Back to Journals » Cancer Management and Research » Volume 12
Circ_0000885 Enhances Osteosarcoma Progression by Increasing FGFR1 Expression via Sponging MiR-1294
Authors Chen Y, Zhang S, Bai C, Guan Z, Chen W
Received 31 December 2019
Accepted for publication 19 June 2020
Published 28 July 2020 Volume 2020:12 Pages 6441—6452
DOI https://doi.org/10.2147/CMAR.S244382
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Sanjeev K. Srivastava
Yinxian Chen, Sicheng Zhang, Chuanqing Bai, Zhiye Guan, Wenjian Chen
Department of Orthopedic, Children’s Hospital of Anhui Medical University, Hefei, Anhui 230032, People’s Republic of China
Correspondence: Wenjian Chen
Department of Orthopedic, Children’s Hospital of Anhui Medical University, Baohe District, Hefei, Anhui 230032, People’s Republic of China
Tel +86-13721021060
Email [email protected]
Background: As a malignant tumor, the progression of osteosarcoma (OS) is mediated by multiple regulators, including circular RNAs (circRNAs). However, the role of circ_0000885 in OS is unclear.
Materials and Methods: Quantitative real-time polymerase chain reaction (qRT-PCR) was applied to detect the expression of circ_0000885, miR-1294 and fibroblast growth factor receptor 1 (FGFR1). Cell proliferation was evaluated using 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyltetrazolium bromide (MTT) assay and colony formation assay. Flow cytometry and transwell assay were employed to determine the cell cycle distribution, cell migration and invasion, respectively. Moreover, the relationship between miR-1294 and circ_0000885 or FGFR1 was confirmed by dual-luciferase reporter assay. The protein level of FGFR1 was assessed via Western blot (WB) analysis. Animal experiments were used to verify the effect of circ_0000885 silencing on OS tumor growth in vivo.
Results: Circ_0000885 level was increased in OS tissues and cells. Knockdown of circ_0000885 repressed the proliferation, migration, invasion and induced cell cycle arrest in OS cells. There was a binding relationship between miR-1294 and circ_0000885, and miR-1294 inhibitor could reverse the inhibitory effect of silenced circ_0000885 on OS progression. MiR-1294 could target FGFR1, and overexpressed FGFR1 could invert the suppression effect of miR-1294 mimic on OS progression. Silencing of circ_0000885 hindered FGFR1 expression, while this effect could be recovered by miR-1294 inhibitor. In addition, circ_0000885 knockdown reduced OS tumor growth via regulating the FGFR1 expression by sponging miR-1294 in vivo.
Conclusion: Circ_0000885 played an active role in OS progression, indicating that it might be a potential target for OS therapy.
Keywords: OS, progression, circ_0000885, miR-1294, FGFR1
Introduction
Osteosarcoma (OS) is a malignant tumor with high mortality that often occurs in children and adolescents.1,2 OS is characterized by pain and lumps in the tumor site and can lead to limited joint movement and muscle atrophy as the disease progresses.3,4 Early detection and early treatment is the treatment criterion for OS, but advanced tumor metastasis and invasion to soft tissue seriously hinder the treatment process of OS patients.5,6 Therefore, exploring the molecules that influence OS progression may provide new strategies for the treatment of OS.
Non-coding RNAs (ncRNAs) are a class of RNAs that are directly transcribed from the genome but do not encode proteins, including the well-studied circular RNAs (circRNAs) and microRNAs (miRNAs) in recent years.7,8 CircRNAs are believed to play an important regulatory role in OS progression, and many circRNA functions have been clarified.9 For instance, circ_0000285 expression is thought to be associated with OS proliferation and migration,10 and upregulated circ_0001658 can promote the proliferation and metastasis of OS.11 Circ_0000885 is a newly discovered circRNA. Li et al reported that circ_0000885 was highly expressed in hepatocellular carcinoma and was involved in the regulation of cell proliferation and cell cycle.12 Zhu et al showed that circ_0000885 was upregulated in the tissues and serum of OS patients, so it was speculated that it might be a good indicator of poor prognosis in OS patients,13 but its role in OS progression had not been studied yet.
MiR-1294 is considered a tumor suppressor in many cancers, such as clear cell renal cell carcinoma and oral squamous cell carcinoma.14,15 Besides, miR-1294 can be sponged by circ_0005198 to participate in the mediation of circ_0005198 on the proliferation, apoptosis and metastasis of glioma.16 In OS, Zhang et al found that miR-1294 could restrain the proliferation and invasion of OS cells,17 which provided a basis for exploring the role of miR-1294 in OS. Fibroblast growth factor receptor 1 (FGFR1) is a member of the FGFR family, and its expression is closely related to biological processes, such as cell survival, migration, and proliferation.18 And there is a lot of evidence that FGFR1 plays an essential role in tumorigenesis.19 Many studies have confirmed that FGFR1 has significant high expression in OS and is related to the proliferation and metastasis of OS.20,21
At present, the possibility that circRNA can act as a “miRNA sponge” has been proved as a candidate mechanism for circRNA.22,23 Therefore, in this study, in addition to exploring the role of circ_0000885 in OS progression, we also explored the molecular mechanism of circ_0000885 based on the circRNA-miRNA-messenger RNA (mRNA) regulatory network. These studies might provide references for exploring new therapeutic targets for OS.
Materials and Methods
Collection of Tissues
Thirty pairs of OS tumor tissues and matched normal tissues were collected from 30 OS patients who underwent treatment and surgery in Children’s Hospital of Anhui Medical University and were saved at −80°C. All patients had written informed consent, and our study was approved by the Ethics Committee of Children’s Hospital of Anhui Medical University.
Cell Culture
OS cell line (Saos-2) and normal human osteoblastic cell line (hFOB) were bought from American Type Culture Collection (ATCC, Manassas, VA, USA) and cultured in McCoy’s 5A medium (Invitrogen, Carlsbad, CA, USA) and Dulbecco’s modified Eagle medium (DMEM, Invitrogen), respectively. OS cell line (SOSP-9607) was purchased from the Department of orthopedics, fourth military medical university (Xian, China) and was approved by the Ethics Committee of Children’s Hospital of Anhui Medical University. SOSP-9607 cells were cultured in RPMI-1640 medium (Invitrogen). All medium contained 10% fetal bovine serum (FBS, Invitrogen) and 1% penicillin/streptomycin (Invitrogen). All cells were placed at 37°C with 5% CO2 incubator.
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Total RNA was extracted using Trizol reagent (Invitrogen). After measuring the concentration, the RNA was reversely converted to cDNA using cDNA Synthesis Kit (Thermo Fisher Scientific, Waltham, MA, USA), and then qRT-PCR was performed via Fast SYBR Green (Thermo Fisher Scientific). Data were normalized using glyceraldehyde 3-phosphate dehydrogenase (GAPDH) or U6. The primer sequences were listed as below: circ_0000885, F 5ʹ-ACTGCCAGAAAGTGTGTCCC-3ʹ, R 5ʹ-CGGGCCTCGTTTTGAACATC-3ʹ; FGFR1, F 5ʹ-CCTCCTCCCTTCCCAAGTAA-3ʹ, R 5ʹ-GGACTGATACCCCAGCTCAG-3ʹ; GAPDH, F 5ʹ-CCACCCATGGCAAATTCCATGGCA-3ʹ, R 5ʹ-TCTAGACGGCAGGTCAGGTCCACC-3ʹ; miR-1294, F 5ʹ-TATGATCTCACCGAGTCCT-3ʹ, R 5ʹ-CACCTTCCTAATCCTCAGTT-3ʹ; U6, F 5ʹ-CTCGCTTCGGCAGCACA-3ʹ, R 5ʹ-AACGCTTCACGAATTTGCGT-3ʹ. All data were calculated by the 2−ΔΔCt method.
Cell Transfection
Saos-2 and SOSP-9607 cells (5 × 105) were seeded in 6-well plates and cultured for 12 h until cells reached 70% confluence. Circ_0000885 small interfering RNA (si-circ_0000885), miR-1294 mimic (miR-1294), miR-1294 inhibitor (anti-miR-1294), FGFR1 overexpression vector (pcDNA-FGFR1) and circ_0000885 lentiviral short hairpin RNA (sh-circ_0000885) or their negative controls (si-NC, miR-NC, anti-miR-NC, pcDNA-NC and sh-NC) were obtained from RiboBio (Guangzhou, China). According to the manufacture’s protocols, 50 ng of the above plasmids or oligonucleotides were transfected into Saos-2 and SOSP-9607 cells using 1 μL Lipofectamine 3000 (Invitrogen). After 48 h, the cells were harvested for the follow-up experiments.
Cell Proliferation Assay
3-(4,5-dimethyl-2-thiazolyl)-2, 5-diphenyltetrazolium bromide (MTT) assay and colony formation assay were used to evaluate the proliferation of Saos-2 and SOSP-9607 cells. For MTT assay, Saos-2 and SOSP-9607 cells were seeded in 96-well plates for 24 h after transfection. Then, MTT solution (Beyotime, Shanghai, China) was added into cells. After 4 h, cells were treated with dimethylsulfoxide (DMSO) and the absorbance at 490 nm was detected to assess the proliferation of cells. For colony formation assay, Saos-2 and SOSP-9607 cells were seeded in 6-well plates after transfection. Cell medium was changed every 2 days, and cells were cultured for 2 weeks. Finally, the cells were fixed with methanol and stained with crystal violet, and then the colony number of cells (>50 cells) was counted using a microscope.
Cell Cycle Distribution
After transfection, Saos-2 and SOSP-9607 cells were seeded in 6-well plates. Then, cells were digested with trypsin and collected into a centrifuge tube after cultured for 48 h. After fixation and RNase A (Thermo Fisher Scientific) treatment, the cells were stained with propidium iodide (PI; Amyjet, Wuhan, China) for 20 min. The cell cycle distribution was finally detected by flow cytometer (Thermo Fisher Scientific).
Transwell Assay
For cell migration, Saos-2 and SOSP-9607 cells were seeded in the upper chamber (Corning Inc., Corning, NY, USA) containing serum-free medium, while the lower chamber was filled with serum medium. For cell invasion, Matrigel (Corning Inc.) was pre-coated on the upper chamber, and other procedures were consistent with cell migration assay. After 24 h, the lower chamber cells were fixed and stained, and the number of migrated or invaded cells was counted.
Dual-Luciferase Reporter Assay
The wild-type (WT) or mutant-type (MUT) sequences of circ_0000885 or FGFR1 were cloned into the pGL3-basic vector (Promega, Madison, WI, USA) and built as circ_0000885-WT/MUT or FGFR1 3ʹUTR-WT/MUT reporter vector. The reporter vector and miR-1294 mimic or miR-NC were co-transfected into Saos-2 and SOSP-9607 cells using Lipofectamine 3000. Luciferase activity was assessed by the Dual-Luciferase Reporter Assay System (Promega) after 48 h transfection.
Western Blot (WB) Analysis
Saos-2 and SOSP-9607 cells were lysed using RIPA buffer (Beyotime) and the concentration was detected using BCA Kit (Beyotime). Then, the protein sample was isolated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) gel and transferred to polyvinylidene fluoride (PVDF) membranes (Thermo Fisher Scientific). After that, the membranes were blocked using skimmed milk, incubated with primary antibody FGFR1 (1:400, Boster, Wuhan, China) or GAPDH (1:400, Boster), and interacted with secondary antibody (1:1,000, Boster). The protein blots were determined using SuperSignal Chemiluminescent Substrates (Thermo Fisher Scientific).
OS Mice Models
BALB/c nude mice (5 weeks old) were obtained from the Guangdong provincial experimental animal center for medicine (Guangdong, China) and randomly divided into 2 experimental groups (n = 6 per/group). Animal procedures were approved by the Committee of Children’s Hospital of Anhui Medical University and performed in accordance with the guidelines of Animal Protection Law of the People’s Republic of China-2009. Saos-2 cells were stably transfected with sh-circ_0000885 or sh-NC. Next, transfected Saos-2 cells (5 × 106) were subcutaneously injected into nude mice. The length and width of the tumor were measured using a caliper from day 7 after injection, and then every 4 days until day 27. Then, mice were euthanized, and the tumors were collected for further experiments. Tumor weight was calculated using width2 × length/2.
Statistical Analysis
Data were analyzed using GraphPad Prism 5.0 software (GraphPad, La Jolla, CA, USA) and were shown as the mean ± standard deviation. Student’s t-test was used for the statistical differences between the two groups, and one-way analysis of variance was used for the differences among the multiple group, followed by Tukey post hoc test. P < 0.05 was regarded as statistically significant. All experiments were performed in triplicate.
Results
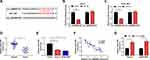
Circ_0000885 Had High Expression in OS Tissues and Cells
We first explored the expression status of circ_0000885 in both OS tissues and cells using qRT-PCR. The results revealed that circ_0000885 was markedly upregulated in OS tumor tissues compared with that in matched normal tissues (Figure 1A). The correlation between circ_0000885 expression and the clinical pathological characteristics of OS patients showed that high circ_0000885 expression was positively correlated with the TNM stage of OS patients (P < 0.05, Table 1). Next, circ_0000885 expression in OS cell lines (Saos-2 and SOSP-9607) was also higher than in hFOB cells (Figure 1B). These results suggested that circ_0000885 might play a vital role in OS.
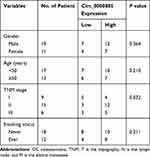 |
Table 1 Correlation Between Relative Circ_0000885 Expression and the Clinical Pathological Characteristics of OS Patients |
Knockdown of Circ_0000885 Decreased Proliferation, Arrested Cell Cycle, - Inhibited Migration and Invasion in OS Cells
To investigate the role of circ_0000885 on OS malignant phenotypes, we used si-circ_0000885 to silence the expression of endogenous circ_0000885 and confirmed the transfection efficiency of si-circ_0000885 by detecting the expression of circ_0000885 in Saos-2 and SOSP-9607 cells (Figure 2A). The effect of circ_0000885 silencing on OS cell proliferation was assessed via MTT assay and colony formation assay. And the results showed that circ_0000885 knockdown inhibited the OD values and colony numbers of Saos-2 and SOSP-9607 cells, indicating that the proliferation of OS cells could be suppressed by circ_0000885 silencing (Figure 2BD). Besides, through detecting the cell cycle distribution, we found that silenced circ_0000885 could induce cell cycle arrest in G0/G1 phase to reduce the number of Saos-2 and SOSP-9607 cells in S phase (Figure 2E and F). Further, the migration and invasion of Saos-2 and SOSP-9607 cells transfected with si-circ_0000885 were significantly reduced (Figure 2G and H). Therefore, we speculated that circ_0000885 might play a pro-cancer role in OS.
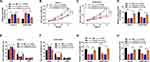
Circ_0000885 Directly Targeted MiR-1294 in OS
Then, we searched the potential miRNAs bound to circ_0000885 using the Starbase tool and found that miR-1294 could bind with circ_0000885 (Figure 3A). Dual-luciferase activity assay results suggested that miR-1294 mimic could remarkably reduce the luciferase activity of circ_0000885-WT reporter vector but not the circ_0000885-MUT reporter vector in Saos-2 and SOSP-9607 cells (Figure 3B and C). Moreover, we also detected the expression of miR-1294 in OS tissues and cells, and the results showed that miR-1294 was significantly down-regulated in OS tumor tissues compared with matched normal tissues (Figure 3D). Similarly, we found same expression trends in OS cells (Figure 3E). More importantly, we observed a negative correlation between miR-1294 and circ_0000885 expression in OS (Figure 3F). And miR-1294 expression could be increased by circ_0000885 knockdown in Saos-2 and SOSP-9607 cells (Figure 3G). Hence, our data indicated that miR-1294 could be sponged by circ_0000885 in OS.
Circ_0000885 Regulated the Progression of OS via Targeting MiR-1294
To investigate whether the regulation of circ_0000885 on OS progression was mediated by miR-1294, Saos-2 and SOSP-9607 cells were co-transfected with si-circ_0000885 and anti-miR-1294. The detection of the miR-1294 expression results showed that miR-1294 inhibitor could abolish the promotion effect of circ_0000885 knockdown on miR-1294 expression (Figure 4A). As exhibited in Figure 4BD, the inhibition effect of circ_0000885 silencing on the OD values and colony numbers of Saos-2 and SOSP-9607 cells could be partially reversed by miR-1294 inhibitor. At the same time, the increased number of G0/G1 phase of cells caused by circ_0000885 knockdown also could be recovered by miR-1294 inhibitor, which could be reflected by increasing the number of Saos-2 and SOSP-9607 cells in S phase (Figure 4E and F). As expected, the migration and invasion abilities of Saos-2 and SOSP-9607 cells inhibited by silenced circ_0000885 could be inverted by miR-1294 inhibitor (Figure 4G and H). These results revealed that miR-1294 participated in the regulation of circ_0000885 on the progression of OS.
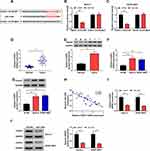
FGFR1 Was a Target of MiR-1294 in OS
Also, we used the Targetscan tool to predict the binding targets of miR-1294. The predicted target binding site between FGFR1 3ʹUTR and miR-1294 is shown in Figure 5A. Further, we found that miR-1294 overexpression could suppress the luciferase activity of FGFR1 3ʹUTR-WT reporter vector but had no significant effects on FGFR1 3ʹUTR-MUT reporter vector (Figure 5B and C). By detecting the mRNA and protein levels of FGFR1, we observed that FGFR1 had elevated expression in OS tissues and cells compared with matched normal tissues and hFOB cells, respectively (Figure 5DG). Besides, FGFR1 expression was negatively correlated with miR-1294 expression in OS, as demonstrated by Pearson correlation analysis (Figure 5H). To evaluate the effect of miR-1294 on FGFR1 expression, we measured FGFR1 expression in Saos-2 and SOSP-9607 cells transfected with miR-1294 mimic, and the results indicated that miR-1294 overexpression could remarkably repress the FGFR1 expression (Figure 5I and J). All data suggested that miR-1294 directly targeted FGFR1 in OS.
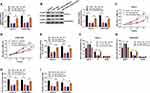
FGFR1 Was Involved in the Regulation of MiR-1294 on OS Progression
For confirming FGFR1 was targeted by miR-1294, we co-transfected with miR-1294 mimic and pcDNA-FGFR1 into Saos-2 and SOSP-9607 cells. Through detecting the mRNA and protein levels of FGFR1 in Saos-2 and SOSP-9607 cells, we discovered that pcDNA-FGFR1 restored the inhibition effect of miR-1294 mimic on FGFR1 expression (Figure 6A and B). The detection of the proliferation ability of Saos-2 and SOSP-9607 cells results indicated that overexpressed miR-1294 hindered the OD values and colony numbers of cells, while FGFR1 overexpression could reverse this effect (Figure 6CE). Meanwhile, the arresting effect of miR-1294 overexpression on the G0/G1 phase of the cell cycle also could be recovered by FGFR1 overexpression in Saos-2 and SOSP-9607 cells, which could also be confirmed by increasing the number of cells in S phase (Figure 6F and G). Besides, overexpressed FGFR1 partially inverted the suppression effect of miR-1294 mimic on the migration and invasion abilities of Saos-2 and SOSP-9607 cells (Figure 6H and I). So, we concluded that miR-1294 regulated the progression of OS by targeting FGFR1.
FGFR1 Expression Was Regulated by Circ_0000885 and MiR-1294
Furthermore, correlation analysis revealed that there was a positive correlation between circ_0000885 and FGFR1 expression in OS (Figure 7A). To further explore whether FGFR1 was regulated by circ_0000885, we tested the FGFR1 expression in Saos-2 and SOSP-9607 cells transfected with si-circ_0000885 and anti-miR-1294. As presented in Figure 7B, silenced circ_0000885 had a significant inhibitory effect on the expression of FGFR1, and this effect could be recovered by miR-1294 inhibitor. The results of the FGFR1 protein level were consistent with the mRNA level results (Figure 7C). Hence, we confirmed that circ_0000885 regulated the expression of FGFR1 through miR-1294.
Interference of Circ_0000885 Inhibited OS Tumor Growth in vivo
To identify the phenotypes of circ_0000885 silencing, we carried out the animal experiments using sh-circ_0000885. As could be seen from Figure 8A, the growth rate of tumor volume in the circ_0000885 knockdown group was significantly lower than that in the sh-NC group, and the tumor weight was markedly reduced in the sh-circ_0000885 group (Figure 8B). To confirm the knockdown efficiency of the sh-circ_0000885, we examined the expression of the circ_0000885 and found that circ_0000885 expression was indeed significantly inhibited in the sh-circ_0000885 group (Figure 8C). In addition, we also uncovered that compared with the sh-NC group, the expression of miR-1294 was markedly increased (Figure 8D), while the expression of FGFR1 was obviously decreased in the sh-circ_0000885 group (Figure 8E and F).
Discussion
In recent years, circRNA has been recognized by researchers as an important regulator and biomarker of cancer because of its ability to mediate the carcinogenesis of many cancers.24,25 For example, hsa_circRNA_103809 was considered to be a biomarker for lung cancer prognosis because of its ability to regulate cell proliferation, invasion, and mediate tumor formation.26 Kun-Peng et al reported that circPVT1 was associated with drug resistance in OS patients, suggesting that it might be a diagnostic and therapeutic biomarker for OS.27 Also, Nie et al showed that circ-NT5C2 might act as a potential prognosis biomarker for OS.28 Agreed with the results of Zhu et al,13 circ_0000885 had increased expression in OS in our study. More importantly, through the loss-of-function experiments, we obtained the results that circ_0000885 knockdown could inhibit the proliferation, cell cycle and metastasis of OS cells in vitro and OS tumor growth in vivo, which confirmed the necessity of circ_0000885 expression in the progression of OS. At the same time, in terms of molecular mechanism, we verified that circ_0000885 mediated OS progression through the regulation of FGFR1 expression by sponging miR-1294.
The role of miR-1294 in many cancers has been elucidated. Shi et al showed that the down-regulation of miR-1294 expression could be used as an indicator in the poor prognosis of gastric cancer.29 In addition to poor prognosis, the low expression of miR-1294 was also associated with the proliferation and cell cycle of epithelial ovarian cancer.30 Although there were few studies on miR-1294 in OS, Zhang et al found that the results of significantly lower expression of miR-1294 in OS also provided a reference for studying the role of miR-1294 in OS,17 which was also confirmed in our study. Moreover, in order to confirm the interaction between miR-1294 and circ_0000885, we conducted the rescue experiments. The partial reversal effect of miR-1294 inhibitor on si-circ_0000885 function proved that miR-1294 was one of the downstream targets of circ_0000885, and there might be other miRNAs regulated by circ_0000885, which needed further investigation. Also, the suppression effect of miR-1294 mimic on the proliferation, cell cycle and metastasis also indicated that miR-1294 could function as a tumor suppressor in OS.
Fe et al reported an abnormal amplification of FGFR1 in OS patients with poor response to chemotherapy, suggesting that its expression was associated with adverse reactions to chemotherapy.31 Also, previous studies had shown that FGFR1 inhibitors had been approved as anti-OS drugs, indicating that the presence of FGFR1 was important for OS progression.32,33 Here, we also revealed that FGFR1 was remarkably highly expressed in OS, and the partial reversal effect of its overexpression on miR-1294 mimic confirmed that it could interact with miR-1294 in OS. Besides, the inhibition effect of circ_0000885 silencing on FGFR1 expression in vivo and in vitro confirmed that FGFR1 expression was indirectly regulated by circ_0000885.
Conclusion
Overall, our study suggested that high circ_0000885 expression promoted OS progression, including proliferation, cell cycle, migration and invasion. In terms of mechanism, we demonstrated that circ_0000885 targeted miR-1294 to promote FGFR1 expression and thus regulate OS progression. Our results confirmed the importance of circ_0000885 as an oncogene in OS, which might provide new insights into treatment options for OS.
Abbreviations
OS, osteosarcoma; qRT-PCR, quantitative real-time polymerase chain reaction; FGFR1, fibroblast growth factor receptor 1; WB, Western blot; ncRNAs, non-coding RNAs; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; MTT, 3-(4,5-dimethyl-2-thiazolyl)-2,5-diphenyltetrazolium bromide; si, small interfering RNA; sh, lentiviral short hairpin RNA; NC, negative control.
Highlights
1. Circ_0000885 knockdown restrains proliferation, migration, invasion, and induces cell cycle arrest in OS.
2. Circ_0000885 can serve as a sponge of miR-1294 in OS.
3. MiR-1294 directly targets FGFR1 in OS.
Disclosure
The authors declare that they have no financial conflicts of interest.
References
1. Lin YH, Jewell BE, Gingold J, et al. Osteosarcoma: molecular Pathogenesis and iPSC modeling. Trends Mol Med. 2017;23(8):737–755. doi:10.1016/j.molmed.2017.06.004
2. Anderson ME. Update on survival in osteosarcoma. Orthop Clin North Am. 2016;47(1):283–292. doi:10.1016/j.ocl.2015.08.022
3. Biazzo A, De Paolis M. Multidisciplinary approach to osteosarcoma. Acta Orthop Belg. 2016;82(4):690–698.
4. Savant D, Kenan S, Kenan S, Kahn L. Extraskeletal osteosarcoma arising in myositis ossificans: a case report and review of the literature. Skeletal Radiol. 2017;46(8):1155–1161. doi:10.1007/s00256-017-2674-x
5. Luetke A, Meyers PA, Lewis I, Juergens H. Osteosarcoma treatment where do we stand? A state of the art review. Cancer Treat Rev. 2014;40(4):523–532. doi:10.1016/j.ctrv.2013.11.006
6. Ahmed G, Zamzam M, Kamel A, et al. Effect of timing of pulmonary metastasis occurrence on the outcome of metastasectomy in osteosarcoma patients. J Pediatr Surg. 2019;54(4):775–779. doi:10.1016/j.jpedsurg.2018.06.019
7. Quan G, Li J. Circular RNAs: biogenesis, expression and their potential roles in reproduction. J Ovarian Res. 2018;11(1):9. doi:10.1186/s13048-018-0381-4
8. Smith CM, Catchpoole D, Hutvagner G. Non-coding RNAs in pediatric solid tumors. Front Genet. 2019;10:798. doi:10.3389/fgene.2019.00798
9. Wang C, Ren M, Zhao X, Wang A, Wang J. Emerging roles of circular RNAs in osteosarcoma. Med Sci Monit. 2018;24:7043–7050. doi:10.12659/MSM.912092
10. Zhang Z, Pu F, Wang B, Wu Q, Liu J, Shao Z. Hsa_circ_0000285 functions as a competitive endogenous RNA to promote osteosarcoma progression by sponging hsa-miRNA-599. Gene Ther. 2019.
11. Wang L, Wang P, Su X, Zhao B. Circ_0001658 promotes the proliferation and metastasis of osteosarcoma cells via regulating miR-382-5p/YB-1 axis. Cell Biochem Funct. 2019.
12. Li XQ, Song JY, Lv W, Zhang D, Wu JZ. Circular circ_0000885 promotes hepatocellular carcinoma proliferation by epigenetically upregulating Caprin1. Eur Rev Med Pharmacol Sci. 2019;23(18):7848–7854. doi:10.26355/eurrev_201909_18994
13. Zhu K, Niu L, Wang J, et al. Circular RNA hsa_circ_0000885 levels are increased in tissue and serum samples from patients with osteosarcoma. Med Sci Monit. 2019;25:1499–1505. doi:10.12659/MSM.914899
14. Pan W, Pang LJ, Cai HL, Wu Y, Zhang W, Fang JC. MiR-1294 acts as a tumor suppressor in clear cell renal cell carcinoma through targeting HOXA6. Eur Rev Med Pharmacol Sci. 2019;23(9):3719–3725. doi:10.26355/eurrev_201905_17797
15. Wang Z, Yan J, Zou T, Gao H. MicroRNA-1294 inhibited oral squamous cell carcinoma growth by targeting c-Myc. Oncol Lett. 2018;16(2):2243–2250. doi:10.3892/ol.2018.8967
16. Wang J, Li J, Wang H, Lv L, Sun J. Overexpression of circ_0005198 sponges miR-1294 to regulate cell proliferation, apoptosis, migration, and invasion in glioma. J Cell Biochem. 2019;120(9):15538–15545. doi:10.1002/jcb.28820
17. Zhang ZF, Li GR, Cao CN, Xu Q, Wang GD, Jiang XF. MicroRNA-1294 targets HOXA9 and has a tumor suppressive role in osteosarcoma. Eur Rev Med Pharmacol Sci. 2018;22(24):8582–8588. doi:10.26355/eurrev_201812_16621
18. Turner N, Grose R. Fibroblast growth factor signalling: from development to cancer. Nat Rev Cancer. 2010;10(2):116–129. doi:10.1038/nrc2780
19. Ma F, Zhang L, Ma L, Zhang Y, Zhang J, Guo B. MiR-361-5p inhibits glycolytic metabolism, proliferation and invasion of breast cancer by targeting FGFR1 and MMP-1. J Exp Clin Cancer Res. 2017;36(1):158. doi:10.1186/s13046-017-0630-1
20. Zhou W, Zhu Y, Chen S, Xu R, Wang K. Fibroblast growth factor receptor 1 promotes MG63 cell proliferation and is associated with increased expression of cyclin-dependent kinase 1 in osteosarcoma. Mol Med Rep. 2016;13(1):713–719. doi:10.3892/mmr.2015.4597
21. Weekes D, Kashima TG, Zandueta C, et al. Regulation of osteosarcoma cell lung metastasis by the c-Fos/AP-1 target FGFR1. Oncogene. 2016;35(22):2852–2861. doi:10.1038/onc.2015.344
22. Panda AC. Circular RNAs act as miRNA sponges. Adv Exp Med Biol. 2018;1087:67–79.
23. Su Q, Lv X. Revealing new landscape of cardiovascular disease through circular RNA-miRNA-mRNA axis. Genomics. 2019.
24. Zhao ZJ, Shen J. Circular RNA participates in the carcinogenesis and the malignant behavior of cancer. RNA Biol. 2017;14(5):514–521. doi:10.1080/15476286.2015.1122162
25. Chen B, Huang S. Circular RNA: an emerging non-coding RNA as a regulator and biomarker in cancer. Cancer Lett. 2018;418:41–50. doi:10.1016/j.canlet.2018.01.011
26. Liu W, Ma W, Yuan Y, Zhang Y, Sun S. Circular RNA hsa_circRNA_103809 promotes lung cancer progression via facilitating ZNF121-dependent MYC expression by sequestering miR-4302. Biochem Biophys Res Commun. 2018;500(4):846–851. doi:10.1016/j.bbrc.2018.04.172
27. Kun-Peng Z, Xiao-Long M, Chun-Lin Z. Overexpressed circPVT1, a potential new circular RNA biomarker, contributes to doxorubicin and cisplatin resistance of osteosarcoma cells by regulating ABCB1. Int J Biol Sci. 2018;14(3):321–330. doi:10.7150/ijbs.24360
28. Nie WB, Zhao LM, Guo R, Wang MX, Ye FG. Circular RNA circ-NT5C2 acts as a potential novel biomarker for prognosis of osteosarcoma. Eur Rev Med Pharmacol Sci. 2018;22(19):6239–6244. doi:10.26355/eurrev_201810_16030
29. Shi YX, Ye BL, Hu BR, Ruan XJ. Expression of miR-1294 is downregulated and predicts a poor prognosis in gastric cancer. Eur Rev Med Pharmacol Sci. 2018;22(17):5525–5530. doi:10.26355/eurrev_201809_15813
30. Guo TY, Xu HY, Chen WJ, Wu MX, Dai X. Downregulation of miR-1294 associates with prognosis and tumor progression in epithelial ovarian cancer. Eur Rev Med Pharmacol Sci. 2018;22(22):7646–7652. doi:10.26355/eurrev_201811_16381
31. Fernanda Amary M, Ye H, Berisha F, et al. Fibroblastic growth factor receptor 1 amplification in osteosarcoma is associated with poor response to neo-adjuvant chemotherapy. Cancer Med. 2014;3(4):980–987. doi:10.1002/cam4.268
32. Kallus S, Englinger B, Senkiv J, et al. Nanoformulations of anticancer FGFR inhibitors with improved therapeutic index. Nanomedicine. 2018;14(8):2632–2643. doi:10.1016/j.nano.2018.08.001
33. Jurek PM, Zablocki K, Wasko U, Mazurek MP, Otlewski J, Jelen F. Anti-FGFR1 aptamer-tagged superparamagnetic conjugates for anticancer hyperthermia therapy. Int J Nanomedicine. 2017;12:2941–2950. doi:10.2147/IJN.S125231
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.