Back to Journals » Cancer Management and Research » Volume 12
Circ_0000376, a Novel circRNA, Promotes the Progression of Non-Small Cell Lung Cancer Through Regulating the miR-1182/NOVA2 Network
Authors Li C, Liu H, Niu Q, Gao J
Received 15 April 2020
Accepted for publication 24 July 2020
Published 24 August 2020 Volume 2020:12 Pages 7635—7647
DOI https://doi.org/10.2147/CMAR.S258340
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Chien-Feng Li
Cui Li,1 Hai Liu,2 Qin Niu,3 Jia Gao3
1Department of Pharmacy, The First People’s Hospital of Lianyungang, Lianyungang, Jiangsu, People’s Republic of China; 2Department of Clinical Laboratory, The First People’s Hospital of Lianyungang, Lianyungang, Jiangsu, People’s Republic of China; 3Department of Oncology, The First People’s Hospital of Lianyungang, Lianyungang, Jiangsu, People’s Republic of China
Correspondence: Hai Liu Email [email protected]
Background: Hypoxia has been shown to induce the malignant progression of cancer, including non-small cell lung cancer (NSCLC). Circular RNA (circRNA) is considered to be an important regulator of cancer progression. However, the role of a newly discovered circRNA, circ_0000376, in the progression of NSCLC is unclear.
Methods: The relative expression levels of circ_0000376, miR-1182 and neuro-oncological ventral antigen 2 (NOVA2) were detected via quantitative real-time polymerase chain reaction (qRT-PCR). Glucose consumption and lactate production were determined using Glucose Assay Kit and Lactate Assay Kit, respectively. Moreover, the protein levels of glycolysis markers and NOVA2 were measured using Western blot (WB) analysis. Furthermore, 3-(4, 5-dimethyl-2 thiazolyl)-2, 5-diphenyl-2-H-tetrazolium bromide (MTT) assay was performed to assess cell viability, and transwell assay was employed to evaluate cell migration and invasion. The interaction between miR-1182 and circ_0000376 or NOVA2 was confirmed by dual-luciferase reporter assay and RNA immunoprecipitation (RIP) assay. In addition, animal experiments were conducted to assess the influence of circ_0000376 silencing on NSCLC tumor growth in vivo.
Results: Circ_0000376 was upregulated in NSCLC, and its high expression was related to the poor overall survival of NSCLC patients. Hypoxia could enhance circ_0000376 expression and promote the glycolysis, viability, migration, and invasion of NSCLC cells. However, silencing of circ_0000376 could inhibit the glycolysis, viability, migration, and invasion of hypoxia-induced NSCLC cells. Additionally, circ_0000376 could sponge miR-1182, and miR-1182 could target NOVA2. MiR-1182 silencing could reverse the inhibitory effect of circ_0000376 knockdown on NSCLC progression, and NOVA2 overexpression also could reverse the suppressive effect of miR-1182 overexpression on NSCLC progression. Meanwhile, miR-1182 inhibitor could invert the negative regulation effect of circ_0000376 silencing on NOVA2 expression. In addition, circ_0000376 knockdown inhibited the NSCLC tumor growth via regulating the miR-1182 and NOVA2 expression in vivo.
Conclusion: Circ_0000376 promoted NSCLC progression by regulating the miR-1182/NOVA2 axis, suggesting that circ_0000376 might be a potential biomarker for NSCLC treatment.
Keywords: NSCLC, circ_0000376, miR-1182, NOVA2
Introduction
Non-small cell lung cancer (NSCLC) is a common pathological type of lung cancer.1,2 About 50% of NSCLC patients have reached the advanced stage when diagnosed, so the treatment of NSCLC is difficult.3 Therefore, it is important to explore new biomarkers for the treatment of NSCLC. Hypoxia is an important feature of solid tumors, and some studies have shown that cancer itself is a chronic disease with severe hypoxia.4,5 Numerous studies have confirmed that hypoxia can induce the glycolysis of cells to drive the malignant progression of cancers, including NSCLC.6–8
As a special non-coding RNA, circular RNA (circRNA) has been shown to play a vital role in the occurrence of cancers.9,10 CircRNA is considered to be an important regulator of cancer progression and mainly plays anti-cancer or pro-cancer role.11,12 At present, many circRNAs are found in NSCLC, and their functions are illuminated. For instance, hsa_circ_0011780 has been proved to be an anti-cancer circRNA of NSCLC, which can inhibit the proliferation and metastasis of NSCLC cells.13 CircCCDC66 is confirmed to play a pro-cancer role in NSCLC to accelerate cell growth and metastasis.14 Therefore, circRNA is the key target to explore new biomarkers for cancer treatment.
Using the GEO2R tool, we identified the differentially expressed circRNA in NSCLC tissues and adjacent normal tissues. Among the 149 differentially expressed circRNAs, we selected a circRNA (circ_0000376) with a large differentially expressed multiple for this study to explore its role in NSCLC progression. Our study is expected to provide new therapeutic targets for the treatment of NSCLC.
Materials and Methods
Differentially Expressed circRNAs
Online analysis tool, GEO2R, was used to screen the differentially expressed circRNAs in 3 paired human NSCLC tissues (Tumor) and adjacent normal tissues (Normal). In GEO database (GSE112214), the cut-off criteria of values were as follows: |log2 fold change| (|log2 FC|) >1 and P < 0.05.
Tissue Samples
Fifty paired NSCLC tissues (Tumor) and adjacent normal tissues (Normal) were available from 50 NSCLC patients who underwent surgery at The First People’s Hospital of Lianyungang. The clinicopathologic features of NSCLC patients are shown in Table 1. All patients signed written informed consent, and all tissues were stored at −80°C. Our research was approved by the Ethics Committee of The First People’s Hospital of Lianyungang and was carried out according to the guidelines of Declaration of Helsinki.
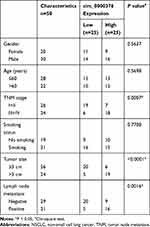 |
Table 1 Relationship Between circ_0000376 Expression and the Clinicopathologic Features of NSCLC Patients |
Cell Culture and Transfection
NSCLC cell lines (H522 and H1975) and human normal bronchial epithelial cell line (BEAS-2B) were bought from American Type Culture Collection (ATCC, Manassas, VA, USA). H522 and H1975 cells were cultured in RPMI-1640 medium (Sigma-Aldrich, St. Louis, MO, USA) and BEAS-2B cells were maintained in BEGM Bullet Kit (Lonza, Basel, Switzerland). Both mediums were supplemented with 10% fetal bovine serum (Sigma-Aldrich) and 1% penicillin/streptomycin solution (Sangon Biotech, Shanghai, China). Under the normoxia (Nor) conditions, all cells were grown at 37°C in a 21% O2, 5% CO2 and 74% N2 incubator. Under the hypoxia (Hyp) conditions, H522 and H1975 cells were cultured at 37°C in a 1% O2, 5% CO2 and 94% N2 incubator.
When cells reached 60–70% confluence, cell transfection could be carried out. Circ_0000376 small interference RNA (si-circ_0000376: 5ʹ-TCCATATGAGAGTTGGATTCT-3ʹ) and lentiviral short hairpin RNA (sh-circ_0000376: 5ʹ-TATCCATATGAGAGTTGGATT-3ʹ) were designed by us and synthesized by General Biosystems (Anhui, China). MiR-1182 mimic and inhibitor (miR-1182 and anti-miR-1182), neuro-oncological ventral antigen 2 (NOVA2) overexpression plasmid (NOVA2), and their negative controls (miR-NC, anti-miR-NC, and vector) were obtained from General Biosystems. According to the protocol of manufacturers, Lipofectamine 3000 reagent (Invitrogen, Carlsbad, CA, USA) was used to transfect all the oligonucleotides (50 nM) and plasmids (2 mg/mL) into cells.
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Total RNA was extracted using TRIzol reagent (Invitrogen), and then the quantity of RNA was determined using an ultraviolet spectrophotometer. The cDNA was obtained using SuperScript III Reverse Transcriptase Kit (Invitrogen) (the RT primers for circ_0000376, PRH1-PRR4, and NOVA2 were used the random primers, and for miR-1182 was used the specific primers). Subsequently, qRT-PCR was performed using PowerUp SYBR Master Mix (Applied Biosystems, Foster City, CA, USA). All primers were shown as below: circ_0000376, F 5ʹ-AGGTTCTCCAGCATTTGGCT-3ʹ, R 5ʹ-TGAATGAAAAAGTTAGTCAGGCAT-3ʹ; PRH1-PRR4, F 5ʹ-TCCCAGCACAAAGTTGGGAG-3ʹ, R 5ʹ-CCACTATCACCAGGGGGTCT-3ʹ; miR-1182, F 5ʹ-ACTTGTCACTGCCTGTCTCC-3ʹ, R 5ʹ-CCAACAGTCACATCCCTCCC-3ʹ; NOVA2, F 5ʹ-GGGTTCCCATAGACCTGGAC-3ʹ, R 5ʹ-CGCTCAGTAGTACCTGGGTAA-3ʹ; glyceraldehyde-3-phosphate dehydrogenase (GAPDH), F 5ʹ-GGTCTCCTCTGACTTCAACA-3ʹ, R 5ʹ-GTGAGGGTCTCTCTCTTCCT-3ʹ; U6, F 5ʹ-GTAGATACTGCAGTACG-3ʹ, R 5ʹ-ATCGCATGACGTACCTGAGC-3ʹ. GAPDH and U6 were regarded as the inner control, and all data were standardized by 2−ΔΔCt method.
Ribonuclease R (RNase R) Digestion Assay
RNase R was purchased from Epicentre (Madison, WI, USA). After extracted RNA from H522 and H1975 cells, the RNA was treated with RNase R for 15 min. Next, qRT-PCR was used to examine the circ_0000376 and PRH1-PRR4 expression. PRH1-PRR4 was used as the linear RNA control.
Detection of Glucose Consumption and Lactate Production
Glucose Assay Kit and Lactate Assay Kit were bought from BioVision (San Francisco, CA, USA). In brief, H522 and H1975 cells were lysed, and cell lysates were collected. According to the protocol of manufacturers, glucose consumption and lactate production were detected using corresponding kits, respectively.
Western Blot (WB) Analysis
H522 and H1975 cells were lysed using RIPA buffer (Beyotime, Shanghai, China) and then centrifuged to collect total proteins. Proteins were separated on SDS-PAGE gels and transferred to PVDF membranes (LaiboBio, Shanghai, China). Next, the membranes were blocked with 5% non-fat milk and incubated with primary antibodies and secondary antibody in turn. Finally, protein signals were detected using BeyoECL Plus Kit (Beyotime). Primary antibodies (glucose transporter (GLUT2, 1:1,000), hexokinase 2 (HK2; 1:1,000), NOVA2 (1:2,000), GAPDH (1:10,000)) and secondary antibody (Goat Anti-Rabbit IgG H&L (HRP) (1:50,000)) were obtained from Abcam (Cambridge, MA, USA).
3-(4, 5-Dimethyl-2 Thiazolyl)-2, 5-Diphenyl-2-H-Tetrazolium Bromide (MTT) Assay
MTT Assay Kit (Trevigen, Gaithersburg, MD, USA) was used for detecting cell viability. H522 and H1975 (2 × 103) cells were cultured in 96-well plates. After 48 h, MTT solution was added into each well for 4 h. Then, dimethyl sulfoxide (DMSO) was added into cells for 10 min. The absorbance at 490 nm was measured using a microplate reader to assess the cell viability.
Transwell Assay
Transwell chambers (Corning Inc., Corning, NY, USA) coated with or without Matrigel (BD Biosciences, San Jose, CA, USA) were used to measure cell invasion and migration, respectively. After transfection for 48 h, H522 and H1975 cells were collected and re-suspended with serum-free medium. Then, cells were seeded in the upper chambers. Complete medium was added to the lower chambers. After incubation for 24 h, the cells were fixed with methanol and stained with crystal violet. The number of migrated and invaded cells were counted in 5 random fields (100 ×).
Dual-Luciferase Reporter Assay
The sequences of circ_0000376 and NOVA2 3ʹUTR containing the predicted or mutated binding sites of miR-1182 were sub-cloned into the psiCHECK-2 reporter vector (Hanbio Biotechnology, Shanghai, China) to generate the wild-type (WT) and mutant-type (MUT) reporter vectors (circ_0000376 WT/MUT and NOVA2 3ʹUTR WT/MUT). H522 and H1975 cells were co-transfected with miR-1182 mimic or miR-NC and the reporter vectors using Lipofectamine 3000 reagent. After transfection for 48 h, the cells were harvested, and the luciferase activities of Firefly and Renilla were determined using Dual-Luciferase Assay Kit (Bioaolaibo, Beijing, China). Relative luciferase activity was the ratio of Firefly luciferase activity to Renilla luciferase activity.
RNA Immunoprecipitation (RIP) Assay
RIP Kit was obtained from Sigma-Aldrich. After transfected with miR-1182 mimic or miR-NC for 48 h, H522 and H1975 cells were lysed to collect cell lysates. Then, the cell lysates were incubated with magnetic beads conjugated with human immunoglobulin G (IgG) antibody (IgG) or argonaute 2 (Ago2) antibody in RIP buffer. The immunoprecipitated RNAs were treated with TRIzol reagent to extract total RNAs, and then the circ_0000376 expression and NOVA2 expression were detected via qRT-PCR.
Animal Experiments
These experiments were approved by the Animal Research Committee of The First People’s Hospital of Lianyungang and was performed according to the Guide for the Care and Use of Laboratory Animals. The Guide for the Care and Use of Laboratory Animals is widely acknowledged and used. Male BALB/c mice were obtained from Cavens (Changzhou, China) and randomly divided into 2 groups (n = 6). After transfected with sh-circ_0000376 or sh-NC, H522 cells were subcutaneously injected into the flank of mice. The tumor length and width were measured every 7 days until 35 days. After that, the mice were sacrificed, and the tumors were removed for other experiments.
Statistical Analysis
All statistical data were expressed as the mean ± standard deviation. Student’s t-test or one-way analysis of variance (ANOVA) followed by Tukey’s post hoc tests was employed to compare two or multiple groups, respectively. The F value of ANOVA was shown in Supplementary material. Log-rank test was used for Kaplan-Meier method. The correlation analysis was performed using Pearson correlation analysis. P < 0.05 was used to analyze statistical significance in GraphPad Prism 6.0 software (GraphPad, La Jolla, CA, USA). All experiments were performed in triplicate.
Results
High Expression of Circ_0000376 Was Found in NSCLC Tissues and Cells
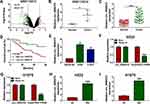
Volcanic map analysis showed that there were 149 differentially expressed circRNAs in NSCLC tissues and adjacent normal tissues, of which 16 were upregulated and 133 were downregulated (Figure 1A). Then, we selected the significantly upregulated circ_0000376 in NSCLC with high difference multiple as the research object (GSE112214: log2 FC = 1.1569625 and P = 0.00867486) (Figure 1B). In our study, by detecting the expression of circ_0000376, we confirmed that it was markedly highly expressed in NSCLC tissues compared to adjacent normal tissues (Figure 1C). Based on the median of circ_0000376 expression (2.33) in NSCLC tumor tissues, NSCLC tissues were divided into the high circ_0000376 expression group (>2.33) and the low circ_0000376 expression group (<2.33). Through analyzing the relationship between circ_0000376 expression and the clinicopathologic features of NSCLC patients, we discovered that high circ_0000376 expression was positively correlated with the TNM stage, tumor size, and lymph node metastasis of NSCLC patients (P < 0.05, Table 1). The results of Kaplan-Meier analysis suggested that the overall survival of NSCLC patients with high circ_0000376 expression was often poor (Figure 1D). In NSCLC cell lines (H522 and H1975), we also found an increased expression of circ_0000376 compared to BEAS-2B cells (Figure 1E). To confirm the circular characteristic of circ_0000376, RNase R assay was performed and the results revealed that circ_0000376 was more resistant to RNase R digestion than linear PRH1-PRR4 in H522 and H1975 cells (Figure 1F and G). Additionally, we also uncovered that circ_0000376 expression was obviously promoted in H522 and H1975 cells after treated with hypoxia for 48 h (Figure 1H and I).
Circ_0000376 Knockdown Inhibited the Glycolysis, Viability and Metastasis of Hypoxia-Treated NSCLC Cells
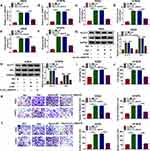
To investigate the function of circ_0000376 on NSCLC progression, we transfected with si-circ_0000376 in hypoxia-treated NSCLC cells. The decreased circ_0000376 expression in hypoxia-treated H522 and H1975 cells revealed that the transfection of si-circ_0000376 was successful (Figure 2A and B). Through measuring the glucose consumption and lactate produce of H522 and H1975 cells, we found that hypoxia could promote the glycolysis of NSCLC cells, while circ_0000376 silencing could reverse this effect (Figure 2C–F). Besides, the increasing effect of hypoxia on the protein levels of glycolysis markers (GLUT1 and HK2) also could be inhibited by circ_0000376 knockdown (Figure 2G and H). Furthermore, the results of MTT assay and transwell assay results revealed that hypoxia could enhance the viabilities, migration, and invasion of H522 and H1975 cells, while this effect could be recovered by the addition of si-circ_0000376 (Figure 2I–L). Therefore, all data indicated that knocking down of circ_0000376 might be an important measure to inhibit the progression of NSCLC.
Circ_0000376 Acted as a Sponge of miR-1182 in NSCLC
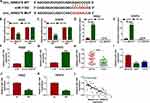
CircRNAs have been reported to act as competing endogenous RNAs (ceRNAs) of microRNAs (miRNAs) to regulate gene expression.15,16 To explore the molecular mechanism by which circ_0000376 regulated NSCLC progression, we used the Circinteractome tool to analyze the targeted miRNAs of circ_0000376. As presented in Figure 3A, miR-1182 was found to have the complementary sequences for circ_0000376. To confirm the binding relationship between miR-1182 and circ_0000376, dual-luciferase reporter assay and RIP assay were carried out. The results revealed that miR-1182 mimic markedly repressed the luciferase activity of circ_0000376 WT reporter, while no effect on the luciferase activity of circ_0000376 MUT reporter in H522 and H1975 cells (Figure 3B and C). Besides, miR-1182 mimic also could significantly enrich circ_0000376 in RIP Ago2 compared with that in RIP IgG (Figure 3D and E). Meanwhile, circ_0000376 knockdown remarkably accelerated the expression of miR-1182 in H522 and H1975 cells (Figure 3F and G). In addition, we detected the miR-1182 expression in NSCLC tissues and cells and found that miR-1182 was downregulated in NSCLC tissues and cells (Figure 3H and I). Importantly, hypoxia also could suppress the miR-1182 expression in H522 and H1975 cells (Figure 3J and K). Additionally, Pearson correlation analysis revealed that there had a negative correlation between miR-1182 and circ_0000376 in NSCLC (Figure 3L).
MiR-1182 Inhibitor Reversed the Effect of Circ_0000376 Silencing on NSCLC Progression
To confirm that circ_000376 regulated NSCLC progression via targeting miR-1182, si-circ_0000376 and anti-miR-1182 were co-transfected into H522 and H1975 cells to perform the rescue experiments. The detection of miR-1182 expression confirmed that the transfection efficiencies of si-circ_0000376 and anti-miR-1182 were excellent, and following experiments could be carried out (Figure 4A and B). We discovered that miR-1182 inhibitor could reverse the inhibition effect of circ_0000376 knockdown on the glucose consumption and lactate produce, as well as the protein levels of GLUT1 and HK2 in hypoxia-induced H522 and H1975 cells (Figure 4C–H). Furthermore, the suppressive effect of circ_0000376 silencing on the viabilities, migration and invasion of hypoxia-treated H522 and H1975 cells also could be reversed by miR-1182 inhibitor (Figure 4I–N). Hence, we confirmed that circ_0000376 regulated NSCLC progression by sponging miR-1182.
NOVA2 Was Targeted by miR-1182 in NSCLC
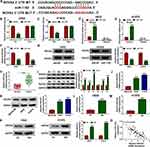
The Targetscan tool was used to predict the targeted mRNAs of miR-1182 and the results revealed that NOVA2 3ʹUTR had binding sites with miR-1182 (Figure 5A). The results of dual-luciferase reporter assay and RIP assay indicated that miR-1182 mimic could special inhibit the luciferase activity of NOVA2 3ʹUTR WT reporter in H522 and H1975 cells, and could remarkably enrich NOVA2 in RIP Ago2 (Figure 5B–E). Moreover, miR-1182 overexpression also could restrain the mRNA and protein levels of NOVA2 in H522 and H1975 cells (Figure 5F and G). In NSCLC tissues and cells, we uncovered that the NOVA2 mRNA and protein levels were significantly improved compared to adjacent normal tissues and BESA-2B cells, respectively (Figure 5H–K). Meanwhile, we also found that hypoxia could promote the mRNA and protein levels of NOVA2 in H522 and H1975 cells (Figure 5L–N). And the results of correlation analysis indicated that NOVA2 expression was negatively correlated with miR-1182 expression in NSCLC (Figure 5O).
Overexpressed NOVA2 Could Reverse the Effect of miR-1182 on NSCLC Progression
For confirming that miR-1182 regulated NSCLC progression via targeting NOVA2, we co-transfected with miR-1182 mimic and NOVA2 overexpression plasmid into H522 and H1975 cells. WB analysis results suggested that miR-1182 mimic could inhibit the protein level of NOVA2 in hypoxia-treated H522 and H1975 cells, while NOVA2 overexpression plasmid could reverse this effect, which confirmed that the transfection efficiency of both was good (Figure 6A and B). By detecting the glucose consumption, lactate produce, and the protein levels of GLUT1 and HK2 in hypoxia-treated H522 and H1975 cells, we found that overexpressed NOVA2 could partially reverse the inhibitory effect of miR-1182 overexpression on the glycolysis of NSCLC cells (Figure 6C–H). In addition, we also discovered that miR-1182 mimic suppressed the viabilities, migration, and invasion of hypoxia-induced H522 and H1975 cells, while the addition of NOVA2 could reverse the effect of miR-1182 (Figure 6I–N). These results revealed that NOVA2 participated in the regulation of miR-1182 on NSCLC progression.
Circ_0000376 Positively Regulated NOVA2 Expression by Sponging miR-1182
The results of Pearson correlation analysis suggested that circ_0000376 expression was positively correlated with NOVA2 expression in NSCLC (Figure 7A). To confirm the regulation of circ_0000376 on NOVA2, we determined the NOVA2 expression in H522 and H1975 cells transfected with si-circ_0000376 and anti-miR-1182. QRT-PCR results indicated that circ_0000376 silencing markedly suppressed the mRNA expression of NOVA2 in H522 and H1975 cells, and this effect could be revered by miR-1182 inhibitor (Figure 7B and C). Also, the detection of NOVA2 protein level got the similar results with the NOVA2 mRNA level in H522 and H1975 cells (Figure 7D and E). All data showed that circ_0000376 sponged miR-1182 to regulate NOVA2 expression in NSCLC.
Silenced Circ_0000376 Repressed NSCLC Tumor Growth in vivo
For further confirming the function of circ_0000376 on NSCLC progression, we performed the animal experiments. By measuring the volume and weight of the tumor, we found that silenced circ_0000376 could inhibit the growth of NSCLC-transplanted tumors (Figure 8A and B). In the sh-circ_0000376 group, the reduction of circ_0000376 expression determined that the knockdown of circ_0000376 in the sh-circ_0000376 group was successful (Figure 8C). Additionally, we also measured the expression of miR-1182 and NOVA2 in the tumors and found that miR-1182 expression was markedly increased (Figure 8D), and the mRNA and protein levels of NOVA2 were remarkably repressed in the tumors (Figure 8E and F). Therefore, we concluded that circ_0000376 knockdown inhibited NSCLC tumor growth through regulating the expression of miR-1182 and NOVA2.
Discussion
Despite significant advances in diagnosis and treatment, NSCLC remains a leading cause of death worldwide.17 Therefore, it is of great clinical significance to explore new therapeutic biomarkers for NSCLC. In our study, we focused on circ_0000376 through screening. We found that circ_0000376 was highly expressed in NSCLC, which was consistent with our predicted results. In previous studies, Jiang et al predicted that circ_0000376 was a significantly upregulated circRNA for gastric cancer.18 In addition, we also discovered that high circ_0000376 expression was related to the poor prognosis of NSCLC patients, so we speculated that circ_0000376 might play a positive role in the malignant progression of NSCLC.
Growing amount of evidence has suggested that hypoxia is one of the important causes of cancer progression, and targeted treatment against hypoxia is an important measure to alleviate cancer progression.19 In view of the vital role of circRNA in cancer progression, we investigated the effect of circ_0000376 on the biological function of NSCLC cells induced by hypoxia. In this study, we discovered that hypoxia could promote circ_0000376 expression and enhance the glycolysis, viability, and metastasis of NSCLC cells, while knockdown of circ_0000376 could reverse the effect of hypoxia on NSCLC cell progression. Additionally, animal experiments also revealed that the interference of circ_0000376 repressed the tumor growth of NSCLC in vivo. These data suggested that circ_0000376 silencing might be a feasible measure to inhibit the progression of NSCLC. In order to perfect the molecular mechanism of circ_0000376 as a ceRNA, we made the bioinformatics prediction and discovered that circ_0000376 could sponge miR-1182 to regulate NOVA2.
In many cancer types, miR-1182 has been considered to be a tumor suppressor regulating cancer progression, such as colorectal cancer, ovarian cancer, and prostate cancer.20–22 Recent studies by Li et al have shown that miR-1182 can be absorbed by circ_0000735 and then participates in the regulation of circ_0000735 on NSCLC.23 Therefore, miR-1182 mainly plays an anti-cancer role in NSCLC. In our study, we found that miR-1182 was underexpressed in NSCLC, which was similar with the previous results.23 The rescue experiments confirmed that miR-1182 participated in the regulation of circ_0000376 in NSCLC. Furthermore, we also suggested that miR-1182 expression was negatively regulated by circ_0000376 in vitro and in vivo.
As a member of NOVA family, NOVA2 has been identified as an oncogene that regulates the malignant progression of cancer.24 The results of Tang et al showed that NOVA2 could act as a β-catenin RNA-binding protein to accelerate the epithelial–mesenchymal transition of breast cancer.25 And Gallo et al reported that NOVA2 was upregulated in colorectal cancer and might be a therapeutic target.26 In NSCLC, Xiao et al suggested that miR-7-5p inhibited NOVA2 expression to restrain the metastasis of cancer.27 Consistent with this, our study showed that NOVA2 was highly expressed in NSCLC. Besides, the reversal effect of NOVA2 overexpression on miR-1182 mimic confirmed that miR-1182 regulated NSCLC progression by targeting NOVA2. In addition, we also suggested that NOVA2 expression was positively regulated by circ_0000376 and negatively regulated by miR-1182. All data revealed that circ_0000376 regulated NOVA2, an oncogene, to enhance NSCLC progression by sponging miR-1182.
However, the current research still has some limitations. In the rescue experiment, we found that the reversal effect of miR-1182 inhibitor on circ_0000376 knockdown is partially, which suggests that there may be other miRNAs involved in the regulation of NSCLC progression by circ_0000376. Similarly, the reversal of NOVA2 on miR-1182 mimic is not complete, suggesting that miR-1182 may also have multiple targets in NSCLC. This requires us to further confirm.
In summary, our results presented that a novel circRNA, circ_0000376, accelerated the glycolysis, viability, and metastasis of NSCLC cells by regulating the miR-1182/NOVA2 axis. Our study is the first to reveal the role and mechanism of circ_0000376 in the progression of NSCLC, which is instructive for exploring the role of circ_0000376 in other cancers. In addition, the positive effect of circ_0000376 on the malignant progression of NSCLC suggests that circ_0000376 may be a new therapeutic target for NSCLC.
Disclosure of Interest
The authors declare that they have no conflicts of interest for this work.
References
1. Ginsberg MS, Grewal RK, Heelan RT. Lung cancer. Radiol Clin North Am. 2007;45(1):21–43. doi:10.1016/j.rcl.2006.10.004
2. Stankovic B, Bjorhovde HAK, Skarshaug R, et al. Immune cell composition in human non-small cell lung cancer. Front Immunol. 2018;9:3101. doi:10.3389/fimmu.2018.03101
3. Cappuzzo F, Soo R, Hochmair M, et al. Global named patient use program of afatinib in advanced non-small-cell lung carcinoma patients who progressed following prior therapies. Future Oncol. 2018;14(15):1477–1486. doi:10.2217/fon-2017-0666
4. Liu Y, Jiang Y, Zhang M, Tang Z, He M, Bu W. Modulating hypoxia via nanomaterials chemistry for efficient treatment of solid tumors. Acc Chem Res. 2018;51(10):2502–2511. doi:10.1021/acs.accounts.8b00214
5. Najafi M, Farhood B, Mortezaee K, Kharazinejad E, Majidpoor J, Ahadi R. Hypoxia in solid tumors: a key promoter of cancer stem cell (CSC) resistance. J Cancer Res Clin Oncol. 2020;146(1):19–31. doi:10.1007/s00432-019-03080-1
6. Xu X, Liu C, Bao J. Hypoxia-induced hsa-miR-101 promotes glycolysis by targeting TIGAR mRNA in clear cell renal cell carcinoma. Mol Med Rep. 2017;15(3):1373–1378. doi:10.3892/mmr.2017.6139
7. Park JS, Lee S, Jeong AL, et al. Hypoxia-induced IL-32β increases glycolysis in breast cancer cells. Cancer Lett. 2015;356(2Pt B):800–808. doi:10.1016/j.canlet.2014.10.030
8. Shi J, Wang H, Feng W, et al. Long non-coding RNA HOTTIP promotes hypoxia-induced glycolysis through targeting miR-615-3p/HMGB3 axis in non-small cell lung cancer cells. Eur J Pharmacol. 2019;862:172615. doi:10.1016/j.ejphar.2019.172615
9. Lei M, Zheng G, Ning Q, Zheng J, Dong D. Translation and functional roles of circular RNAs in human cancer. Mol Cancer. 2020;19(1):30. doi:10.1186/s12943-020-1135-7
10. Zhang Q, Wang W, Zhou Q, et al. Roles of circRNAs in the tumour microenvironment. Mol Cancer. 2020;19(1):14. doi:10.1186/s12943-019-1125-9
11. Jiang Y, Zhang Y, Chu F, Xu L, Wu H. Circ_0032821 acts as an oncogene in cell proliferation, metastasis and autophagy in human gastric cancer cells in vitro and in vivo through activating MEK1/ERK1/2 signaling pathway. Cancer Cell Int. 2020;20:74. doi:10.1186/s12935-020-1151-0
12. Jie M, Wu Y, Gao M, et al. CircMRPS35 suppresses gastric cancer progression via recruiting KAT7 to govern histone modification. Mol Cancer. 2020;19(1):56. doi:10.1186/s12943-020-01160-2
13. Liu Y, Yang C, Cao C, Li Q, Jin X, Shi H. Hsa_circ_RNA_0011780 represses the proliferation and metastasis of non-small cell lung cancer by decreasing FBXW7 via targeting miR-544a. Onco Targets Ther. 2020;13:745–755. doi:10.2147/OTT.S236162
14. Wang Y, Zhao W, Zhang S. STAT3-induced upregulation of circCCDC66 facilitates the progression of non-small cell lung cancer by targeting miR-33a-5p/KPNA4 axis. Biomed Pharmacother. 2020;126:110019. doi:10.1016/j.biopha.2020.110019
15. Qiu Y, Pu C, Li Y, Qi B. Construction of a circRNA-miRNA-mRNA network based on competitive endogenous RNA reveals the function of circRNAs in osteosarcoma. Cancer Cell Int. 2020;20:48. doi:10.1186/s12935-020-1134-1
16. Zhong Y, Du Y, Yang X, et al. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol Cancer. 2018;17(1):79. doi:10.1186/s12943-018-0827-8
17. Sui H, Ma N, Wang Y, et al. Anti-PD-1/PD-L1 therapy for non-small-cell lung cancer: toward personalized medicine and combination strategies. J Immunol Res. 2018;2018:6984948. doi:10.1155/2018/6984948
18. Jiang F, Shen XB. miRNA and mRNA expression profiles in gastric cancer patients and the relationship with circRNA. Neoplasma. 2019;66(6):879–886. doi:10.4149/neo_2018_181211N952
19. Salem A, Asselin MC, Reymen B, et al. Targeting hypoxia to improve non-small cell lung cancer outcome. J Natl Cancer Inst. 2018;110(1). doi:10.1093/jnci/djx160
20. Li Y, Li C, Xu R, Wang Y, Li D, Zhang B. A novel circFMN2 promotes tumor proliferation in CRC by regulating the miR-1182/hTERT signaling pathways. Clin Sci. 2019;133(24):2463–2479. doi:10.1042/CS20190715
21. Zong ZH, Du YP, Guan X, Chen S, Zhao Y. CircWHSC1 promotes ovarian cancer progression by regulating MUC1 and hTERT through sponging miR-145 and miR-1182. J Exp Clin Cancer Res. 2019;38(1):437. doi:10.1186/s13046-019-1437-z
22. Huang C, Deng H, Wang Y, et al. Circular RNA circABCC4 as the ceRNA of miR-1182 facilitates prostate cancer progression by promoting FOXP4 expression. J Cell Mol Med. 2019;23(9):6112–6119. doi:10.1111/jcmm.14477
23. Li W, Jiang W, Liu T, Lv J, Guan J. Enhanced expression of circ_0000735 forecasts clinical severity in NSCLC and promotes cell progression via sponging miR-1179 and miR-1182. Biochem Biophys Res Commun. 2019;510(3):467–471. doi:10.1016/j.bbrc.2019.01.134
24. Angiolini F, Belloni E, Giordano M, et al. A novel L1CAM isoform with angiogenic activity generated by NOVA2-mediated alternative splicing. Elife. 2019;8:e44305. doi:10.7554/eLife.44305
25. Tang S, Zhao Y, He X, et al. Identification of NOVA family proteins as novel β-catenin RNA-binding proteins that promote epithelial-mesenchymal transition. RNA Biol. 2020;17(6):881–891. doi:10.1080/15476286.2020.1734372
26. Gallo S, Arcidiacono MV, Tisato V, et al. Upregulation of the alternative splicing factor NOVA2 in colorectal cancer vasculature. Onco Targets Ther. 2018;11:6049–6056. doi:10.2147/OTT.S171678
27. Xiao H. MiR-7-5p suppresses tumor metastasis of non-small cell lung cancer by targeting NOVA2. Cell Mol Biol Lett. 2019;24:60. doi:10.1186/s11658-019-0188-3
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.