Back to Journals » Cancer Management and Research » Volume 12
Circ_0000517 Contributes to Hepatocellular Carcinoma Progression by Upregulating TXNDC5 via Sponging miR-1296-5p
Authors Zang H, Li Y, Zhang X, Huang G
Received 28 December 2019
Accepted for publication 17 April 2020
Published 14 May 2020 Volume 2020:12 Pages 3457—3468
DOI https://doi.org/10.2147/CMAR.S244024
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Antonella D'Anneo
Hongliang Zang, Yuhui Li, Xue Zhang, Guomin Huang
Department of General Surgery, China-Japan Union Hospital of Jilin University, Jilin, Changchun 130012, People’s Republic of China
Correspondence: Guomin Huang
Department of General Surgery, China-Japan Union Hospital of Jilin University, No. 829 Xinmin Street, Jilin, Changchun 130012, People’s Republic of China
Tel +86-431-84997753
Email [email protected]
Background: Circular RNAs (circRNAs) function as essential regulators in diverse human cancers, including hepatocellular carcinoma (HCC). However, the function of circ_0000517 in HCC was unknown. We aimed to explore the roles and mechanisms of circ_0000517 in HCC.
Materials and Methods: The levels of circ_0000517, RPPH1 mRNA and microRNA-1296-5p (miR-1296-5p) were measured using quantitative real-time polymerase chain reaction (qRT-PCR). The characteristics of circ_0000517 were explored by RNase R digestion and actinomycin D assays. Cell proliferation was evaluated by Cell Counting Kit-8 (CCK-8) and colony formation assays. Cell cycle process and cell apoptosis were analyzed by flow cytometry analysis. The function of circ_0000517 in vivo was explored by a murine xenograft model. The association between miR-1296-5p and circ_0000517 or thioredoxin domain containing 5 (TXNDC5) was determined by dual-luciferase reporter assay and RNA immunoprecipitation (RIP) assay. The protein level of TXNDC5 was detected by Western blot assay.
Results: Circ_0000517 was upregulated in HCC tissues and cells. Silencing of circ_0000517 suppressed HCC cell viability and colony formation and promoted cell cycle arrest and apoptosis in vitro and hampered tumor growth in vivo. MiR-1296-5p was a target of circ_0000517 and the effects of circ_0000517 silencing on HCC cell viability, cell cycle, colony formation and apoptosis were abolished by miR-1296-5p inhibition. TXNDC5 functioned as a target gene of miR-1296-5p, and the inhibitory effect of miR-1296-5p on HCC cell progression was rescued by TXNDC5 overexpression. Moreover, circ_0000517 promoted TXNDC5 expression via targeting miR-1296-5p.
Conclusion: Circ_0000517 accelerated HCC progression by upregulating TXNDC5 through sponging miR-1296-5p.
Keywords: HCC, circ_0000517, miR-1296-5p, TXNDC5
Highlights
- High expression of circ_0000517 and TXNDC5 and low expression of miR-1296-5p are observed in HCC tissues and cells.
- Circ_0000517 knockdown suppresses HCC cell viability, cell cycle process and colony formation and promotes apoptosis in vitro and blocks tumorigenesis in vivo.
- MiR-1296-5p is target of circ_0000517 and circ_0000517 knockdown represses HCC cell progression by targeting miR-1296-5p.
- MiR-1296-5p overexpression suppresses HCC cell progression via downregulating TXNDC5.
- Circ_0000517 promotes TXNDC5 expression by sponging miR-1296-5p in HCC cells.
Introduction
As a common primary liver cancer, hepatocellular carcinoma (HCC) has been the fourth major cause of cancer-related death in the world.1,2 About three-quarters of HCC are caused by chronic HBV and HCV infections.3 Unfortunately, the survival rate of HCC patients is still unfavorable due to high metastasis and recurrence, though the diagnosis and treatment strategies have been improved.4,5 Thus, it is imperative to explore the mechanisms and new effective therapeutic targets for HCC.
Recently, several non-coding RNAs (ncRNAs), such as circular RNAs (circRNAs) and microRNAs (miRNAs), have been documented to play crucial roles in human cancers.6,7 CircRNAs possess closed-loop structures without 5ʹ cap structure and 3ʹ poly (A) structure and have been proved to be widespread in various eukaryotes.8 Multiple circRNAs have been identified to be involved in the development of HCC. For example, circ_0091570 knockdown facilitated HCC cell growth and motility and repressed apoptosis.9 Circ_0101432 deficiency decelerated HCC progression via suppressing tumor cell growth and invasion and inducing apoptosis.10 Circ_0016788 contributed to HCC cell viability and motility in vitro and tumorigenesis in vivo.11 These reports suggested that circRNAs played dual roles in HCC. Circ_0000517 has been demonstrated to be abnormally high expressed in HCC and related to poor prognosis.12 However, the exact roles and possible molecular mechanisms of circ_0000517 remain unclear.
MiRNAs, another type of ncRNAs (about 22 nucleotides), act as vital regulators in diverse biological processes at the posttranscriptional level through recognizing the 3ʹ untranslated region (3ʹUTR) of target mRNA.13,14 It has been increasingly validated that the aberrant expression of miRNAs plays vital roles in HCC. For example, Lu et al claimed that miR-1470 facilitated cell growth and hampered apoptosis in HCC via interacting with ALX4.15 Yao et al declared that miR-3194-3p was reduced in HCC and its elevation repressed the motility and epithelial–mesenchymal transition (EMT) of HCC by binding to BCL9.16 Moreover, miR-1296 could lessen HCC metastasis and EMT via targeting the SRPK1/AKT pathway.17 However, the underlying mechanisms of miR-1296-5p remain far insufficient. Thioredoxin domain-containing 5 (TXNDC5) belongs to the protein disulfide isomerase family and has been verified to be a positive regulator in diverse cancers, including HCC.18,19 But whether TXNDC5 can be targeted by miR-1296-5p has not been elucidated yet.
In this research, we firstly identified the expression of circ_0000517 in HCC. Then, we explored the possible biological functions and mechanisms of circ_0000517 in HCC cell progression.
Materials and Methods
Tissues Collection
After the work was permitted by the Ethics Committee of China–Japan Union Hospital of Jilin University and written informed consents were signed by all participants, 60 HCC tissues and adjacent normal tissues were harvested from patients diagnosed with HCC at China–Japan Union Hospital of Jilin University. None of the patients received any treatment prior to surgery in our study. The collected samples were saved at −80°C until use.
Cell Culture
Normal liver cells (THLE-2) were bought from the American Type Culture Collection (ATCC, Manassas, VA, USA) and HCC cells (SNU-387 and Huh7) were bought from the Type Culture Collection of the Chinese Academy of Sciences (Shanghai, China). And Dulbecco’s modified Eagle’s medium (DMEM; Invitrogen, Carlsbad, CA, USA) containing 10% fetal bovine serum (FBS; Invitrogen) and 1% penicillin-streptomycin (Invitrogen) was used to culture the cells. The medium was placed at 37°C in an incubator with 5% CO2.
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
The tissues and cells were lysed in TRIzol reagent (Invitrogen) to isolate total RNA. Then, the RNAs were determined on a NanoDrop 2000c spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). Next, circ_0000517 was reversely transcribed into complementary DNA (cDNA) with the PrimeScript™ RT reagent Kit (Takara, Dalian, China) and Random primers or Oligo (dt)18 primers. MiRNA and mRNA were reversely transcribed into cDNAs with mirVanaTM qRT-PCR miRNA Detection Kit (Ambion, Austin, TX, USA) and PrimeScript™ RT reagent Kit (Takara), respectively. QRT-PCR was performed using SYBR Premix Ex Taq II (Takara). The expression was calculated using the 2−ΔΔCt method and glyceraldehyde 3-phosphate dehydrogenase (GAPDH) or U6 was utilized as an internal reference. The primers were: circ_0000517: (F: 5ʹ-GGGAGGTGAGTTCCCAGAGA-3ʹ and R: 5ʹ-TGGCCCTAGTCTCAGACCTC-3ʹ); RPPH1: (F: 5ʹ-GTCACTCCACTCCCATGTCC-3ʹ and R: 5ʹ-CAGCCATTGAACTCACTTCG-3ʹ); miR-1296-5p: (F: 5ʹ-GTTAGGGCCCTGGCTCC-3ʹ and R: 5ʹ-CAGTGCGTGTCGTGGAGT-3ʹ); TXNDC5: (F: 5ʹ-TCACTGAGGGAGTACGTGGA-3ʹ and R: 5ʹ-AGCAGTGCAGTCTACTTCGG-3ʹ); GAPDH: (F: 5ʹ-ACAACTTTGGTATCGTGGAAGG-3ʹ and R: 5ʹ-GCCATCACGCCACAGTTTC-3ʹ); U6: (F: 5ʹ-CTCGCTTCGGCAGCACATATACTA-3ʹ and R: 5ʹ-ACGAATTTGCGTGTCATCCTTGCG-3ʹ).
RNase R Digestion Assay
Twenty μg total RNA was incubated with or without RNase R (20 mg/mL; Solarbio, Beijing, China) for 15 min at 37°C. Then, qRT-PCR was utilized to examine the levels of circ_0000517 and linear RPPH1 mRNA.
Actinomycin D Assay
SNU-387 and Huh7 cells were placed into 24-well plates and incubated overnight. Then, cells were treated with 2 μg/mL Actinomycin D (Abcam, Cambridge, MA, USA) to block transcription for 0 h, 4 h, 8 h, 12 h and 24 h. Finally, the cells were harvested and qRT-PCR was employed to measure circ_0000517 and linear RPPH1 mRNA levels in cells.
Cell Transfection
Small interfering RNA targeting circ_0000517 (si-circ_0000517) and its control (si-NC), short hairpin RNA targeting circ_0000517 (sh-circ_0000517) and its control (sh-NC), miR-1296-5p mimic and control mimic (miRNA NC), miR-1296-5p inhibitor and inhibitor NC, the overexpression vector of TXNDC5 (pc-TXNDC5) and its control (pc-NC), the overexpression vector of circ_0000517 (circ_0000517) and its control (pLVX) were synthesized by GeneCopoeia (Guangzhou, China). Cell transfection was conducted using Lipofectamine 2000 (Invitrogen).
Cell Counting Kit-8 (CCK-8) Assay
The viability of SNU-387 and Huh7 cells was evaluated through CCK-8 assay kit (Beyotime, Shanghai, China) after indicated transfection. Firstly, cells were seeded into 96-well plates (2×103 cells/well) and incubated for 72 h. Next, CCK-8 (5 mg/mL; Beyotime) was added to each well following incubation for an additional 4 h. At last, the optical density value was measured at 450 nm with a microplate reader (Bio-Rad, Hercules, CA, USA).
Flow Cytometry Analysis
To analyze the cell cycle, transfected SNU-387 and Huh7 cells were collected and fixed with ice-cold 75% ethanol at 4°C overnight. Next, cells were washed with PBS (Solarbio), resuspended in binding buffer and then incubated with propidium iodide (PI; Beyotime) and RNase (Solarbio) at room temperature for 30 min. Thereafter, the cell cycle was analyzed with a flow cytometer (BD Biosciences, San Jose, CA, USA).
For the detection of cell apoptosis, transfected SNU-387 and Huh7 cells were collected, washed and resuspended. Then, 5 μL Annexin V-fluorescein isothiocyanate (Beyotime) and 10 μL PI (Beyotime) were added into the cells and maintained in the dark for 15 min. The apoptosis rate was examined using a flow cytometer (BD Biosciences).
Colony Formation Assay
Transfected SNU-387 and Huh7 cells were placed into 12-well plates (600 cells/well) and cultured in a complete medium at a condition of 37°C and 5% CO2. About 2 weeks later, cell clones were washed with PBS (Solarbio) and stained with 0.1% crystal violet (Solarbio). Next, the clones were imaged and counted by a microscope (Olympus, Tokyo, Japan).
Murine Xenograft Model
Approximately 5×105 Huh7 cells transfected with lentivirus-mediated sh-circ_0000517 or sh-NC were subcutaneously injected into the BALB/c nude mice (6-week-old; Shanghai Laboratory Animal Center, Shanghai, China). Then, tumor length (L) and width (W) were monitored weekly and tumor volume was calculated using the formula: (L×W2)/2. Tumors were harvested and weighed after inoculation for 4 weeks. The harvested samples were saved at −80°C for qRT-PCR assay. The animal experiments were conducted after approval was obtained from the Ethics Committee of Animal Research of China–Japan Union Hospital of Jilin University. All experimental procedures involving animals were conducted in accordance with the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health and were performed according to the institutional ethical guidelines for animal experiments.
RNA Immunoprecipitation (RIP) Assay
RIP assay was conducted using Magna RIPTM RNA Binding Protein Immunoprecipitation Kit (Millipore, Bedford, MA, USA). Transfected SNU-387 and Huh7 cells were lysed in RIP buffer and cell lysates were maintained with magnetic beads conjugated with Anti-Argonaute2 (Anti-Ago2; Abcam) or Anti-immunoglobulin G (Anti-IgG; Abcam). Then, the samples were interacted with proteinase K (Solarbio) and immunoprecipitated RNAs were isolated. The RNAs were extracted and the levels of circ_0000517 and miR-1296-5p were measured via qRT-PCR.
Subcellular Fraction Assay
The separation of nuclear and cytosolic fractions was carried out with the PARIS Kit (Thermo Fisher Scientific) based on the protocols. The expression levels of GAPDH, U6, circ_0000517 and miR-1296-5p in the nuclear and cytoplasm of SNU-387 and Huh7 cells were measured by qRT-PCR. GAPDH and U6 were utilized as a control of cytoplasm transcript and nuclear transcript, respectively.
Dual-Luciferase Reporter Assay
Circ_0000517 or TXNDC5 3ʹTUR fragments including the predicted wild-type or mutant miR-1296-5p complementary sequences were synthesized and inserted into pGL3 plasmid (Promega, Madison, WI, USA), and the luciferase plasmids were named as WT-circ_0000517, MUT-circ_0000517, WT-TXNDC5-3ʹUTR and MUT-TXNDC5-3ʹUTR, respectively. Then, SNU-387 and Huh7 cells were transfected with miRNA NC or miR-1296-5p mimic together with the corresponding vector. After 48 h, firefly or renilla activity was examined by using the Dual-Luciferase Reporter Assay Kit (Promega).
Western Blot Assay
Total protein in tissues and cells were extracted with RIPA buffer (Beyotime) and determined by a BCA Protein Quantification Kit (Vazyme, Nanjing, China). Then, 30 μg proteins were separated by 10% sodium dodecyl sulfonate-polyacrylamide gel (SDS-PAGE; Solarbio) and transferred onto polyvinylidene difluoride membranes (Millipore). Next, the proteins were blocked in skim milk at room temperature for 1 h and incubated with primary antibody TXNDC5 (ab13820; Abcam) or GAPDH (ab9485; Abcam) at 4°C overnight. Following incubation with indicated secondary antibody (ab205719; Abcam) at room temperature for 2 h, the protein bands were visualized with an ECL Western blot kit (Beyotime).
Statistical Analysis
Each experiment in our study was conducted in triplicate. The data were estimated using software GraphPad Prism 7 (GraphPad Inc., La Jolla, CA, USA) and presented as mean ± standard deviation (SD). Statistical differences were analyzed by Student’s t-test or one-way analysis of variance (ANOVA). It was defined to be significant if P<0.05.
Results
Circ_0000517 Was Upregulated in HCC Tissues and Cells
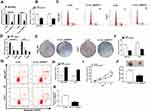
To explore the potential role of circ_0000517, the expression of circ_0000517 in HCC tissues and adjacent normal tissues was firstly tested by qRT-PCR assay. The data showed that circ_0000517 was highly expressed in HCC tissues in reference to normal tissues (Figure 1A). Moreover, we found that circ_0000517 expression in the patients with worse tumor-node-metastasis (TNM) stage was higher than in the better TNM stage (Figure 1B). In the meanwhile, we determined the expression level of circ_0000517 in HCC cells (SNU-387 and Huh7) and normal cells (THLE-2). The results of qRT-PCR exhibited that circ_0000517 was conspicuously elevated in HCC cells compared to THLE cells (Figure 1C). Next, the circular characteristics of circ_0000517 were confirmed by RNase R assay and Actinomycin D assay. The results of RNase R digestion assay exhibited that circ_0000517 was resistant to RNase R treatment in SNU-387 and Huh7 cells, while the level of its corresponding linear species (RPPH1 mRNA) was markedly reduced by RNase R digestion (Figure 1D). Actinomycin D assay indicated that circ_0000517 had a longer half-life compared to RPPH1 mRNA in SNU-387 and Huh7 cells (Figure 1E). In addition, total RNA in SNU-387 and Huh7 cells was reversely transcribed into cDNA with Random primers or Oligo (dt)18 primers and the qRT-PCR assay was conducted to measure the expression levels of circ_0000517 and linear RPPH1 mRNA. As we observed in (Figure 1F), Oligo (dt)18 primers reversely transcribed cDNA could barely amplify circ_0000517 when compared with Random primers reversely transcribed cDNA. Circ_0000517 was located in chromosome 14 and transcribed from the exon1 of RPPH1 gene (Figure 1G). All these data indicated that circ_0000517 was upregulated in HCC and more stable than linear RPPH1 mRNA.
Circ_0000517 Knockdown Suppressed Cell Viability, Cell Cycle Process and Cell Colony Formation and Promoted Apoptosis in HCC Cells in vitro and Blocked Tumor Growth in vivo
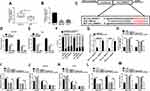
In order to explore the exact functions of circ_0000517 in HCC progression, loss-of-function experiments were carried out by transfecting si-circ_0000517 or si-NC into SNU-387 and Huh7 cells. As presented in (Figure 2A), si-circ_0000517 transfection remarkably decreased circ_0000517 expression but did not affect linear RPPH1 mRNA expression in SNU-387 and Huh7 cells. The results of CCK-8 assay showed that compared to si-NC control groups, the viability of SNU-387 and Huh7 cells was evidently inhibited in si-circ_0000517 transfected groups (Figure 2B). Flow cytometry analysis indicated that the percentage of cells in G1/G0 phase was increased and the percentage of cells in S phase was decreased in SNU-387 and Huh7 cells after circ_0000517 knockdown compared to control group (Figure 2C and D). Colony formation assay displayed that circ_0000517 silencing markedly repressed the colony numbers of SNU-387 and Huh7 cells compared to si-NC control groups (Figure 2E and F). Compared to control groups, cell apoptosis in SNU-387 and Huh7 cells was remarkably promoted by circ_0000517 deficiency (Figure 2G and H). Subsequently, to investigate the effect of circ_0000517 on tumor growth in vivo, we constructed a murine xenograft model by injecting sh-NC or sh-circ_0000517 transfected Huh7 cells into the mice. Tumor volume was examined every 7 days and tumor weight was measured after 28 days of inoculation. The results implied that tumor volume and weight were distinctly blocked by circ_0000517 deficiency compared to control groups (Figure 2I and J). Moreover, we measured the expression level of circ_0000517 in the collected tumors and found that circ_0000517 was evidently reduced in the tumor samples of sh-circ_0000517 groups compared to sh-NC group (Figure 2K). To sum up, circ_0000517 could suppress HCC cell progression in vitro and tumorigenesis in vivo.
Circ_0000517 Regulated HCC Cell Viability, Cell Cycle Process, Colony Formation and Apoptosis by Targeting miR-1296-5p
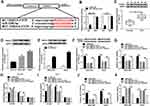
Additionally, we found that miR-1296-5p was weakly expressed in HCC tissues and cells relative to corresponding normal tissues and cells (Figure 3A and B). Thus, we wondered if there was a targeting relationship between circ_0000517 and miR-1296-5p. By searching software starBase 2.0, miR-1296-5p was found to contain the potential binding sites of circ_0000517 (Figure 3C). RIP assay displayed that circ_0000517 and miR-1296-5p were all notably enriched in SNU-387 and Huh7 cells after Anti-Ago2 RIP compared to Anti-IgG control group (Figure 3D and E). The data of the subcellular fraction assay presented that circ_0000517 and miR-1296-5p were mainly enriched in the cytoplasm of SNU-387 and Huh7 cells, suggesting the possibility of circ_0000517 and miR-1296-5p combination in space (Figure 3F). As illustrated by dual-luciferase reporter assay, the luciferase activity in SNU-387 and Huh7 cells co-transfected with miR-1296-5p mimic and WT-circ_0000517 was obviously inhibited compared to that in miRNA NC and WT-circ_0000517 co-transfected cells, while the luciferase activity was not changed in MUT-circ_0000517 groups, indicating the combination between circ_0000517 and miR-1296-5p (Figure 3G). Thereafter, SNU-387 and Huh7 cells were divided into four groups: si-NC, si-circ_0000517, si-circ_0000517+inhibitor NC and si-circ_0000517+miR-1296-5p inhibitor. As we observed in (Figure 3H), circ_0000517 knockdown markedly increased the expression of miR-1296-5p in SNU-387 and Huh7 cells, whereas miR-1296-5p inhibitor transfection partially reversed the increase. Moreover, we found that the effects of circ_0000517 deficiency on cell viability, cell cycle process, colony formation ability and cell apoptosis were remarkably abrogated following miR-1296-5p inhibition in SNU-387 and Huh7 cells (Figure 3I–M). All these data demonstrated that circ_0000517 repressed HCC cell progression by binding to miR-1296-5p.
MiR-1296-5p Inhibited HCC Cell Progression by Interacting with TXNDC5
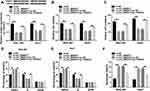
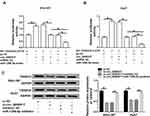
To further explore the potential regulatory mechanism of circ_0000517 in HCC, we searched starBase 2.0 and found TXNDC5 might be a target gene of miR-1296-5p (Figure 4A). Dual-luciferase reporter assay indicated that the co-transfection of WT-TXNDC5-3ʹUTR and miR-1296-5p mimic led to a distinct suppression in the luciferase activity in SNU-387 and Huh7 cells compared to WT-TXNDC5-3ʹUTR and miRNA NC co-transfected groups, while no change was observed in MUT-TXNDC5-3ʹUTR groups, confirming the combination between miR-1296-5p and TXNDC5 (Figure 4B). Next, we examined the protein level of TXNDC5 in HCC tissues and cells via Western blot assay. As expected, TXNDC5 protein level was drastically raised in HCC tissues and cells relative to normal tissues and cells (Figure 4C and D). Then, we knocked known TXNDC5 expression by transfecting si-TXNDC5 into SNU-387 and Huh7 cells. We found that TXNDC5 knockdown suppressed cell viability, colony formation and cell cycle process and facilitated apoptosis in SNU-387 and Huh7 cells (Figure S1). Moreover, we observed that TXNDC5 expression was elevated in SNU-387 and Huh7 cells following pc-TXNDC5 transfection compared to pc-NC groups (Figure 4E). Subsequently, SNU-387 and Huh7 cells were transfected with miRNA NC, miR-1296-5p mimic, miR-1296-5p mimic+pc-NC or miR-1296-5p mimic+pc-TXNDC5 to explore the relationship between miR-1296-5p and TXNDC5. As shown in (Figure 4F), miR-1296-5p mimic transfection markedly reduced the protein level of TXNDC5 in SNU-387 and Huh7 cells, while pc-TXNDC5 transfection partly reversed the reduction. CCK-8 assay indicated that miR-1296-5p resulted in an obvious suppression in the viability of SNU-387 and Huh7 cells, while TXNDC5 overexpression abolished the effect (Figure 4G). Flow cytometry analysis exhibited that the cell cycle was arrested by miR-1296-5p in SNU-387 and Huh7 cells, while the impact was ameliorated following the elevation of TXNDC5 (Figure 4H and I). The colony formation ability of SNU-387 and Huh7 cells was strikingly repressed by miR-1296-5p, but TXNDC5 overexpression partly abolished the repression, as demonstrated by colony formation assay (Figure 4J). Moreover, TXNDC5 overexpression effectively attenuated the promotional effect of miR-1296-5p on cell apoptosis in SNU-387 and Huh7 cells (Figure 4K). Collectively, TXNDC5 overexpression rescued the inhibitory impact of miR-1296-5p on HCC cell progression.
Upregulation of TXNDC5 Abrogated the Effects of Circ_0000517 Silencing on Cell Viability, Cell Colony Formation, Cell Cycle Process and Apoptosis in HCC Cells
To further investigate the relationship between circ_0000517 and TXNDC5 in HCC cells, SNU-387 and Huh7 cells were assigned to si-NC, si-circ_0000517, si-circ_0000517+pc-NC and si-circ_0000517+pc-TXNDC5 groups. The analysis of TXNDC5 protein level showed that circ_0000517 deficiency triggered a marked downregulation of TXNDC5 level in SNU-387 and Huh7 cells, whereas this effect was abolished following TXNDC5 overexpression (Figure 5A). Moreover, we found that circ_0000517 knockdown evidently repressed cell viability and cell colony formation and induced cell cycle arrest and cell apoptosis in SNU-387 and Huh7 cells, while TXNDC5 overexpression effectively weakened these impacts (Figure 5B–F). The outcomes implicated that circ_0000517 knockdown impeded HCC cell progression by downregulating TXNDC5 expression.
Circ_0000517 Positively Regulated TXNDC5 Expression by Targeting miR-1296
In order to explore the association among circ_0000517, miR-1296-5p and TXNDC5, luciferase reporter vector WT-TXNDC5-3ʹUTR together with indicated vectors or miRNA mimics were transfected into SNU-387 and Huh7 cells and then the luciferase activity was examined. The data showed that circ_0000517 overexpression apparently increased the luciferase activity, indicating TXNDC5 level was increased; however, miR-1296-5p mimic transfection reversed the increase of the luciferase activity. Moreover, we found the luciferase activity in SNU-387 and Huh7 cells transfected with WT-TXNDC5-3ʹUTR+control vector of circ_0000517 (pLVX)+miR-1296-5p mimic was reduced compared to cells transfected with WT-TXNDC5-3ʹUTR+overexpression vector of circ_0000517 (circ_0000517)+miR-1296-5p mimic or WT-TXNDC5-3ʹUTR+ control vector of circ_0000517 (pLVX)+miRNA NC (Figure 6A and B). Additionally, Western blot assay indicated that circ_0000517 knockdown strikingly decreased the protein level of TXNDC5 in SNU-387 and Huh7 cells, while miR-1296-5p deletion restored the effect (Figure 6C). The above data suggested that circ_0000517 could upregulate TXNDC5 expression via targeting miR-1296-5p in HCC cells.
Discussion
As more and more circRNAs were discovered, the effects of circRNAs in cancers gradually attracted researchers’ attention. Herein, we mainly investigated the functional roles of circ_0000517 in HCC. We observed that circ_0000517 was elevated in HCC. Circ_0000517 silencing impeded HCC cell viability, cell cycle process and colony formation and facilitated apoptosis. Moreover, a novel regulatory network circ_0000517/miR-1296-5p/TXNDC5 axis was established in HCC.
It is acknowledged that various circRNAs are dysregulated and closely related to HCC development.20 Yao et al suggested that circ_0001955 level was raised in HCC and circ_0001955 deficiency hampered HCC cell proliferation and colony formation in vivo and tumorigenesis in vivo.21 Li et al revealed that circ_0085616 was elevated in HCC and accelerated cell growth, motility and cell cycle process and hampered apoptosis in HCC cells.22 Though Wang et al suggested that circ_0000517 was conspicuously increased in HCC,12 the cellular functions of circ_0000517 have not been identified. In this research, a high expression of circ_0000517 in HCC tissues and cells was observed. By loss-of-function experiments, we demonstrated that circ_0000517 knockdown led to noteworthy repression in cell viability, colony number and cell cycle process and a marked enhancement in cell apoptosis in HCC cells in vitro. Additionally, circ_0000517 knockdown blocked tumorigenesis in vivo. All these data indicated that circ_0000517 played a tumorigenic role in HCC.
Accumulating evidence suggests that circRNAs can act as competitive endogenous RNAs to sponge miRNAs and then alter their functions.23 Using bioinformatic software, dual-luciferase reporter assay and RIP assay, we proved that miR-1296-5p was a target of circ_0000517. We observed that miR-1296-5p was weakly expressed in HCC tissues and cells. MiR-1296-5p overexpression impeded HCC cell growth and enhanced apoptosis. Moreover, miR-1296-5p deletion rescued the suppressive role of circ_0000517 silencing on HCC cell progression, indicating that circ_0000517 could take part in the regulation of HCC development via targeting miR-1296-5p.
MiR-1296-5p has been confirmed to function as a tumor suppressor by interacting with targeted genes. For example, Jia et al unraveled that miR-1296-5p level was diminished in gastric cancer (GC) and its elevation repressed GC cell proliferation and motility by binding to CDK6 and EGFR.24 Chen et al claimed that miR-1296-5p impeded breast cancer cell growth via targeting ERBB2/mTORC1 pathway.25 Moreover, Xu et al proved that miR-1296 was reduced in HCC and overexpression of miR-1296 repressed HCC motility through interacting with SRPK1/AKT pathway.17 In the research, TXNDC5 was a target gene of miR-1296-5p. Zhang et al uncovered that TXNDC5 overexpression suppressed GC cell apoptosis and facilitated colony formation and metastasis.26 Yu et al disclosed that miR-218-5p hampered HCC cell proliferation and motility and accelerated apoptosis, whereas TXNDC5 elevation overturned the impacts.27 Herein, we found that TXNDC5 was elevated in HCC. TXNDC5 elevation abrogated the impacts of miR-1296-5p overexpression or circ_0000517 deletion on cell viability, cell cycle, colony formation and apoptosis in HCC cells. Moreover, circ_0000517 could upregulate TXNDC5 expression via targeting miR-1296-5p.
Taken together, our findings illuminated that circ_0000517 was drastically upregulated in HCC and it could sponge miR-1296-5p to alter TXNDC5 expression. Moreover, circ_0000517 contributed to HCC progression via regulating miR-1296-5p/TXNDC5 axis, indicating that circ_0000517 could be a potential new target for HCC treatment.
Disclosure
The authors declare that they have no conflicts of interest in this work.
References
1. Gomaa AI, Khan SA, Toledano MB, et al. Hepatocellular carcinoma: epidemiology, risk factors and pathogenesis. World J Gastroenterol. 2008;14(27):4300–4308. doi:10.3748/wjg.14.4300
2. Yang JD, Roberts LR. Hepatocellular carcinoma: a global view. Nat Rev Gastroenterol Hepatol. 2010;7(8):448–458. doi:10.1038/nrgastro.2010.100
3. Bosetti C, Turati F, La Vecchia C. Hepatocellular carcinoma epidemiology. Best Pract Res Clin Gastroenterol. 2014;28(5):753–770. doi:10.1016/j.bpg.2014.08.007
4. Cidon EU. Systemic treatment of hepatocellular carcinoma: past, present and future. World J Hepatol. 2017;9(18):797–807. doi:10.4254/wjh.v9.i18.797
5. Maluccio M, Covey A. Recent progress in understanding, diagnosing, and treating hepatocellular carcinoma. CA Cancer J Clin. 2012;62(6):394–399. doi:10.3322/caac.21161
6. Zhang HD, Jiang LH, Sun DW, et al. CircRNA: a novel type of biomarker for cancer. Breast Cancer. 2018;25(1):1–7. doi:10.1007/s12282-017-0793-9
7. Reddy KB. MicroRNA (miRNA) in cancer. Cancer Cell Int. 2015;15:38. doi:10.1186/s12935-015-0185-1
8. Qu S, Yang X, Li X, et al. Circular RNA: a new star of noncoding RNAs. Cancer Lett. 2015;365(2):141–148. doi:10.1016/j.canlet.2015.06.003
9. Wang YG, Wang T, Ding M, et al. hsa_circ_0091570 acts as a ceRNA to suppress hepatocellular cancer progression by sponging hsa-miR-1307. Cancer Lett. 2019;460:128–138. doi:10.1016/j.canlet.2019.06.007
10. Zou H, Xu X, Luo L, et al. Hsa_circ_0101432 promotes the development of hepatocellular carcinoma (HCC) by adsorbing miR-1258 and miR-622. Cell Cycle. 2019;18(19):2398–2413. doi:10.1080/15384101.2019.1618120
11. Guan Z, Tan J, Gao W, et al. Circular RNA hsa_circ_0016788 regulates hepatocellular carcinoma tumorigenesis through miR-486/CDK4 pathway. J Cell Physiol. 2018;234(1):500–508. doi:10.1002/jcp.26612
12. Wang X, Wang X, Li W, et al. Up-regulation of hsa_circ_0000517 predicts adverse prognosis of hepatocellular carcinoma. Front Oncol. 2019;9:1105. doi:10.3389/fonc.2019.01105
13. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. cell. 2004;116(2):281–297. doi:10.1016/S0092-8674(04)00045-5
14. Hua Y, Duan S, Murmann AE, et al. miRConnect: identifying effector genes of miRNAs and miRNA families in cancer cells. PLoS One. 2011;6(10):e26521. doi:10.1371/journal.pone.0026521
15. Lu Y, Yang L, Qin A, et al. miR-1470 regulates cell proliferation and apoptosis by targeting ALX4 in hepatocellular carcinoma. Biochem Biophys Res Commun. 2019.
16. Yao B, Li Y, Wang L, et al. MicroRNA-3194-3p inhibits metastasis and epithelial-mesenchymal transition of hepatocellular carcinoma by decreasing Wnt/beta-catenin signaling through targeting BCL9. Artif Cells Nanomed Biotechnol. 2019;47(1):3885–3895. doi:10.1080/21691401.2019.1670190
17. Xu Q, Liu X, Liu Z, et al. MicroRNA-1296 inhibits metastasis and epithelial-mesenchymal transition of hepatocellular carcinoma by targeting SRPK1-mediated PI3K/AKT pathway. Mol Cancer. 2017;16(1):103. doi:10.1186/s12943-017-0675-y
18. Horna-Terron E, Pradilla-Dieste A, Sanchez-De-Diego C, et al. TXNDC5, a newly discovered disulfide isomerase with a key role in cell physiology and pathology. Int J Mol Sci. 2014;15(12):23501–23518. doi:10.3390/ijms151223501
19. Park MS, Kim SK, Shin HP, et al. TXNDC5 gene polymorphism contributes to increased risk of hepatocellular carcinoma in the Korean male population. Anticancer Res. 2013;33(9):3983–3987.
20. Huang X, Zhang W, Shao Z. Prognostic and diagnostic significance of circRNAs expression in hepatocellular carcinoma patients: a meta-analysis. Cancer Med. 2019;8(3):1148–1156. doi:10.1002/cam4.1939
21. Yao Z, Xu R, Yuan L, et al. Circ_0001955 facilitates hepatocellular carcinoma (HCC) tumorigenesis by sponging miR-516a-5p to release TRAF6 and MAPK11. Cell Death Dis. 2019;10(12):945. doi:10.1038/s41419-019-2176-y
22. Li W, Zhou X, Wu X, et al. The role of circular RNA hsa_circ_0085616 in proliferation and migration of hepatocellular carcinoma cells. Cancer Manag Res. 2019;11:7369–7376. doi:10.2147/CMAR.S211020
23. Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388. doi:10.1038/nature11993
24. Jia Y, Zhao LM, Bai HY, et al. The tumor-suppressive function of miR-1296-5p by targeting EGFR and CDK6 in gastric cancer. Biosci Rep. 2019;39(1).
25. Chen G, He M, Yin Y, et al. miR-1296-5p decreases ERBB2 expression to inhibit the cell proliferation in ERBB2-positive breast cancer. Cancer Cell Int. 2017;17:95. doi:10.1186/s12935-017-0466-y
26. Zhang L, Hou Y, Li N, et al. The influence of TXNDC5 gene on gastric cancer cell. J Cancer Res Clin Oncol. 2010;136(10):1497–1505. doi:10.1007/s00432-010-0807-x
27. Yu J, Yang M, Zhou B, et al. CircRNA-104718 acts as competing endogenous RNA and promotes hepatocellular carcinoma progression through microRNA-218-5p/TXNDC5 signaling pathway. Clin Sci. 2019;133(13):1487–1503. doi:10.1042/CS20190394
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.