer (GC) is one of the leading deadly cancers worldwide.1,2 Even though the surgery, chemotherapy, and targeted drugs offer symptomatic relief and modest improvement in survival for GC patients, the overall survival rate of GC patients still remains unfavorable.3–5 Hence, an in-depth investigation into the underlying mechanism of GC is highly desirable to explore the optimized therapy for GC.
Circular RNA (circRNA) is a newly-discovered class of endogenous non-coding RNA, which exists widely and stably in mammalian cells without the function of encoding proteins.6,7 In cancer biology, circRNA has been gradually recognized as a new clinical biomarker and potential therapeutic target.8–10 Mounting studies indicate that circRNA plays important roles in multiple cancers, including GC.11,12 For instance, circ-SFMBT2 promotes the proliferation of GC cells by increasing the expression of CREB1, and circ-PDSS1 facilitates GC progression.13,14 On the other side, circ-LMTK2 functions as a tumor suppressor in GC, whose low expression predicts unfavorable prognosis of GC patients.15 However, the defined role of circ_0001023, a novel circRNA, in cancer remains poorly understood.
MicroRNA (miRNA) is a class of small non-coding RNA, containing 19 to 25 nucleotides. Abnormally expressed miRNA is a frequent event in the tumorigenesis and progression of diverse cancers.16 miRNA is involved in promoting or inhibiting a variety of biological behaviors of cells.17 Accumulating pieces of evidence have verified that miRNA can interact with other non-coding RNA by base complementary pairing (also called “sponging”). For example, circ_PSMC3 impedes the proliferation of GC cells via sponging miR-296-5p.18 Circ_NRIP1 sponges miR-149-5p to facilitate GC progression.19 miR-409-3p is proven to be lowly expressed in GC tissues, thus playing a tumor-suppressive role.20 However, the mechanism of miR-409-3p dysregulation in GC remains poorly clarified. It is worth noting that bioinformatics analysis (https://circinteractome.nia.nih.gov/) suggests the existence of potential binding sites between circ_0001023 and miR-409-3p (context score percentile = 99, which implied a high possibility of regulatory relationship between them), indicating that circ_0001023 can probably function as the molecular sponge of miR-409-3p. Such a regulatory relationship in the tumorigenesis and progression of GC remains to be further verified.
Plant homeodomain finger 10 (PHF10), belonging to the zinc finger protein family, can repress the transcription of caspase-3 and impaired the apoptosis pathway in GC cells, so as to promote the growth of tumor.21 PHF10 was significantly overexpressed in GC and colonic cancer tissues,22–24 indicating that PHF10 plays an oncogenic role in the progression of cancers. In addition, it is reported that PHF10 is negatively regulated miR-409-3p in GC.20 However, the underlying mechanism of PHF10 dysregulation in GC remains far from fully elucidated.
In the present study, for the first time, it was demonstrated that circ_0001023 was overexpressed in GC tissues and cells, and it promoted the malignant biological behaviors of GC cells, including proliferation, anti-apoptosis, migration, and invasion. In terms of mechanism, this work proved that circ_0001023 could promote GC progression via miR-409-3p/PHF10 axis.
Materials and Methods
Tissue Collection
This study was conducted in accordance with the ethical standards in the Declaration of Helsinki. All patients involved gave informed consent to the study and signed a written consent form. Thirty-three cases of GC tissues and matched adjacent tissues were collected under the approval of Ethics Review Committee of Zhejiang Provincial People’s Hospital. The resected tissues were immediately stored in −196°C liquid nitrogen for further analysis. No patients received neoadjuvant therapy (chemotherapy or radiotherapy) before the surgery.
Cell Culture
Human GC cell lines (AGS, BGC-823, MGC-803, MKN-28, and SGC-7901 cells) were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA). The immortalized gastric epithelial cell line (GES-1) was a gift from the Shanghai Institute of Digestive Disease (Shanghai, China). All cells were cultured in Dulbecco’s Modified Essential Medium (DMEM) (Hyclone, Logan, UT, USA) containing 10% heat-inactivated fetal bovine serum (FBS; Invitrogen, Grand Island, NY, USA), 100U/mL penicillin and 100 μg/mL streptomycin (Invitrogen, Grand Island, NY, USA) in 5% CO2 at 37°C. The medium was replaced at an interval of 3 to 4 days.
Cell Transfection
Circ_0001023 overexpression plasmid pcDNA3.1‐circ_0001023 (pcDNA-circ_0001023), pcDNA3.1 vector (pcDNA-NC), siRNA control (si-con), circ_0001023 targeting siRNAs (si-circ_0001023), miRNA control (miR-con), miR-409-3p mimics and its corresponding inhibitor were procured from RiboBio (Guangzhou, China). pcDNA-circ_0001023 was transfected into AGS cells, and si-circ_0001023 was transfected into MKN-28 and SGC-7901 cells by lipofectamine ®3000 (Thermo Fisher Scientific, Rockford, IL, USA). Quantitative real-time polymerase chain reaction (qRT-PCR) was then conducted to measure the transfection efficiency. The sequences of siRNA circ_0001023 were shown as follows: siRNA-1: 5ʹ-ACCATCCTGTTATAGATAACA-3ʹ; siRNA-2: 5ʹ-TCCTGTTATAGATAACAGACA-3ʹ; siRNA-3: 5ʹ-CATCCTGTTATAGATAACAGA-3ʹ.
RNA Isolation and qRT-PCR
Total RNA was isolated from GC tissues and cells with TRIzol Reagent (Invitrogen, Grand Island, NY, USA). One microgram total RNA was reverse transcribed to cDNA with SuperScript First Strand cDNA System (Invitrogen, Grand Island, NY, USA). Then, real-time PCR was performed with SYBR Green Premix Ex Taq II (TaKaRa, Dalian, China) and ABI 7500 real-time PCR system (Applied Biosystems, Foster City, CA, USA). The relative expressions of circ_0001023 and miR-409-3p were calculated using 2−ΔΔCT method. The primer sequences used in this work were shown as follows: circ_0001023: 5ʹ‐GAGGTGTACAAATCGTGGGC‐3′ (forward) and 5′‐CCACCTGCCACCTCAAGAAT‐3′ (reverse); miR-409-3p: 5ʹ-GAATGTTGCTCGGTGA-3ʹ (forward) and 5ʹ-GTGCAGGGTCCGAGGT-3ʹ (reverse); U6: 5ʹ-CTCGCTTCGGCAGCACA-3ʹ (forward) and 5ʹ-AACGCTTCACGAATTTGCGT-3ʹ (reverse); β-actin: 5ʹ-TTCGAGCAAGAGATGGCCA-3ʹ (forward) and 5ʹ-TACATGGTGGTGCCGCC-3ʹ (reverse).
Cell Counting Kit-8 Assay
The proliferation of GC cells was tested with Cell Counting Kit-8 (CCK-8) (Dojindo, Kumamoto, Japan). In brief, AGS, MKN-28, and SGC-7901 cells were harvested 48 h after transfection. A total of 1×103 cells resuspended in 100 μL medium were then inoculated in each well of a 96-well plate. Following that, the 96-well plate was placed in the incubator. Twenty-four hours later, 10 μL enhanced CCK-8 solution was then added to continue the culture in an incubator for 1 hour. After the culture was terminated, the 96-well plate was placed in the microplate reader (Bio-Tek, USA), and the absorbance (OD value) of each well at 450 nm wavelength was determined. Afterward, the absorbance of cells in each group was measured at 24, 48, 72, and 96 hours.
Colony Formation Assay
A total of 1×103 GC cells were inoculated in each 6.0 cm culture dish. The cells were maintained in a humidified incubator (5% CO2, 37°C). After 2 weeks of culture, the medium was discarded and the colonies were carefully washed with phosphate-buffered saline (PBS) twice. Cells were then fixed with 10% paraformaldehyde for 10 min and stained with 0.1% crystal violet for 15 min. After the colonies were gently washed and dried, the number of colonies formed was recorded under a microscope.
Apoptosis Assay
Cells in each group were washed and centrifuged. After that, binding buffer (Invitrogen, Carlsbad, CA, USA) was used to resuspend cells, and the cell density was adjusted to 1×104/mL. After stained with Annexin V-FITC solution and propidium iodide (PI) (Invitrogen, Carlsbad, CA, USA), the cells were fully mixed and incubated in dark at room temperature for 15 min. After that, the apoptotic rate of cells in each group was measured by flow cytometry (BD Biosciences, Franklin Lakes, NJ, USA).
Transwell Assay
According to the manufacturer’s protocol, GC cells in each group were harvested and resuspended with serum-free medium, and the density of cells was adjusted to 1×105/mL. After that, 200μL single-cell suspension was added in each Transwell chamber (8 μm pore diameter, BD Biosciences, CA, USA) placed in a 24-well plate. Six hundred microliters complete medium was added to each well of the 24-well plate. Then, the 24-well plate was placed in the incubator. Twenty-four hours later, the cells on the upper surface of the Transwell membrane were gently wiped off with a cotton swab. The migrated cells were fixed with 95% ethanol for 20 min and then stained with 0.1% crystal violet for 10 min. After that, the Transwell chamber was inverted and the cells were counted under a microscope. For invasion assays, before the cells were inoculated, the membrane of the Transwell chambers was coated with a layer of Matrigel (1:8 dilution, BD Biosciences, CA, USA), and the other steps were the same as the migration experiment.
Western Blot
The cells were lysed with RIPA buffer (Beyotime Biotech, Shanghai, China). After high-speed centrifugation (12,000× g, 4°C, 15 min), the supernatant was collected and heated in a water bath at 100°C for 10 min to denature the protein. The concentration of the protein was then determined by bicinchoninic acid (BCA) kit (Beyotime biotech, Shanghai, China). Following that, the protein samples were separated with 12% SDS-PAGE and transferred to PVDF membranes (Millipore, Schwalbach, Germany). After the PVDF membrane was washed with TBST (20mM Tris–HCl, 150mM NaCl, and 0.1% Tween 20, pH 7.5) solution, the primary antibody was added to incubate the PVDF membrane overnight. Following that, the PVDF membrane was then rinsed with TBST solution and incubated with the secondary antibody (Abcam, ab205718, 1:2000) at room temperature for 1 hour. After that, TBST was used to wash the membrane again. Ultimately, color rendering was performed using hypersensitive ECL (Hubei Biossci Biotechnology Co, Ltd.). The primary antibodies included Anti-PHF10 antibody (Abcam, ab154637, 1:1000), Anti-Bax antibody (Abcam, ab32503, 1:1000), Anti-Bcl-2 antibody (Abcam, ab185002, 1:1000), and internal reference anti-β-actin antibody (Abcam, ab20272, 1:3000).
Dual-Luciferase Reporter Gene Assay
Wild type (WT) or mutant type (MUT) circ_0001023 sequences were subcloned into pmiRGLO dual-luciferase miRNA target expression reporter (Promega, Madison, WI, USA). MiR-409-3p or miR-con were co-transfected with WT or MUT reporters into GC cells, respectively. Luciferase activity was measured 48 hours after transfection. Dual-luciferase reporter assay system (Promega, Madison, WI, USA) was adopted to determine the luciferase activity, and firefly luciferase activity was normalized to renilla luciferase activity.
In vivo Experiment
The procedures of in vivo experiment were approved by the Ethics Committee of Animal Model Research Center of Zhejiang Provincial People’s Hospital. The procedures also followed United Kingdom Coordinating Committee on Cancer Research (UKCCCR) Guidelines for the welfare of animals in experimental neoplasia. BALB/c nude mice (4 weeks old, male) were used. In subcutaneous xenotransplantation studies, 1×107 MKN-28 cells were resuspended in 100 mL phosphate buffer (PBS) and injected subcutaneously to the left (si-circ_0001023 group) and right side (si-con group) of nude mice (n=6). The tumor size was monitored every 3 days, and the tumor volume was calculated according to the formula (V=1/2×L×W2).
Statistic Analysis
Data were presented as mean ± standard deviation (SD). GraphPad Prism 8(GraphPad Software, Inc., La Jolla, CA, USA) was adopted for statistical analysis. “t” test was carried out to analyze the difference of data. Chi-square test was performed to analyze the correlation between circ_0001023 expression and clinicopathological indexes. p < 0.05 indicated statistical significance.
Results
Circ_0001023 Was Highly Expressed in GC Cells
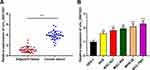
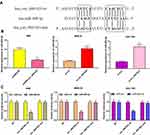
First of all, qRT-PCR was conducted to detect the expressions of circ_0001023 in 33 cases of GC. We found that GC tissues exhibited a higher expression of circ_0001023 than adjacent tissues (Figure 1A). Besides, we detected the expressions of circ_0001023 in five kinds of GC cells including AGS, BGC-823, MGC-803. MKN-28, and SGC-7901. It was discovered that, compared with GES-1 cells, all of the five GC cell lines mentioned above displayed a significant upregulation of circ_000102 expression (Figure 1B).
The Expression of circ_0001023 Was Linked to Multiple Pathological Indexes in Patients with GC
Then, we further analyzed the association between circ_0001023 expression and the clinicopathological parameters of GC patients. It was indicated that highly expressed circ_0001023 in tumor tissues was markedly correlated with local lymph node invasion and higher T stage in GC patients, but had no association with age, gender, tumor size, and degree of differentiation (Table 1).
 |
Table 1 Correlations Between Circ_0001023 Expression and Clinical Characteristics in GC Patients |
Circ_0001023/miR-409-3p Axis Regulated the Proliferation of GC Cells
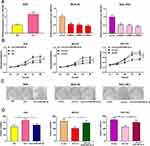
To explore the effect of circ_0001023 on the proliferation of GC cells and its potential mechanism, we transfected AGS cells with pcDNA-circ_0001023 and successfully constructed a model of circ_0001023 overexpression cells. MKN-28 cells and SGC-7901 cells were transfected with si-circ_0001023 to establish circ_0001023 knockdown cell model (Figure 2A). Then, the proliferation of cells in each group was detected by CCK-8 assay. The results suggested that the proliferation of GC cells was notably promoted by overexpression of circ_0001023, and this effect was partially weakened by co-transfection of miR-409-3p mimics; meanwhile, knockdown of circ_0001023 markedly arrested the proliferation of GC cells, while miR-409-3p inhibitors partially reversed it (Figure 2B). Subsequently, colony formation assay showed that upregulated circ_0001023 in GC cells significantly increased the number of colonies, whereas miR-409-3p mimics restrained the colony formation of GC cells; after circ_ 0001023 was knocked down, colonies showed a decline in its number, while miR-409-3p inhibitors partially reversed the inhibitory effect caused by knockdown circ_0001023 (Figure 2C and D). In short, the above data suggested that circ_0001023 could modulate the proliferation of GC cells via regulating miR-409-3p.
Circ_0001023/miR-409-3p Axis Regulated the Apoptosis of GC Cells
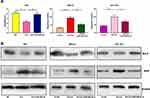
Next, by performing flow cytometry analysis, it was found that overexpressed circ_0001023 restrained the apoptosis of AGS cells, and the inhibitory effect was partially reversed by miR-409-3p mimics; meanwhile, the apoptosis of MKN-28 and SGC-7901 cells was increased significantly after the knockdown of circ_0001023, and such effect was weakened after co-transfection of miR-409-3p inhibitors (Figure 3A). Then, we detected the expression levels of apoptosis-related proteins Bcl-2 and Bax by Western blot, and the results showed that circ_0001023/miR-409-3p axis modulated the expressions of Bcl-2 and Bax (Figure 3B). The above data indicated that circ_0001023/miR-409-3p also regulated the apoptosis of GC cells.
Circ_0001023/miR-409-3p Axis Modulated the Metastasis of GC Cells
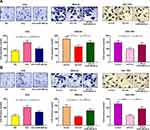
Transwell assay was subsequently performed to detect the role of circ_0001023 in the migration and invasion of GC cells. As shown, overexpressed circ_0001023 promoted the migration of AGS cells, which was partially inhibited by miR-409-3p mimics; on the contrary, after circ_0001023 was knocked down, the migration of MKN-28 and SGC-7901 cells was inhibited, which was reversed after the co-transfection of miR-409-3p inhibitors (Figure 4A). In invasion assays, similar results were observed (Figure 4B). These data suggested that circ_0001023 facilitated the migration and invasion of GC cells via modulating miR-409-3p.
Circ_0001023 Interacted Directly with miR-409-3p and Negatively Regulated Its Expression
As shown, circ_0001023 had a potential binding site with miR-409-3p, which was predicted by Circinteractome database (Figure 5A). qRT-PCR suggested that circ_0001023 overexpression could significantly reduce the expression level of miR-409-3p, while knockdown circ_0001023 exerted the opposite effect (Figure 5B). To further study whether miR-409-3p can bind directly to circ_0001023, dual-luciferase report assay was performed, and it showed that overexpressed miR-409-3p significantly decreased the luciferase activity of circ_0001023-WT reporter, but had no significant impact on circ_0001023-MUT reporter in all GC cell lines (Figure 5C). The above data revealed that circ_0001023 regulated the expression of miR-409-3p by directly targeting it.
Correlations Were Observed Among the Expressions of circ_0001023, miR-409-3p, and PHF10 in GC Samples
The data above suggested that circ_0001023 was involved in the tumorigenesis and progression of GC by regulating miR-409-3p. Subsequently, we analyzed the expression levels of circ_0001023, miR-409-3p, and PHF10 mRNA in GC samples. The results indicated that there was a negative correlation between circ_0001023 and miR-409-3p (Figure 6A), which further validated the regulatory relationship between them. The expression of miR-409-3p was also verified to be negatively correlated with that of PHF10 (Figure 6B), while a positive correlation between the expression of circ_0001023 and the expression of PHF10 was observed (Figure 6C). Taken together, it was concluded that circ_0001023 could function as ceRNA for miR-409-3p, and increase the expression level of PHF10.
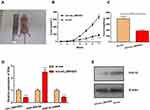
Knockdown of circ_0001023 Could Suppress Tumor Growth in vivo
To determine the effect of circ_0001023 on tumor growth in vivo, MKN-28 cells transfected with si-circ_0001023 or si-NC were subcutaneously injected into nude mice, and the size and weight of tumor formed in the back of mice were observed (Figure 7A). As shown, circ_0001023 knockdown repressed the growth of MKN-28 cells in vivo (Figure 7B). Additionally, the average tumor weight in si-circ_0001023 group was significantly lower than that si-con group (Figure 7C). qRT-PCR showed that, after circ_0001023 was knocked down in MKN-28 cells, the expressions of circ_0001023 and PHF10 in tumor tissues were significantly downregulated, while the expression of miR-409-3p was notably upregulated (Figure 7D). Subsequently, Western blot informed us that the knockdown of circ_0001023 results in a decrease in the expression of PHF10 on protein level (Figure 7E). These results further proved the oncogenic role of circ_0001023 in GC.
Discussion
In recent years, a lot of studies indicate that non-coding RNAs, including circular RNA, play a pivotal role in regulating the progression of tumors.25–28 To explore the mechanism between circRNA and the biological behavior of GC cells will offer novel clues for the treatment of GC.29–31 To our best knowledge, this is the first study to explore the role of circ_0001023 in GC.
Accumulating pieces of evidence have demonstrated that circRNA is a key regulator implicated in the regulation of malignant biological behaviors of GC.32–34 Firstly, circRNA has a covalent closed-loop structure that makes it not easy to be degraded by RNase;35 secondly, circRNA is mainly composed of exons and (or) introns and it contains miRNA reaction element;36 thirdly, circRNA can interact with RNA binding protein (RBPs) to form a larger RNA-protein complex.37 Owing to these special structures, circRNA is endowed with the regulatory functions in the progression of diverse malignancies. In this study, abnormally highly expressed circ_0001023 was observed in GC tissues and cells, and its overexpression was significantly correlated with lymph node metastasis and higher T stage. In addition, it was found that the proliferation, migration, and invasion of GC cells were significantly restrained after circ_0001023 was knocked down, and the proportion of apoptotic cells was markedly increased. On the other hand, upregulation of circ_0001023 could enhance the above malignant phenotypes of GC cells. In vivo experiments further confirmed that knocking down circ_0001023 could repress tumor growth. Collectively, the experimental data demonstrate that circ_0001023 facilitates the tumorigenesis and progression of GC, and circ_0001023 is a promising biomarker and therapy target for GC.
Similar as circRNA, miRNA is also implicated in the development of multiple cancers.16–20,38 It is reported that miR-409-3p exerts a tumor-suppressive role in the proliferation and metastasis of osteosarcoma cells via targeting Zinc finger E-box-binding homeobox-1.39 Moreover, miR-409-3p can inhibit the migration and invasion of bladder cancer cells by targeting c-Met.40 It has been indicated that miR-409-3p, which is lowly expressed in GC tissues, can block the progression of GC by repressing the expression of PHF10.20 It is well known that circRNA can sponge miRNAs to regulate their expression.28,33,34 Their interactions contribute to the progression of cancer. For example, circ_0001368 inhibits the development of GC through regulating miR-6506-5p/FOXO3 axis;26 circ_0067997 facilitates the progression of GC by sponging miR-515-5p and activating X-chromosome-linked inhibitor of apoptosis.32 In this work, bioinformatics analysis was carried out to predict the existence of a potential binding site between circ_0001023 and miR-409-3p, and the predicted binding site was validated by dual-luciferase reporter assay. Additionally, it was also demonstrated that knockdown of circ_0001023 could increase the expression of miR-409-3p in GC cells, and overexpression of circ_0001023 could suppress the expression of miR-409-3p. Furthermore, after miR-409-3p inhibitors were co-transfected into GC cells with circ_0001023 knocked down, the inhibitory effects induced by circ_0001023 knockdown on cell proliferation and metastasis were partially reversed. These data identify miR-409-3p as a downstream target of circ_0001023, and partly explained the reason that miR-409-3p was dysregulated in GC tissues.
Additionally, in the present study, it was demonstrated that the expression level of PHF10 was positively associated with that of circ_0001023 in GC samples, and PHF10 was downregulated in GC cells after circ_0001023 was knocked down. The above data indicated that circ_0001023 might serve as a ceRNA to modulate miR-409-3p/PHF10 axis during GC progression. Considering that PHF10 is a crucial component of the PBAF (SWI/SNF) multiprotein complex, which changes the chromatin structure and dominates multiple eukaryotic gene expression,24 it is possible that circ_0001023 has the potential to indirectly regulate more downstream oncogenes to drive GC progression, which needs to be investigated in the future.
Conclusion
In summary, our findings highlight a novel circRNA-regulated mechanism in GC, that is circ_0001023 facilitates the proliferation, migration, and invasion of GC cells by regulating miR-409-3p/PHF10 axis. Our study explores new mechanisms in the development of GC and provides a novel theoretical basis for the diagnosis and treatment of GC.
Data Sharing Statement
The data used to support the findings of this study are available from the corresponding authors upon request.
Author Contributions
All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors declare that they have no competing interest.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin. 2019;69(1):7–34. doi:10.3322/caac.21551
2. Kather JN, Pearson AT, Halama N, et al. Deep learning can predict microsatellite instability directly from histology in gastrointestinal cancer. Nat Med. 2019;25(7):1054–1056. doi:10.1038/s41591-019-0462-y
3. He H, Li H, Su X, et al. Study on safety of laparoscopic total gastrectomy for clinical stage I gastric cancer: the protocol of the CLASS02-01 multicenter randomized controlled clinical trial. BMC Cancer. 2018;18(1):944. doi:10.1186/s12885-018-4846-z
4. Wang P, Wang J, Tan H, et al. Acid- and reduction-sensitive micelles for improving the drug delivery efficacy for pancreatic cancer therapy. Biomater Sci. 2018;6(5):1262–1270. doi:10.1039/c7bm01051f
5. Yoshioka T, Shien K, Takeda T, et al. Acquired resistance mechanisms to afatinib in HER2-amplified gastric cancer cells. Cancer Sci. 2019;110(8):2549–2557. doi:10.1111/cas.14089
6. Wu J, Qi X, Liu L, et al. Emerging epigenetic regulation of circular RNAs in human cancer. Mol Ther Nucleic Acids. 2019;16:589–596. doi:10.1016/j.omtn.2019.04.011
7. Sang Y, Chen B, Song X, et al. circRNA_0025202 regulates tamoxifen sensitivity and tumor progression via regulating the miR-182-5p/FOXO3a axis in breast cancer. Mol Ther. 2019;27(9):1638–1652. doi:10.1016/j.ymthe.2019.05.011
8. Zhang Y, Liu H, Li W, et al. CircRNA_100269 is downregulated in gastric cancer and suppresses tumor cell growth by targeting miR-630. Aging (Albany NY). 2017;9(6):1585–1594. doi:10.18632/aging.101254
9. Gu W, Sun Y, Zheng X, et al. Identification of gastric cancer-related circular RNA through microarray analysis and bioinformatics analysis. Biomed Res Int. 2018;2018:2381680. doi:10.1155/2018/2381680
10. Zhou LH, Yang YC, Zhang RY, Wang P, Pang MH, Liang LQ. CircRNA_0023642 promotes migration and invasion of gastric cancer cells by regulating EMT. Eur Rev Med Pharmacol Sci. 2018;22(8):2297–2303. doi:10.26355/eurrev_201804_14818
11. Li P, Chen S, Chen H, et al. Using circular RNA as a novel type of biomarker in the screening of gastric cancer. Clin Chim Acta. 2015;444:132–136. doi:10.1016/j.cca.2015.02.018
12. Fang J, Hong H, Xue X, et al. A novel circular RNA, circFAT1(e2), inhibits gastric cancer progression by targeting miR-548g in the cytoplasm and interacting with YBX1 in the nucleus. Cancer Lett. 2019;442:222–232. doi:10.1016/j.canlet.2018.10.040
13. Handong S, Pengcheng X, Sun Z, et al. Circ-SFMBT2 promotes the proliferation of gastric cancer cells through sponging miR-182-5p to enhance CREB1 expression. Cancer Manag Res. 2018;10:5725–5734. doi:10.2147/cmar.S172592
14. Ouyang Y, Li Y, Huang Y, et al. CircRNA circPDSS1 promotes the gastric cancer progression by sponging miR-186-5p and modulating NEK2. J Cell Physiol. 2019;234(7):10458–10469. doi:10.1002/jcp.27714
15. He J, Chen J, Ma B, Jiang L, Zhao G. CircLMTK2 acts as a novel tumor suppressor in gastric cancer. Biosci Rep. 2019;39(5):
16. Dong L, Zhang Z, Xu J, et al. Consistency analysis of microRNA-arm expression reveals microRNA-369-5p/3p as tumor suppressors in gastric cancer. Mol Oncol. 2019;13(7):1605–1620. doi:10.1002/1878-0261.12527
17. Ruggieri V, Russi S, Zoppoli P, et al. The role of microRNAs in the regulation of gastric cancer stem cells: a meta-analysis of the current status. J Clin Med. 2019;8(5):
18. Rong D, Lu C, Zhang B, et al. CircPSMC3 suppresses the proliferation and metastasis of gastric cancer by acting as a competitive endogenous RNA through sponging miR-296-5p. Mol Cancer. 2019;18(1):25. doi:10.1186/s12943-019-0958-6
19. Zhang X, Wang S, Wang H, et al. Circular RNA circNRIP1 acts as a microRNA-149-5p sponge to promote gastric cancer progression via the AKT1/mTOR pathway. Mol Cancer. 2019;18(1):20. doi:10.1186/s12943-018-0935-5
20. Li C, Nie H, Wang M, et al. MicroRNA-409-3p regulates cell proliferation and apoptosis by targeting PHF10 in gastric cancer. Cancer Lett. 2012;320(2):189–197. doi:10.1016/j.canlet.2012.02.030
21. Wei M, Liu B, Su L, et al. A novel plant homeodomain finger 10-mediated antiapoptotic mechanism involving repression of caspase-3 in gastric cancer cells. Mol Cancer Ther. 2010;9(6):1764–1774. doi:10.1158/1535-7163.Mct-09-1162
22. Wet M, Liu JY, Lv X, et al. Preparation of PHF10 antibody and analysis of PHF10 expression gastric cancer tissues. Xi Bao Yu Fen Zi Mian Yi Xue Za Zhi. 2010;26(9):874–876.
23. Cavalieri D, Dolara P, Mini E, et al. Analysis of gene expression profiles reveals novel correlations with the clinical course of colorectal cancer. Oncol Res. 2007;16(11):535–548. doi:10.3727/096504007783438376
24. Tatarskiy EV, Georgiev GP, Soshnikova NV. Oncogene c-MYC controls the expression of PHF10 subunit of PBAF chromatin remodeling complex in SW620 cell line. Dokl Biochem Biophys. 2019;484(1):66–68. doi:10.1134/s1607672919010204
25. Guarnerio J, Zhang Y, Cheloni G, et al. Intragenic antagonistic roles of protein and circRNA in tumorigenesis. Cell Res. 2019;29(8):628–640. doi:10.1038/s41422-019-0192-1
26. Lu J, Zhang PY, Li P, et al. Circular RNA hsa_circ_0001368 suppresses the progression of gastric cancer by regulating miR-6506-5p/FOXO3 axis. Biochem Biophys Res Commun. 2019;512(1):29–33. doi:10.1016/j.bbrc.2019.02.111
27. Shao Y, Li J, Lu R, et al. Global circular RNA expression profile of human gastric cancer and its clinical significance. Cancer Med. 2017;6(6):1173–1180. doi:10.1002/cam4.1055
28. Ding L, Zhao Y, Dang S, et al. Circular RNA circ-DONSON facilitates gastric cancer growth and invasion via NURF complex dependent activation of transcription factor SOX4. Mol Cancer. 2019;18(1):45. doi:10.1186/s12943-019-1006-2
29. Tan H, Gan L, Fan X, Liu L, Liu S. Diagnostic value of circular RNAs as effective biomarkers for cancer: a systematic review and meta-analysis. Onco Targets Ther. 2019;12:2623–2633. doi:10.2147/ott.S197537
30. Li Y, Zhao J, Yu S, et al. Extracellular vesicles long RNA sequencing reveals abundant mRNA, circRNA, and lncRNA in human blood as potential biomarkers for cancer diagnosis. Clin Chem. 2019;65(6):798–808. doi:10.1373/clinchem.2018.301291
31. Jiang F, Hong F, Shah MW, Shen X. Circular RNAs as diagnostic biomarkers in gastric cancer: a meta-analysis review. Pathol Res Pract. 2019;215(6):152419. doi:10.1016/j.prp.2019.04.011
32. Zhang H, Wang X, Huang H, Wang Y, Zhang F, Wang S. Hsa_circ_0067997 promotes the progression of gastric cancer by inhibition of miR-515-5p and activation of X chromosome-linked inhibitor of apoptosis (XIAP). Artif Cells Nanomed Biotechnol. 2019;47(1):308–318. doi:10.1080/21691401.2018.1553787
33. Huang X, Li Z, Zhang Q, et al. Circular RNA AKT3 upregulates PIK3R1 to enhance cisplatin resistance in gastric cancer via miR-198 suppression. Mol Cancer. 2019;18(1):71. doi:10.1186/s12943-019-0969-3
34. Suzuki H, Tsukahara T. A view of pre-mRNA splicing from RNase R resistant RNAs. Int J Mol Sci. 2014;15(6):9331–9342. doi:10.3390/ijms15069331
35. Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495(7441):333–338. doi:10.1038/nature11928
36. Hentze MW, Preiss T. Circular RNAs: splicing’s enigma variations. EMBO J. 2013;32(7):923–925. doi:10.1038/emboj.2013.53
37. Zhang L, Song X, Chen X, et al. Circular RNA CircCACTIN promotes gastric cancer progression by sponging MiR-331-3p and regulating TGFBR1 expression. Int J Biol Sci. 2019;15(5):1091–1103. doi:10.7150/ijbs.31533
38. Jiao Y, Yang H, Qian J, et al. miR‑3664‑5P suppresses the proliferation and metastasis of gastric cancer by attenuating the NF‑κB signaling pathway through targeting MTDH. Int J Oncol. 2019;54(3):845–858. doi:10.3892/ijo.2019.4680
39. Wu L, Zhang Y, Huang Z, et al. MiR-409-3p inhibits cell proliferation and invasion of osteosarcoma by targeting zinc-finger E-box-binding homeobox-1. Front Pharmacol. 2019;10:137. doi:10.3389/fphar.2019.00137
40. Xu X, Chen H, Lin Y, et al. MicroRNA-409-3p inhibits migration and invasion of bladder cancer cells via targeting c-Met. Mol Cells. 2013;36(1):62–68. doi:10.1007/s10059-013-0044-7
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.