Back to Journals » OncoTargets and Therapy » Volume 13
Circ-ELF2 Acts as a Competing Endogenous RNA to Facilitate Glioma Cell Proliferation and Aggressiveness by Targeting MiR-510-5p/MUC15 Signaling
Authors Zhang W, Xu CM , Guo J, Guo L, Huo H, Wang H
Received 16 August 2020
Accepted for publication 17 September 2020
Published 9 October 2020 Volume 2020:13 Pages 10087—10096
DOI https://doi.org/10.2147/OTT.S275218
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Gaetano Romano
Weixin Zhang,1 ChunMiao Xu,2 Jianyu Guo,3 Limei Guo,2 Hongliang Huo,4 Huan Wang5
1Department of Nursing, Qiqihar Medical University, Qiqihar, 161000, China; 2Department of Nursing of Medicine and Surgery, Qiqihar Medical University, Qiqihar, 161000, China; 3Department of Orthopedics Surgery, The First Affiliated Hospital of Qiqihar Medical University, Qiqihar, 161000, China; 4Department of Nursing, The Second Affiliated Hospital of Qiqihar Medical University, Qiqihar, 161000, China; 5Department of Community Nursing, Qingdao Health School of Shandong Province, Qingdao, 266000, China
Correspondence: ChunMiao Xu Tel +86 452 2664572
Email [email protected]
Objective: Glioma (GM) is a common type of malignant and aggressive tumor in brain with poor prognosis. Circular RNAs (circRNAs) are well-known regulators in cancer progression. However, its molecular basis in GM remains to be investigated.
Materials and Methods: CircRNA microarray was used to detect differentially expressed circRNAs in GM and matched noncancerous tissues. qRT-PCR was applied to detect the expression profile of circ-ELF2 in GM tissue specimens and cell lines. CCK-8, clone formation, AO/EB staining, flow cytometry, wound healing, and transwell assays were performed to identify the functions of circ-ELF2 in GM cells. The distribution of circ-ELF2 was analyzed by RNA-FISH and subcellular fractionation assay. Dual-luciferase reporter assay was applied to verify the predicted binding sites between miR-510-5p and circ-ELF2/MUC15 3ʹ-UTR. Rescue assay was finally conducted to explore whether the oncogenic role of circ-ELF2 was partially attributed to miR-510-5p/MUC15 signaling.
Results: We observed that circ-ELF2 was significantly upregulated in GM tissues, which was analyzed by circRNA microarray and qRT-PCR. Upregulation of circ-ELF2 was associated with poor prognosis and high recurrence rate for GM patients after surgery. The collapse of circ-ELF2 caused growth arrest and downregulation of cell migratory and invasive potential of GM cells and promoted cell apoptosis. In contrast, elevated expression of circ-ELF2 led to the opposite effect. Mechanistically, circ-ELF2 acted as a competing endogenous RNA (ceRNA) for miR-510-5p to positive modulate MUC15 expression at posttranscriptional level. Circ-ELF2 upregulated MUC15 by sponging miR-510-5p, thus promoting GM growth and aggressiveness.
Conclusion: This study indicates that circ-ELF2/miR-510-5p/MUC15 signaling plays a key role in promoting the occurrence and development of GM.
Keywords: glioma, circRNA, circ-ELF2, miR-510-5p, MUC15
Introduction
Glioma (GM) is a common type of malignant and aggressive tumor in brain, which can be divided into astrocytomas, oligodendrogliomas, and glioblastomas according to morphological criteria.1,2 Although a combination of surgical resection, radiotherapy and adjuvant chemotherapy provides survival benefit to selected patients,3 the prognosis of GM remains dismal. Due to the blooming of genomic, transcriptomic and epigenetic profiling, molecular understandings concerning the initiation and progression of GM will help to provide novel and potential therapeutic targets.4
Circular RNAs (circRNAs) are not simply splicing products but a new member of noncoding RNAs (ncRNAs) derived from exons by nonclassical back-splicing, hence producing the covalent closed-loop structures that endow circRNAs higher stability than linear RNAs.5,6 Previous publications have largely highlighted the regulatory functions of circRNAs in cancers, such as circRNA_0084043 being a growth-promoting molecule in malignant melanoma7 and hsa_circ_0007059 being a tumor suppressor in lung cancer.8 In this study, circRNA microarray was used to detect the differentially expressed circRNAs in four pairs of GM tissues and adjacent noncancerous samples. The biological role of circ-ELF2 (hsa_circRNA_103741) in GM was detailly analyzed. Circ-ELF2 is mapped to chr4:139,981,477–139,994,721. The spliced length of circ-ELF2 is 919 nt. The detailed functions and mechanisms of circ-ELF2 have not been studied yet.
Materials and Methods
Ethics and Samples
All the experiments were strictly followed by the Helsinki Declaration concerning the biomedical principles of human subjects and all operating protocols were authorized by the Ethical Committee of Qiqihar Medical University. Ninety-six patients who were diagnosed with GM have enrolled in our study and signed the written informed consent for this research purpose. According to the median expression of circ-ELF2, these patients were classified into high/low expression groups. These specimens were snap-frozen in liquid nitrogen for 5 min and then stably saved in a −80°C ultra-low temperature freezer.
Cell Lines and Culture
GM cells (U87MG, U118, U251, and LN229) and NHA cells were acquired from American Type Cell Collection (Manassas, VA, USA). All the cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; HyClone, Logan City, UT, USA) containing 10% fetal bovine serum (FBS; HyClone) at 37°C in a humidified atmosphere.
qRT-PCR and Cell Transfection
Total RNA was extracted from cell lines or tissues by Trizol. The content of RNA was determined by ultraviolet spectrophotometry. The total RNA was synthesized into cDNA by reverse transcription kit and PCR reaction was carried out according to the instructions of qRT-PCR kit. U6 was taken as the internal reference for the expression level of miRNAs and the circ-ELF2/MUC15 mRNA expression level was taken GAPDH as the internal reference, and the qRT-PCR results were expressed as 2−ΔΔCT. PCR primers for circ-ELF2 are listed below: Forward, 5ʹ-CACTGCATCTGTGTCAGCAAC-3ʹ and Reverse, 5ʹ-CTCAAGCAGGTAGGAGATTCCA-3ʹ.
Twenty-four hours before cell transfection, the cells in good growth condition were inoculated in a 6-well plate (5×105 cells per well), and the cells were transfected according to the instructions of LipofectamineTM 3000. The targeted sequences of the shRNAs specifically targeting to circ-ELF2 are listed as follows: sh-circ-ELF2-1, 5ʹ-AGCAGCTCCAAGTGGAAGCAT-3ʹ and sh-circ-ELF2-2, 5ʹ-CAGCAGCTCCAAGTGGAAGCA-3ʹ.
Cell Counting Kit-8 (CCK-8) and Clone-Forming Assay
CCK-8 (Sigma-Aldrich) was employed for cell proliferation detection. The transfected GM cells were supplemented with CCK-8 solution with 10 μL per well, followed by the absorbance analysis via the microplate reader at 450 nm. Human GM cells (LN229 and U87MG cells) were seeded in 25-mm plates, incubated at 37°C for 12 d, fixed with paraformaldehyde for 20 min and stained with crystal violet for 15 min. The number of colonies was counted manually.
Cell Apoptosis Determination
To detect cell apoptosis, we conducted AO/EB staining assay in GM cells. GM cells were seeded in 12-wells plates. Then, the cells were stained by a mixed AO/EB solution for 5 min as per the manufacturer’s instructions. Images were acquired and cell counting was performed by Image Pro Plus software. For flow cytometric assay, 1 × 105 transfected cells were collected to perform the double staining of FITC-Annexin and PI via FITC-Annexin V Apoptosis Detection Kit (BD Biosciences) in accordance with the users’ manual.
Wound Healing Assay
The cells were cultivated in 6-well plates (Corning, NY, USA) until 100% confluence. A tip was used to create a cell-free region, and the medium was discarded. The cells were further cultured for 1 d with serum-free DMEM medium. Five fields at the lesion border were randomly selected and observed under an inverted microscope (Leica, Germany).
Transwell Experiments
Boyden chambers (Corning, NY, USA) were coated with or without Matrigel and used to study the cell migration and invasion. LN229 and U87MG cells were suspended in 200 μL serum-free DMEM medium, and seeded in the top of each chamber insert. The bottom chambers contained complete medium with 10% FBS. After 24 hours of culturing, paraformaldehyde was fixed, and the staining solution was 0.2% crystal violet solution. Five fields were randomly selected and observed under an inverted microscope (Leica, Germany).
Subcellular Fractionation Assay
LN229 and U87MG cell lines were collected in cell fractionation buffer using PARIS™ Kit (Invitrogen, USA). PBS washed the cell fragments. Cell fractionation buffer was used to place the cell lysates and centrifuged. The isolated RNA was used for PCR amplification.
Dual-Luciferase Reporter Gene Assay
The wild-type circ-ELF2 (circ-ELF2-wt) and the wild-type MUC15 (MUC15-wt) with predicted binding site of miR-510-5p and the promoter region of circ-ELF2-mut and MUC15-mut were synthesized and cloned into luciferase reporter vector pGL3 (Promega, Madison, WI). 293T cells were cotransfected with corresponding reporter plasmids and miR-510-5p mimics or mimics-NC by Lipofectamine 3000, and were detected by dual-luciferase reporter system.
RNA Immunoprecipitation (RIP)
EZ-Magna RIP™ RNA-Binding Protein Immunoprecipitation Kit (Millipore) was used for RIP assay. Cells were lysed in RIP lysis buffer, and incubated with magnetic beads conjugated with anti-Ago2 or anti-IgG. The immunoprecipitated RNA samples were then subjected to qRT-PCR analysis.
Immunoblotting
Total protein was isolated from human GM cells (LN229 and U87MG) by using a protein purification kit (Biochain, Hayward, CA, USA). Forty μg of protein was separated by SDS-PAGE and transferred onto polyvinylidene fluoride membranes (PVDF, Millipore, Bradford, MA, USA). The membrane was blocked in TBST buffer for 2 h and then reacted with primary antibodies against MUC15 or GAPDH. The membrane was then washed three times in TBST buffer and incubated with secondary antibody for 2 h at 37°C. The bands were viewed using enhanced chemiluminescence (ECL, Pierce, USA) and protein intensity was evaluated by ImageJ software (Madison, WI, USA).
Data Analysis
All data were expressed as mean ± standard deviation (SD). Data analysis was performed according to the overall characteristics of each group via the t-test. One-way analysis of variance (ANOVA) was used to analyze more than two groups. Kaplan–Meier analysis with Log rank test was used to analyze the clinical value of circ-ELF2. Pearson’s correlation coefficient test was applied to explore the association between the expression levels of RNAs. P < 0.05 was deemed statistically significant.
Results
Circ-ELF2 is Overexpressed in GM and is Correlated with Poor Prognosis
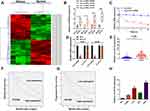
To identify the abnormally expressed circRNAs in GM, we performed circRNA microarray (Table S1) to detect the differentially expressed circRNAs in GM tissues and the clustered heatmap is shown in Figure 1A. Hsa_circRNA_000552, hsa_circRNA_104798, hsa_circRNA_000543, and hsa_circRNA_103741 were identified as four most elevated circRNAs in GM tissue samples. qRT-PCR was used to investigate the levels of these circRNAs in 20 pairs of GM/adjacent normal samples. We identified hsa_circRNA_103741 (circ-ELF2) as the most upregulated one (Figure 1B). Compared to the RNA abundance at 0 h post-treatment of Actinomycin D, ELF2 mRNA level was quickly decreased 70% while circ-ELF2 was almost unchanged at 24 h (Figure 1C). After the exposure of RNA to RNase R, circ-ELF2 was shown to have higher resistance to exonucleolytic activity than ELF2 mRNA (Figure 1D). Preliminarily, we affirmed the upregulation of circ-ELF2 and its high stability in GM cells. In addition, qRT-PCR analysis demonstrated that circ-ELF2 expression was higher in GM specimens than in non-tumorous samples (Figure 1E, P < 0.01). According to the median expression of circ-ELF2, these patients were classified into high and low expression groups. Kaplan–Meier analysis indicated that the patients with high circ-ELF2 expression correlated with worse overall survival (Figure 1F) and high recurrence rate (Figure 1G). Circ-ELF2 expression was generally elevated in GM cells than NHA cell line (Figure 1H). Taken together, these results indicated that high circ-ELF2 expression is associated with poor prognosis, highlighting the potential of circ-ELF2 as an oncogene in GM.
Circ-ELF2 Accelerates GM Cell Progression in vitro
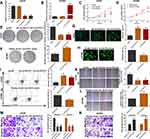
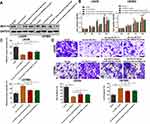
In the knockdown of circ-ELF2 mediated by shRNA vectors in LN229 cells, sh-circ-ELF2-1 had a better knockdown efficiency than sh-circ-ELF2-2 (Figure 2A). Overexpression study was performed on U87MG cells because of its lowest expression of circ-ELF2 among the enrolled GM cells (Figure 2B). The inhibition of cell viability (Figure 2C) and colony formation (Figure 2E) suggested that the downregulated circ-ELF2 impeded cell growth. In contrast, overexpression of circ-ELF2 significantly enhanced the proliferation of U87MG cells (Figure 2D and F). AO/EB double staining (Figure 2G) and flow cytometric analysis (Figure 2I) showed that decreased circ-ELF2 led to more apoptotic cells in LN229 cells. However, cell apoptosis was inhibited after circ-ELF2 overexpression in U87MG cells (Figure 2H and J). Additionally, transfection of sh-circ-ELF2-1 or sh-circ-ELF2-2 blocked the migration of LN229 cells analyzed by wound healing and transwell migration assays (Figure 2K and M). Conversely, ectopic expressed circ-ELF2 significantly contributed to cell migration in U87MG cells (Figure 2L and N). Moreover, transwell invasion assay manifested that downregulated circ-ELF2 attenuated LN229 cell invasion (Figure 2M). Upregulation of circ-ELF2 aggravated the invasive potential of U87MG cells (Figure 2N).
Circ-ELF2 Acts as MiRNA Sponge for MiR-510-5p in GM
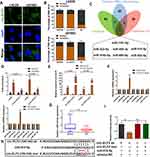
CircRNAs could recruit miRNAs to function as ceRNAs during oncogenesis.9 To verify whether circ-ELF2 had a similar function in GM cells, RNA-FISH and subcellular fractionation assay was used to predict its localization in cells. Data showed that circ-ELF2 was mainly localized to the cytoplasm (Figure 3A and B), indicating that circ-ELF2 might regulate target protein expression at posttranscriptional level. Next, we used the bioinformatics databases including starBase (http://starbase.sysu.edu.cn/), circBank (http://www.circbank.cn), and circular RNA interactome (https://circinteractome.nia.nih.gov) to predict the potential miRNAs. According to the predicted result, there were seven miRNAs (miR-432-5p, miR-495-3p, miR-510-5p, miR-516b-5p, miR-660-5p, miR-942-5p, and miR-1251-5p) that may be interacted with circ-ELF2 (Figure 3C). RIP assay indicated that circ-ELF2 could be co-immunoprecipitated with Ago2 in LN229 and U87MG cells. In addition, this phenotype was abrogated upon circ-ELF2 depletion (Figure 3D). Circ-ELF2 upregulation boosted the efficiency of pulldown using biotin-labeled circ-ELF2 probe (Figure 3E). What is more, only miR-510-5p was enriched in the circ-ELF2 pulldowns from GM cells (Figure 3F). Therefore, we regarded miR-510-5p as the main candidate for further research. We identified the downregulation of miR-510-5p in GM tissues relative to normal specimens (Figure 3G). To verify the direct binding of circ-ELF2 and miR-510-5p at the endogenous level, luciferase reporter analysis was constructed, which included circ-ELF2 binding sites of wild-type (wt) or mutant (mut) syndrome (Figure 3H). The results manifested that miR-510-5p mimics decreased the luciferase intensity of circ-ELF2-wt reporter vector. However, the luciferase activity of circ-ELF2-mut reporter vector did not decrease (Figure 3I).
MUC15 is a Downstream Target of MiR-510-5p
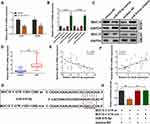
TargetScan database (http://www.targetscan.org/vert_72/) was applied to predict the potential target gene of miR-510-5p. We found that decreased circ-ELF2 strikingly attenuated the expression of MUC15 in LN229 and U87MG cell lines (Figure 4A). qRT-PCR and immunoblotting were applied to evaluate the expression of MUC15 in GM cells upon miR-510-5p silencing/overexpression. As Figure 4B and C exhibited, miR-510-5p negatively modulated MUC15 expression. In addition, we found that MUC15 mRNA expression was elevated in GM samples than their matched noncancerous specimens (Figure 4D). Pearson correlation coefficient test indicated a negative correlation between the expression of miR-510-5p and MUC15 mRNA (Figure 4E). Additionally, a positive association was found in circ-ELF2 and MUC15 mRNA expression (Figure 4F). To verify the direct binding of MUC15 3ʹ-UTR and miR-510-5p at the endogenous level, luciferase reporter analysis was constructed, which included MUC15 binding sites of wt or mut syndrome. The data indicated that miR-510-5p mimics inhibited the luciferase intensity of MUC15 3ʹ-UTR-wt reporter vector. Nevertheless, the luciferase intensity of MUC15 3ʹ-UTR-mut reporter vector did not change (Figure 4G and H).
Circ-ELF2 Facilitates Cell Progression by Targeting MiR-510-5p/MUC15 Signaling
We then determined whether circ-ELF2 could affect the expression of MUC15 through competitive binding with miR-510-5p, rescue experiments were designed. As expected, Western blotting indicated that the silencing of circ-ELF2 decreased the protein levels of MUC15 in LN229 cells, while upregulation of circ-ELF2 enhanced the levels of MUC15 in U87MG cells. Simultaneously, the effects caused by silencing or overexpressing circ-ELF2 were partly reversed by miR-510-5p inhibitor or mimics, respectively. MUC15 vector could also rescue the decreased MUC15 expression induced by sh-circ-ELF2-1. Co-transfection with circ-ELF2 vector and sh-MUC15 could partially reverse the upregulated MUC15 in U87MG cells (Figure 5A). CCK-8, clone formation, and transwell invasion assays showed that miR-510-5p inhibitor or MUC15 vector reversed the proliferation, colony formation, and invasion inhibition effects induced by knockdown of circ-ELF2 in LN229 cells, whereas miR-510-5p mimics/sh-MUC15 counter-acted the promoting effects induced by overexpression of circ-ELF2 in U87MG cells by CCK-8, colony formation, and transwell invasion assays (Figure 5B–D). Collectively, rescue assays illustrated that circ-ELF2 served as a ceRNA for miR-510-5p to upregulate MUC15 expression, thus leading to the progression and development of GM.
Discussion
GM tumorigenesis is induced by a series of genetic changes resulting in the activation of oncogenes and the inactivation of tumor suppressor genes.10 Dysregulated circRNAs have been validated to play the oncogenic or tumor-suppressive roles in cancers.11,12 Through the characteristic analysis in GM cells, we found that circ-ELF2 was expressed more stable than linear ELF2 mRNA and was mainly localized in the cytoplasm. For its clinical significance, the upregulated circ-ELF2 was closely associated with poor prognosis and high recurrence rate for GM patients. All these elementary analyses manifested that circ-ELF2 might be involved in the regulation of cancer development and progression in GM.
Starting from these observations, we conducted gain-/loss-of function experiments to uncover the functions of circ-ELF2 in GM cells. As a result, we found circ-ELF2 contributes to the proliferation, colony formation, migration, and invasion, but inhibits the apoptosis of GM cells. Increasing evidence shows that there are extensive interaction networks involving ceRNA, in which circRNAs could regulate target gene expression by binding to the specific miRNAs.7–9 Evidence suggests that ceRNA regulatory model has been validated in multiple cancers. For example, circ-FAT1(e2) facilitates the progression of osteosarcoma through the miR-181b/HK2 axis.13 Circ-MTO1 suppresses ovarian cancer cell progression and metastasis by sponging miR-182-5p and upregulating KLF15 expression.14 In the present study, miR-510-5p was identified as a target for circ-ELF2 among seven candidate miRNAs. MiR-510-5p was previously identified as a tumor-suppressive miRNA in several human malignancies, such as renal cell carcinoma.15 However, a recent study indicated miR-510-5p as an oncogene in mediating thyroid cancer cell proliferation, migration, and invasion through suppressing SNHG15.16 The above evidence suggests a tissue-specific mechanism employed by miR-510-5p. In this study, miR-510-5p functions as a tumor suppressor in GM cells, which is in line with most studies in human cancers. What is more, we identified MUC15 as the direct target of miR-510-5p. Mucins are high-molecular-weight membrane glycoproteins (>200 kDa) in various types of epithelial cells.17,18 Studies showed that membrane-associated mucin proteins are intracellular receptors involved in signal transduction, leading to coordinated cellular responses.19,20 MUC15 is one of the member of mucins family. It was identified upregulation in multiple malignancies including colorectal carcinoma,21 liver cancer,22 etc. In the current study, the rescue analysis demonstrated circ-ELF2 as an oncogene to accelerate the malignant behaviors of GM depending on the miR-510-5p/MUC15 signaling pathway. However, there are still some limitations to the study. For instance, animal study is needed to further confirm the in vitro data.
To sum up, we showed for the first time that circ-ELF2 exhibited its oncogenic function in GM through sponging miR-510-5p to mediate MUC15 expression. Therefore, the current work uncovered a novel oncogenic pathway in GM tumorigenesis, which will provide a novel insight into investigating the therapeutic target for GM.
Data Sharing Statement
The datasets used in the project are available from the corresponding author.
Funding
Research Foundation of Qiqihar Academy of Medical Sciences (Grant No. QMSI2019M-16).
Disclosure
The authors declare they have no conflicts of interest in the present project.
References
1. Laug D, Glasgow SM, Deneen B. A glial blueprint for gliomagenesis. Nat Rev Neurosci. 2018;19(7):393–403. doi:10.1038/s41583-018-0014-3
2. Shergalis A, Bankhead A, Luesakul U, et al. Current challenges and opportunities in treating glioblastoma. Pharmacol Rev. 2018;70(3):412–445.
3. Stupp R, Hegi ME, Mason WP, et al. Effects of radiotherapy with concomitant and adjuvant temozolomide versus radiotherapy alone on survival in glioblastoma in a randomised Phase III study: 5-year analysis of the EORTC-NCIC trial. Lancet Oncol. 2009;10(5):459–466. doi:10.1016/S1470-2045(09)70025-7
4. Alinezhad A, Jafari F. Novel management of glioma by molecular therapies, a review article. Eur J Transl Myol. 2019;29(3):8209. doi:10.4081/ejtm.2019.8209
5. Hsiao KY, Sun HS, Tsai SJ. Circular RNA-new member of noncoding RNA with novel functions. Exp Biol Med (Maywood). 2017;242(11):1136–1141. doi:10.1177/1535370217708978
6. Eger N, Schoppe L, Schuster S, et al. Circular RNA splicing. Adv Exp Med Biol. 2018;1087:41–52.
7. Luan W, Shi Y, Zhou Z, et al. circRNA_0084043 promote malignant melanoma progression via miR-153-3p/snail axis. Biochem Biophys Res Commun. 2018;502(1):22–29. doi:10.1016/j.bbrc.2018.05.114
8. Gao S, Yu Y, Liu L, et al. Circular RNA hsa_circ_0007059 restrains proliferation and epithelial-mesenchymal transition in lung cancer cells via inhibiting microRNA-378. Life Sci. 2019;233:116692. doi:10.1016/j.lfs.2019.116692
9. Xu Y, Kang P, Leng K, et al. Circ_ASPH promotes cholangiocarcinoma growth and metastasis through the miR-581/ATP-binding cassette transporter G1 signaling pathway. Cancer Commun (Lond). 2020. doi:10.1002/cac2.12083
10. Zhang S, Wang C, Ma B, et al. Mutant p53 drives cancer metastasis via RCP-mediated Hsp90α secretion. Cell Rep. 2020;32(1):107879. doi:10.1016/j.celrep.2020.107879
11. Xu Y, Yao Y, Zhong X, et al. Downregulated circular RNA hsa_circ_0001649 regulates proliferation, migration and invasion in cholangiocarcinoma cells. Biochem Biophys Res Commun. 2018;496(2):455–461. doi:10.1016/j.bbrc.2018.01.077
12. Xu Y, Yao Y, Liu Y, et al. Elevation of circular RNA circ_0005230 facilitates cell growth and metastasis via sponging miR-1238 and miR-1299 in cholangiocarcinoma. Aging (Albany NY). 2019;11(7):1907–1917. doi:10.18632/aging.101872
13. Gu H, Cheng X, Xu J, et al. Circular RNA circFAT1(e2) promotes osteosarcoma progression and metastasis by sponging miR-181b and regulating HK2 expression. Biomed Res Int. 2020;2020:3589871. doi:10.1155/2020/3589871
14. Wang N, Cao QX, Tian J, et al. Circular RNA MTO1 inhibits the proliferation and invasion of ovarian cancer cells through the miR-182-5p/KLF15 axis. Cell Transplant. 2020;29:963689720943613. doi:10.1177/0963689720943613
15. Sun J, Pan S, Cui H, et al. CircRNA SCARB1 promotes renal cell carcinoma progression via Mir- 510-5p/SDC3 axis. Curr Cancer Drug Targets. 2020;20(6):461–470. doi:10.2174/1568009620666200409130032
16. Liu Y, Li J, Li M, et al. microRNA-510-5p promotes thyroid cancer cell proliferation, migration, and invasion through suppressing SNHG15. J Cell Biochem. 2019. doi:10.1002/jcb.28454
17. Allen A, Flemstrom G. Gastroduodenal mucus bicarbonate barrier: protection against acid and pepsin. Am J Physiol Cell Physiol. 2005;288(1):C1–C19. doi:10.1152/ajpcell.00102.2004
18. Corfield AP, Myerscough N, Longman R, et al. Mucins and mucosal protection in the gastrointestinal tract: new prospects for mucins in the pathology of gastrointestinal disease. Gut. 2000;47:589–594. doi:10.1136/gut.47.4.589
19. Carraway KL, Ramsauer VP, Haq B, et al. Cell signaling through membrane mucins. Bioessays. 2003;25(1):66–71. doi:10.1002/bies.10201
20. Meerzaman D, Shapiro PS, Kim KC. Involvement of the MAP kinase ERK2 in MUC1 mucin signaling. Am J Physiol Lung Cell Mol Physiol. 2001;281(1):L86–L91. doi:10.1152/ajplung.2001.281.1.L86
21. Huang J, Che ML, Huang YT, et al. Overexpression of MUC15 activates extracellular signal-regulated kinase 1/2 and promotes the oncogenic potential of human colon cancer cells. Carcinogenesis. 2009;30:1452–1458. doi:10.1093/carcin/bgp137
22. Wang RY, Chen L, Chen HY, et al. MUC15 inhibits dimerization of EGFR and PI3K–AKT signaling and is associated with aggressive hepatocellular carcinomas in patients. Gastroenterology. 2013;145(6):1436–1448. doi:10.1053/j.gastro.2013.08.009
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.