Back to Journals » Cancer Management and Research » Volume 12
Circ-SAR1A Promotes Renal Cell Carcinoma Progression Through miR-382/YBX1 Axis
Authors Zhao X, Zhao Z, Xu W, Liu H, Chang J, Xu W, Li S , Cao S, Hou J
Received 14 January 2020
Accepted for publication 30 June 2020
Published 18 August 2020 Volume 2020:12 Pages 7353—7361
DOI https://doi.org/10.2147/CMAR.S245918
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Beicheng Sun
Xiaolei Zhao,* Zhenhua Zhao,* Wenchao Xu, Hui Liu, Junkai Chang, Weibo Xu, Song Li, Songqiang Cao, Junqing Hou
Department of Urology, Huaihe Hospital of Henan University, Kaifeng 475000, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Junqing Hou
Department of Urology, Huaihe Hospital of Henan University, Kaifeng 475000, People’s Republic of China
Email [email protected]
Background: Accumulating evidence points to a role for circular RNAs (circRNAs) in important regulatory function in tumor advancement. We explored the effect and function of circ-SAR1A in renal cell carcinoma (RCC).
Methods: circ-SAR1A expression in RCC tissues and cell lines was explored by qRT-PCR. The roles of circ-SAR1A on RCC progression were explored by in vitro function assays. Moreover, we determined the underlying mechanism of circ-SAR1A in RCC progression through bioinformatics analysis and dual-luciferase reporter assays.
Results: Our data reveal that circ-SAR1A is significantly high in RCC tissues and cell lines. High circ-SAR1A levels are correlated to advanced Fuhrman grade, and lymph-node metastasis in RCC patients. Functional experiments indicate that circ-SAR1A suppression decreased RCC cell growth and invasion abilities in vitro. Mechanistically, circ-SAR1A upregulated YBX1 expression by sponging miR-382, resulting in promoting the growth and invasion in RCC cells.
Conclusion: Our data indicate that the circ-SAR1A/miR-382/YBX1 axis plays a critical role in RCC progression, which serve as a potential novel treatment strategy of RCC.
Keywords: circ-SAR1A, miR-382, YBX1, renal cell carcinoma
Introduction
Renal cell carcinoma (RCC) is one of the most prevalent malignancies of urological cancer in adults.1,2 Due to its high resistance to chemotherapy and radiotherapy, the surgical strategy is the sole curable method for RCC patients.3 Nevertheless, the 5-year cancer-specific survival rates remain unsatisfied due to postoperative recurrence and metastasis.4,5 Hence, there is a pressing need to investigate the molecular mechanism that underlies RCC progression.
CircRNAs is a category of non-coding RNAs (ncRNAs) that have a vital function in gene levels at post-transcriptional, transcriptional and epigenetic levels.6 Recently, increasing reports showed that dysregulation of circRNAs contributed to multiple pathological states, including cancer.7,8 For example, Zong et al indicated that high circRNA102231 levels in lung cancer promoted the growth and invasion in vitro.9 Ge et al demonstrated that circMTO1 reduced the growth and invasion via Wnt/β-catenin axis in colorectal cancer.10 Xue et al found that hsa_circ_0081143 may regulate the miR-646/CDK6 axis to promote cisplatin resistance in gastric cancer.11 However, the roles of circRNAs on tumor progression remain unclear.
Herein, we discovered that circ-SAR1A is upregulated in RCC and correlated to advanced clinical features. In mechanism, circ-SAR1A encouraged the growth and invasion by sponging miR-382 to increase Y-box binding protein 1 (YBX1) levels in RCC cells. These findings suggested that circ-SAR1A acts as a novel treatment target for RCC.
Patients and Methods
Patients and Samples Collection
The study was granted approval by ethics committee of Huaihe Hospital of Henan University. Informed consent was granted by all patients in this study. We recruited 41 patients, who were clinically and histopathologically diagnosed with RCC in our hospital. No patients were administered therapy prior to surgery. All tissue samples were gathered and immediately kept at −80°C for further studies.
Cell Culture and Transfection
RCC cell lines (786-O, Caki-1, 769-P, ACHN, and A498) and human proximal tubule epithelial cell line (HK-2) were bought from ATCC. Cells were maintained Dulbecco’s Modified Eagle Medium (DMEM) with 10% Fetal bovine serum (FBS; Sigma), penicillin (100 μL/mL), streptomycin (100 mg/mL) in a humidified atmosphere that contains 5% CO2 at 37°C.
SiRNA silencing circ-SAR1A (si-circ-SAR1A), miR-382 mimics, miR-382 inhibitors, and negative controls obtained from GenePharma (Shanghai). Lipofectamine 3000 (Invitrogen) was used for cell transfection. QRT-qPCR was used to verify the transfection efficiency.
Quantitative Real-Time PCR (qRT-PCR)
RNA was extracted through the RNeasy total RNA isolation kit (Invitrogen). cDNA was produced and amplified utilizing the TaqMan miRNA Reverse Transcription Kit (Takara, Dalian, China). circ-SAR1A, miR-382, and YBX1 mRNA were evaluated through qRT-PCR by TaqMan Human miRNA Assay Kit (Applied Biosystems) according to manufacturer’s recommendations. Glyceraldehyde-3-Phosphate Dehydrogenase (GAPDH) and U6 were employed as an endogenous reference. The data were assessed utilizing the 2–ΔΔCT method.
Colony Formation Assay
To explore cell growth, colony formation assay was used. Briefly, transfected cells were placed in 6-well culture plates at 400 cells per well. Two weeks later, the cells were placed in methanol and stained with 0.1% crystal violet. Colonies were imaged and measured.
Transwell Assay
Transwell invasion experiment was conducted as per the previous study.10
Western Blot
The Western blot was conducted as previously published.12 Primary antibodies of YBX1 and GAPDH were purchased from Abcam (NY, USA). Protein expression was identified using enhanced chemiluminescence reagents (Pierce, Rockford, IL, USA).
Luciferase Reporter Assay
The luciferase reporter containing the circ-SAR1A (or YBX1) sequence in the 3′-UTR was created by sub-cloning the circ-SAR1A (or YBX1) fragment to the area downstream of a cytomegalovirus promoter-driven firefly luciferase cassette in a pGL3 vector (Promega, Madison, WI, USA). miRNA-binding site mutations in the pGL3 vector sequence were constructed by utilizing a Mut Express II Fast Mutagenesis Kit (Vazyme, Nanjing, China). Cells were placed into a 12-well plate, co-transfected with WT-circRNA (or WT-YBX1) or Mut-circRNA (Mut-YBX1) and miR-382 mimics or miR-NC. The relative luciferase activity was assessed using a Dual-Luciferase Assay System (Promega, Madison, WI, USA) as per established guidelines. Relative luciferase activity was normalized as per Renilla luciferase activity.
Immunohistochemistry
The paraffin-embedded tumor tissues were cut into 5 μm thick sections. Sections were deparaffinized, rehydrated, and then subjected to retrieve antigen in antigen retrieval buffer for 10 min. Next, sections were incubated with 3% H2O2 for 15 min at room temperature to quench the activity of endogenous peroxidases. Sections were then incubated with goat serum (Solarbio) for 15 min to block non-specific binding. Afterwards, sections were incubated overnight at 4°C with primary antibody. After washing the antigen 3 times with PBS, biotin-labeled secondary antibody was added for 15 minutes. After being washed with PBS, DAB was used to visualize immunoreactivity and counterstained with hematoxylin.
Statistical Analysis
Statistical analyses were carried out by software of SPSS 18.0. Data were represented as mean ± standard deviation (SD), and each assay was repeated three times. All data were analyzed utilizing Student’s t-test or one-way analysis of variance (ANOVA). A p value<0.05 represents statistical significance.
Results
circ-SAR1A is Upregulated in RCC
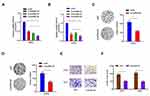
To investigate the function of circRNAs in RCC, we evaluated the differentially expressed circRNAs in GSE100186. Results showed that circRNA-018634 (circ-SAR1A) was one of the most significantly increased circRNAs in ccRCC tissues (Figure 1A). To validate this observation, we investigated circ-SAR1A in 41 paired RCC tissues. QRT-PCR indicated that circ-SAR1A levels were substantially increased in RCC tissues and cell lines (Figure 1B and C). Furthermore, correlation analysis indicated that high circ-SAR1A levels were correlated with advanced Fuhrman grade, and lymph-node metastasis in RCC patients (Figure 1D and E). This data suggests that circ-SAR1A has an oncogenic function in RCC progression.
circ-SAR1A Silencing Reduces the Growth and Invasion of ccRCC Cells
Circ-SAR1A resides on chromosome chr10 with a spliced sequence length of 496 bp. To determine the biological functions of circ-SAR1A in RCC, we reduced circ-SAR1A expression in 786-O and ACHN cells by transfected si-circSAR1A, and the transfection efficiency was determined using qRT-PCR (Figure 2A and B). Subsequently, circ-SAR1A roles in RCC progression were determined. Colony formation assay demonstrated that circ-SAR1A inhibition reduced 786-O and ACHN cells’ viabilities in comparison to si-NC group (Figure 2C and D). Transwell assay indicated that the invasive capability of 786-O and ACHN cells was remarkably suppressed in si-circ-SAR1A group in comparison to the si-NC group (Figure 2E and F).
circ-SAR1A Acts as a Sponge for miR-382
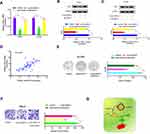
To discover the underlying mechanism of circ-SAR1A in RCC progression, we firstly tested circ-SAR1A location in 786-O and ACHN cells, results showed that the majority of circ-SAR1A was distributed in the cytoplasm (Figure 3A and B). Next, we predicted the possible targets, results showed circ-SAR1A possessed a complementary sequence to miR-382 seed region (Figure 3C–E). QRT-PCR demonstrated that circ-SAR1A knockdown increased miR-382 levels in 786-O and ACHN cells (Figure 3F). Luciferase reporter assay demonstrated that miR-382 mimics decreased the relative luciferase activity of WT-circ-SAR1A group, with no influence on Mut-circ-SAR1A group (Figure 3G).
YBX1 is Directly Targeted by miR-382
According to bioinformatics prediction, miR-382 may target YBX1 mRNA 3′ UTR with a high score (Figure 4A and B). To additionally validate the interaction, luciferase reporter assay was utilized. Results demonstrated that miR-382 mimics substantially decreases the luciferase activity of WT-YBX1 group, with no influence on Mut-YBX1 group (Figure 4C). Western blot demonstrated that miR-382 mimics substantially decrease YBX1 protein in RCC cells (Figure 4D). These results suggested that YBX1 was directly targeted by miR-382.
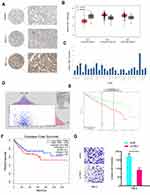
Furthermore, we investigated the function of YBX1 in RCC. IHC results demonstrated that YBX1 was significantly increased and correlated to metastasis (Figure 5A). TCGA database further confirmed the results (Figure 5B). Subsequently, we explored YBX1 expression decreased in normal tissue, including renal tissues (Figure 5C). Kaplan-Meier analysis indicated that high YBX1 expression was correlated to worse overall survival rate and disease-free survival rate in RCC patients (Figure 5D–F). In addition, Transwell assay showed that YBX1 inhibition reduced 786-O cells’ invasion ability in vitro (Figure 5G). These data implied that YBX1 inhibition repressed RCC progression.
circ-SAR1A Regulates RCC Progression by the miR-382/YBX1 Axis
Previous study demonstrated that circRNA can regulate gene expression by performing as a molecular sponge of miRNAs.13,14 Therefore, we explored whether circ-SAR1A regulated YBX1 expression by sponging miR-382 in RCC. As expected, results indicated that YBX1 was substantially decreased by introduction of si-circ-SAR1A in RCC cells, whilst miR-382 inhibitors eliminated the effects (Figure 6A–C). Correlation analysis indicated that circ-SAR1A is associated with YBX1 expression in RCC tissues (Figure 6D). Subsequently, clone formation assay demonstrated that miR-382 inhibitors substantially overturned the effect of si-circ-SAR1A on ACHN cells’ proliferation ability (Figure 6E). Transwell assay revealed that miR-382 inhibitors remarkably abolished the invasion ability of si-circ-SAR1A on 786-O cells (Figure 6F). This data suggests that the circ-SAR1A/miR-382/YBX1 axis plays a critical role in RCC progression (Figure 6G).
Discussion
In recent years, growing evidence emphasized that dysregulation of circRNAs is involved in tumor progression, including RCC.15,16 For example, Wang et al discovered that hsa_circ_0001451 was decreased and suppressed ccRCC tumor growth in vitro.17 Zhou et al showed that circPCNXL2 acted as a sponge of miR-153 to encourage ccRCC advancement by regulating ZEB2 levels.18 Xiong et al showed that circRNA ZNF609 served as a ceRNA in RCC to control FOXP4 expression by sponging miR-138-5p.19 However, the function and underlying mechanism of circRNA in RCC remains largely unknown.
Through the GSE100186 data analysis, we exhibited that circ-SAR1A is the most significantly increased circRNAs in ccRCC. Nevertheless, the function and underlying mechanism of circ-SAR1A is still unknown. In our study, we found that circ-SAR1A expression levels were obviously heightened in RCC tissues and cell lines. High circ-SAR1A levels were correlated to advanced Fuhrman grade, and lymph-node metastasis. In functional experiments, we found that circ-SAR1A suppression decreased the growth and invasion of RCC cells in vitro.
Recently, increasing studies exhibited that circRNA may function as “miRNA sponge” in tumor advancement.14 For example, Shen et al displayed that the hsa_circ_0002577/miR-197/CTNND1 axis acted as an important role in endometrial carcinoma progression.20 Shao et al showed that hsa_circ_0001742 encouraged the growth and invasion by controlling miR-634 expression in tongue squamous cell carcinoma.21 In this study, bioinformatics prediction demonstrated that circ-SAR1A possessed a complementary sequence to miR-382. Subsequently, qRT-PCR and luciferase reporter assays validated the interaction amongst circ-SAR1A and miR-382 in RCC. Previously published studies established that miR-382 may act as a tumor suppressor in certain human cancers, including esophageal squamous cell carcinoma, lung cancer, and colorectal cancer.22–24 In this study, we verified that miR-382 mediated the regulatory effect of circ-SAR1A on RCC cells’ progression.
The Y-box-binding protein 1 (YBX1) is a part of the cold-shock protein superfamily that binds DNA and RNA to coordinate transcription and translation, pre-mRNA splicing, DNA repair, and mRNA packaging.25,26 Recent studies indicated that YBX1 has a critical function in tumor advancement. For example, Xu et al discovered that YBX1 encouraged bladder cancer growth through increasing glycolysis expression.27 Xu et al demonstrated that high YBX1 indicated a poor prognosis in nasopharyngeal carcinoma and promoted tumor metastasis in vitro.27 Lim et al demonstrated that YBX1 suppression decreased breast cancer cell migration and invasion abilities by CORO1C.28 Nevertheless, the role and underlying mechanisms of YBX1 in RCC remains unknown. We found that YBX1 was increased and correlated with metastasis, and poor prognosis in RCC patients. YBX1 suppression inhibited RCC cells’ invasion ability in vitro. Subsequently, we validated that YBX1 is directly targeted by miR-382. Moreover, we demonstrated that miR-382 inhibitors eradicated the influence of si-circ-SAR1A on RCC cells’ growth and invasion in vitro. Thus, we exhibited that circ-SAR1A may function as a ceRNA to control YBX1 by sponging miR-382 in RCC progression.
In conclusion, our data suggested that circ-SAR1A knockdown suppressed RCC cells’ growth and invasion by controlling the miR-382/YBX1 axis. These data suggest that circ-SAR1A may function as a promising treatment target for RCC management.
Disclosure
The authors declare that they have no competing financial or non-financial interests.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2016. CA Cancer J Clin. 2016;66(1):7–30. doi:10.3322/caac.21332
2. Escudier B, Eisen T, Stadler WM, et al. Sorafenib in advanced clear-cell renal-cell carcinoma. N Engl J Med. 2007;356(2):125–134. doi:10.1056/NEJMoa060655
3. Cohen HT, McGovern FJ. Renal-cell carcinoma. N Engl J Med. 2005;353(23):2477–2490. doi:10.1056/NEJMra043172
4. Ljungberg B, Cowan NC, Hanbury DC, et al. EAU guidelines on renal cell carcinoma: the 2010 update. Eur Urol. 2010;58(3):398–406. doi:10.1016/j.eururo.2010.06.032
5. Ljungberg B, Campbell SC, Cho HY, et al. The epidemiology of renal cell carcinoma. Eur Urol. 2011;60(4):615–621. doi:10.1016/j.eururo.2011.06.049
6. Granados-Riveron JT, Aquino-Jarquin G. The complexity of the translation ability of circRNA. BBA-Gene Regul Mech. 2016;1859(10):1245–1251. doi:10.1016/j.bbagrm.2016.07.009
7. Haque S, Harries L. Circular RNAs (circRNAs) in health and disease. Genes. 2017;8(12):353.
8. Patop IL, Kadener S. circRNAs in cancer. Curr Opin Genet Dev. 2018;48:121–127. doi:10.1016/j.gde.2017.11.007
9. Zong L, Sun Q, Zhang H, et al. Increased expression of circRNA_102231 in lung cancer and its clinical significance. Biomed Pharmacother. 2018;102:639–644. doi:10.1016/j.biopha.2018.03.084
10. Ge Z, Li LF, Wang CY, et al. CircMTO1 inhibits cell proliferation and invasion by regulating Wnt/beta-catenin signaling pathway in colorectal cancer. Eur Rev Med Pharmacol Sci. 2018;22(23):8203–8209. doi:10.26355/eurrev_201812_16513
11. Xue M, Li G, Fang X, et al. hsa_circ_0081143 promotes cisplatin resistance in gastric cancer by targeting miR-646/CDK6 pathway. Cancer Cell Int. 2019;19(1):25. doi:10.1186/s12935-019-0737-x
12. Yang F, Zhang H, Chen SJ, et al. MiR-506 is down-regulated in clear cell renal cell carcinoma and inhibits cell growth and metastasis via targeting FLOT1. PLoS One. 2015;10(3):e0120258. doi:10.1371/journal.pone.0120258
13. Chen L, Nan A, Zhang N, et al. Circular RNA 100146 functions as an oncogene through direct binding to miR-361-3p and miR-615-5p in non-small cell lung cancer. Mol Cancer. 2019;18(1):13. doi:10.1186/s12943-019-0943-0
14. Zhong Y, Du Y, Yang X, et al. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol Cancer. 2018;17(1):79. doi:10.1186/s12943-018-0827-8
15. Meng S, Zhou H, Feng Z, et al. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer. 2017;16(1):94. doi:10.1186/s12943-017-0663-2
16. Kristensen LS, Hansen TB, Venø MT, et al. Circular RNAs in cancer: opportunities and challenges in the field. Oncogene. 2018;37(5):555. doi:10.1038/onc.2017.361
17. Wang G, Xue W, Jian W, et al. The effect of Hsa_circ_0001451 in clear cell renal cell carcinoma cells and its relationship with clinicopathological features. J Cancer. 2018;9(18):3269. doi:10.7150/jca.25902
18. Zhou B, Zheng P, Li Z, et al. CircPCNXL2 sponges miR-153 to promote the proliferation and invasion of renal cancer cells through upregulating ZEB2. Cell Cycle. 2018;17(23):2644–2654. doi:10.1080/15384101.2018.1553354
19. Xiong Y, Zhang J, Song C. CircRNA ZNF609 functions as a competitive endogenous RNA to regulate FOXP4 expression by sponging miR‐138‐5p in renal carcinoma. J Cell Physiol. 2019;234(7):10646–10654. doi:10.1002/jcp.27744
20. Shen Q, He T, Yuan H. Hsa_circ_0002577 promotes endometrial carcinoma progression via regulating miR-197/CTNND1 axis and activating Wnt/β-catenin pathway. Cell Cycle. 2019;18(11):1229–1240. doi:10.1080/15384101.2019.1617004
21. Shao B, He L. Hsa_circ_0001742 promotes tongue squamous cell carcinoma progression via modulating miR-634 expression. Biochem Biophys Res Commun. 2019;513(1):135–140. doi:10.1016/j.bbrc.2019.03.122
22. Chen T, Ren H, Thakur A, et al. miR-382 inhibits tumor progression by targeting SETD8 in non-small cell lung cancer. Biomed Pharmacother. 2017;86:248–253. doi:10.1016/j.biopha.2016.12.007
23. Zhou B, Song J, Han T, et al. MiR‐382 inhibits cell growth and invasion by targeting NR2F2 in colorectal cancer. Mol Carcinog. 2016;55(12):2260–2267. doi:10.1002/mc.22466
24. Qi B, Lu JG, Yao WJ, et al. Downregulation of microRNA-382 is associated with poor outcome of esophageal squamous cell carcinoma. World J Gastroenterol. 2015;21(22):6884. doi:10.3748/wjg.v21.i22.6884
25. Kohno K, Izumi H, Uchiumi T, et al. The pleiotropic functions of the Y‐box‐binding protein, YB‐1. Bioessays. 2003;25(7):691–698. doi:10.1002/bies.10300
26. Eliseeva IA, Kim ER, Guryanov SG, et al. Y-box-binding protein 1 (YB-1) and its functions. Biochemistry. 2011;76(13):1402–1433. doi:10.1134/S0006297911130049
27. Xu L, Li H, Wu L, et al. YBX1 promotes tumor growth by elevating glycolysis in human bladder cancer. Oncotarget. 2017;8(39):65946. doi:10.18632/oncotarget.19583
28. Lim JP, Shyamasundar S, Gunaratne J, et al. YBX1 gene silencing inhibits migratory and invasive potential via CORO1C in breast cancer in vitro. BMC Cancer. 2017;17(1):201. doi:10.1186/s12885-017-3187-7
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.