Back to Journals » Infection and Drug Resistance » Volume 12
Characterization of the most common embCAB gene mutations associated with ethambutol resistance in Mycobacterium tuberculosis isolates from Iran
Authors Khosravi AD , Sirous M, Abdi M, Ahmadkhosravi N
Received 2 December 2018
Accepted for publication 1 February 2019
Published 6 March 2019 Volume 2019:12 Pages 579—584
DOI https://doi.org/10.2147/IDR.S196800
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Joachim Wink
Azar Dokht Khosravi,1,2 Mehrandokht Sirous,2 Mahtab Abdi,2 Nazanin Ahmadkhosravi3
1Infectious and Tropical Diseases Research Center, Health Research Institute, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran; 2Department of Microbiology, Faculty of Medicine, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran; 3Khuzestan Tuberculosis Regional Reference Laboratory, Ahvaz Jundishapur University of Medical Sciences, Ahvaz, Iran
Introduction: Ethambutol (Emb) is one of the first-line drugs in the standard combination therapy for tuberculosis; however, due to the rapid increase in Emb resistance among clinical isolates of Mycobacterium tuberculosis (MTB), early detection of Emb resistance is desirable. As the embCAB operon is considered involved in resistance to Emb, this study aimed to analyze the most common mutations within the embCAB operon among MTB isolates from Iran to find any correlations of these mutations with Emb resistance.
Methods: A total of 307 clinical isolates of MTB were screened for Emb resistance by phenotypic drug-susceptibility testing. PCR amplification was performed on extracted DNA from all Emb-resistant and randomly selected Emb-susceptible isolates using sets of primers for various gene loci of embC, embA, and embB, followed by sequencing for the detection of most common alterations.
Results: In total, ten isolates showed resistance to Emb by phenotypic susceptibility testing (3.25%). The mutation rate in ten Emb-resistant MTB strains was 20% (n=2), comprising one mutation in embB (10%), at codon 306 Met–Val and one in embC (10%) at codon 270 Thr–Ile. A nonsynonymous mutation in the embA gene in one of the randomly selected Emb-susceptible isolates located in codon 330 Leu–Leu was also noticed.
Conclusion: The majority of our Emb-resistant isolates (n=8, 80%) did not demonstrate the sequences investigated within the embCAB operon. As such, these mutations solely are insufficient for the development of complete resistance to Emb in MTB isolates. Additional mechanisms of resistance other than mutations in these sequences studied within the embCAB operon should also be considered.
Keywords: Mycobacterium tuberculosis, susceptibility testing, ethambutol, drug resistance, embCAB
Introduction
Tuberculosis (TB), caused by Mycobacterium tuberculosis (MTB), remains a life-threatening disease and is still one of the most important causes of morbidity and mortality worldwide.1 In the latest World Health Organization (WHO) global report on TB,2 there were an estimated 1.3 million TB deaths in 2017 and an additional 300,000 deaths resulting from TB disease among HIV-positive people. The worldwide emergence of drug-resistant MTB (DR-TB), especially multi-DR TB (MDR-TB) and extensive DR-TB, is widely considered to be a serious challenge for TB control programs.3,4 MDR-TB is caused by strains of MTB that do not respond to at least both isoniazid (Inh) and rifampicin (Rif), and extensive DR-TB is defined as an MDR isolate that is also resistant to a fluoroquinolone and at least one second-line injectable agent (amikacin, kanamycin, and capreomycin).3 According to the WHO report, there were an estimated 558,000 cases of MDR Rif-resistant TB in 2017.1 As such, the rapid detection of drug resistance is essential for appropriate treatment of the disease that avoids treatment failure and prevents the spread of DR strains.5
Ethambutol (Emb) is an important first-line drug that is widely used for the treatment of TB and MDR-TB.6,7 The mechanism of action of Emb is inhibition of the synthesis of the arabinan component of lipoarabinomannan and arabinogalactan in the mycobacterial cell wall.8 Arabinosyl transferase is encoded by the embCAB locus, which comprises the three genes of embC, embA and embB. embA and embB genes apparently contribute to the synthesis of arabinogalactan, whereas embC is reserved for the synthesis of lipoarabinomannan.9,10 Mutations in the 10 kb embCAB operon are associated with resistance to Emb, especially the embB gene.11 The most commonly detected mutations have occur at codons 306, 406, and 497 within the embB gene. Therefore, these codons represent promising diagnostic markers for the rapid detection of Emb resistance.12 Five distinct missense mutations are found in codon 306 with changes in the first or third base (ATG, GTG, CTG, ATA, ATC, or ATT), resulting in the replacement of methionine by three different amino acids (Met to Val, Leu, or Ile).13 As the embCAB operon (embC, embA, and embB) of MTB is considered involved in resistance to Emb,14 this study aimed to analyze the most common mutations within he embCAB operon among MTB isolates from Iran to find any correlation of these mutations with Emb resistance.
Methods
Sample collection and bacterial strains
A total of 307 clinical isolates of MTB were detected in 906 samples during a 16-month period from February 2016 to June 2017. The samples were obtained from patients with suspected pulmonary TB diagnosed by infectious-disease specialists and referred to the regional tuberculosis laboratory of Khuzestan province, Iran. The preliminary proposal of the work was approved in joint by the institutional review board and ethics committee of Ahvaz Jundishapur University of Medical Sciences, Iran, and necessary permission was granted for sample collection. Apart from this, as part of the regional center policy, referred patients were requested to sign the informed consent in case their samples were used for research purposes apart from routine clinical investigation, and our study was conducted in accordance with the Declaration of Helsinki. Isolates were identified as MTB on the basis of acid-fast-stain microscopy, growth in modified Löwenstein–Jensen (LJ) medium, and performance of conventional biochemical tests, including niacin accumulation, nitrate reductase, and catalase at 37°C and 68°C.15
Antimicrobial susceptibility testing
Antimicrobial susceptibility testing (AST) for first-line anti-TB drugs was performed on LJ medium using a proportional method according to the Clinical and Laboratory Standard Institute guideline.16 The following critical drug concentrations were used for AST: 0.25 µg/mL for Inh, 0.02 µg/mL for Rif, and 4 µg/mL for Emb (Sigma-Aldrich, St Louis, MO, USA). H37Rv (ATCC 27294) was used as the control for AST. Susceptibility was defined as no or <1% growth on LJ medium-containing drugs compared with the control medium.
DNA extraction
DNA was extracted from MTB grown on LJ medium by a simple boiling method as previously described.17 The concentration of extracted DNA was measured by biophotometry (Eppendorf, Hamburg, Germany) at 260 nm, and samples were stored at −20˚C until use.
PCR amplification
PCR amplification was performed in an Eppendorf thermocycler (Roche, Basel, Switzerland) by application of primers designed for various gene loci of embC, embA, and embB,18,19 as presented in Table 1.
  | Table 1 Oligonucleotide primer sequences employed for amplification of the embCAB genes |
Amplification reactions in a total volume of 50 µL containing 5 µL 10× PCR buffer, 1.5 mM MgCl2, 0.2 mM dNTP mix, 0.2 µM of each primer, 2.5 U Taq polymerase, and 5 µL template DNA (10 ng) were prepared. Amplification conditions were: embB gene – initial denaturation at 94°C for 5 minutes, followed by 30 cycles of denaturation at 94°C for 40 seconds, primer annealing at 48°C for 30 seconds, and extension at 72°C for 30 seconds, followed by a final extension at 72°C for 5 minutes; embA and embC genes – initial denaturation at 95°C for 5 minutes, followed by 30 cycles of denaturation at 95°C for 40 seconds, annealing at 63°C for 30 seconds, and extension at 72°C for 30 seconds, and a final extension at 72°C for 5 minutes. H37Rv (ATCC 27294) was used as a reference strain. PCR products were loaded on a 1.5% (w:v) agarose gel with 0.5 µg/mL ethidium bromide, analyzed by gel electrophoresis, and photographed using the gel-documentation system (ProteinSimple, San Jose, CA, USA). SPSS version 16 was used for data analysis. PCR products were sent to Bioneer (Daejeon, South Korea) for sequencing.
Results
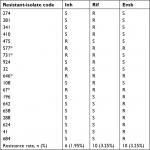
From the total 906 sputa processed, 307 (33.88%) were identified as MTB on the basis of culture and biochemical criteria. These belonged to 233 men and 74 women with a mean age of 33 years. In total, 26 isolates (8.47%) showed resistance to the antimicrobial agents tested. The resistance profiles of the 307 MTB isolates according to AST were Inh 6 (1.95%), Rif 10 (3.25%), and Emb 10 (3.25%). The prevalence of MDR was four (1.30%), three of which showed coresistance to Emb (Table 2).
The ten Emb-resistant and 28 randomly selected Emb susceptible isolates were processed for characterization of embCAB genes. According to the sequencing analyses, the mutation rate in the Emb-resistant MTB strains was 20% (n=2), comprising one mutation in embB (10%) at codon 306 Met–Val (ATG→GTG) and one in embC (10%) at codon 270 Thr–Ile (ACC→ATC). No mutation in embA was demonstrated in Emb-resistant isolates in this study (Table 3). Only one of the MDR strains with coresistance to Emb showed a mutation in embB. Moreover, we noticed a nonsynonymous mutation in the embA gene in one of the randomly selected Emb-susceptible isolates, located in codon 330 Leu–Leu (CTG→TTG). In Table 4, the mutations detected in all Emb-resistant and selected Emb-susceptible isolates are presented.
  | Table 3 Mutation pattern of embCAB genes in ten ethambutol-resistant isolates with different drug-resistance phenotypes Abbreviation: MDR, multidrug-resistant. |
Discussion
Emb is used as an alternative drug to streptomycin in the standard four-drug combination TB therapy; however, due to the rapid increase in Emb resistance among clinical isolates of MTB, early accurate detection of Emb resistance is essential to avoid the risk of adverse reactions, particularly optic neuritis.20 In this study, molecular methods based on embCAB-gene mutations were used to investigate the nature of Emb resistance in MTB isolates. DNA-sequencing analysis of embCAB loci in ten Emb-resistant and 26 Emb-susceptible isolates revealed three polymorphic nucleotide sites, each located in different regions of the three gene loci. We demonstrated only two mutations in Emb-resistant strains (20%), one a common mutation with alteration of methionine–valine at codon 306. Alterations at codon 306 of embB have been demonstrated as the most common alterations in Emb-resistant MTB clinical isolates.19,21 Moreover, high detection rates of mutations at codon embB306 among Emb-resistant MTB isolates have been reported in recent years from China (55%),22 Cuba and the Dominican Republic (70%),23 and Germany (68%).24
The second mutation occurred at codon 270 of embC, with alteration in threonine–isoleucine. This mutation has also been described previously with less frequency among Emb-resistant isolates of MTB. We have also identified a nonsense mutation in one Emb-susceptible isolate in the embA gene as Leu330Leu. The region of EmbB containing residue Met306 is highly conserved in MTB, Mycobacterium avium, Mycobacterium leprae, and M. smegmatis strains,25 and the data generated from the Emb-susceptibility testing indicate a strong nonrandom association between certain amino-acid substitutions at embB position 306 and the level of resistance to Emb.26 We did not find any other mutations in the rest of the Emb-resistant MDR strains, and our results suggest that embB306 and embC270 mutations do not cause Emb resistance in any MTB isolates, and other mutations, including the involvement of less common mutations in codons 406 and 497, should be considered for resistance to this antibiotic in isolates. Moreover, we cannot rule out a contribution of mutations outside the embCAB operon that could have been coselected during growth in the presence of Emb. Gene mutations only modestly increase resistance to Emb in MTB isolates. Our findings were in line with a previous study conducted by Safi et al,27 demonstrating that although embB306 mutations are necessary for Emb resistance, no resistant strains were detected by these mutations.
Although in this study the large number of MTB isolates (n=307) was investigated, the number of Emb-resistant isolates was quite low (n=10, 3.25%). However, according to previous reports from Iran, the resistance rate of MTB to Emb varied widely: from 1% in Tavanaee et al28 and 4.2% in Seif et al29 to as high as 14% in Farazi et al.30 Similarly, the prevalence of MDR in our study was also low (n=4, 1.30%), three of which showed coresistance to Emb as well, and in only one Emb-resistant MDR isolate did we find the 306 embB mutation.
Among the Emb phenotypic resistant isolates, only two were detected by mutations within the embCAB operon. The discordance between phenotypic AST of Emb and mutations in the embCAB operon has also been reported by investigators from other countries and Iran.31–33 In the study from Iran, embB mutations were investigated in a few Iranian cities, and based on their results from four Emb-resistant isolates recovered from Tehran, only one 306 mutation was detected, and in some cities no 306 mutation was demonstrated among the phenotypic Emb-resistant isolates.33 In the current study, we investigated the most common mutation sites within the three genes embA, embB, and embC. Further studies should investigate other mutations within and outside this operon over an extended period, as other investigators have concluded that a few mutations conferring Emb resistance may occurred outside this operon.13
Conclusion
Few of our Emb-resistant isolates (n=8, 80%) were demonstrated by mutations in investigated sequences within the embCAB operon. As such, these mutations alone are insufficient for the development of full resistance to Emb in MTB strains. Additional mechanisms of resistance other than mutations in the current studied sequences within the embCAB operon should also be considered. Although this study demonstrates a low level of Emb resistance in our region, for the control of drug-resistance spread, regular monitoring is necessary to maintain a lower resistance rate, which is crucial for the treatment strategy and management of TB.
Acknowledgments
This work is part of the MSc thesis of Mahtab Abdi, which was approved by the Infectious and Tropical Diseases Research Center, which is appreciated. We are grateful to Research Affairs at the university for financial support of the study (grant OG-95126). We are also greatly thankful to the staff of regional TB reference laboratories for their collaboration in sample collection.
Disclosure
The authors report no conflicts of interest in this work.
References
WHO. Drug resistant tuberculosis. Geneva: World Health Organization; 2018. Available from: https://www.who.int/tb/areas-of-work/drug-resistant-tb/en. Accessed September 26, 2018. | ||
WHO. Global tuberculosis report. Geneva: World Health Organization; 2018. Available from: https://www.who.int//iris/bitstream/handle/10665/274453/9789241565646-eng.pdf. Accessed September 26, 2018. | ||
Lange C, Abubakar I, Alffenaar JW, et al. Management of patients with multidrug-resistant/extensively drug-resistant tuberculosis in Europe: a TBNET consensus statement. Eur Respir J. 2014;44(1):23–63. | ||
Kerantzas CA, Jacobs WR. Origins of combination therapy for tuberculosis: lessons for future antimicrobial development and application. MBio. 2017;8(2):e01586–16. | ||
Brossier F, Veziris N, Aubry A, Jarlier V, Sougakoff W. Detection by GenoType MTBDRsl test of complex mechanisms of resistance to second-line drugs and ethambutol in multidrug-resistant Mycobacterium tuberculosis complex isolates. J Clin Microbiol. 2010;48(5):1683–1689. | ||
Caminero JA, Sotgiu G, Zumla A, Migliori GB. Best drug treatment for multidrug-resistant and extensively drug-resistant tuberculosis. Lancet Infect Dis. 2010;10(9):621–629. | ||
Daley CL, Caminero JA. Management of multidrug resistant tuberculosis. Semin Respir Crit Care Med. 2013;34(1):44–59. | ||
Jackson M, Mcneil MR, Brennan PJ. Progress in targeting cell envelope biogenesis in Mycobacterium tuberculosis. Future Microbiol. 2013;8(7):855–875. | ||
Mikusová K, Slayden RA, Besra GS, Brennan PJ. Biogenesis of the mycobacterial cell wall and the site of action of ethambutol. Antimicrob Agents Chemother. 1995;39(11):2484–2489. | ||
Jankute M, Grover S, Rana AK, Besra GS. Arabinogalactan and lipoarabinomannan biosynthesis: structure, biogenesis and their potential as drug targets. Future Microbiol. 2012;7(1):129–147. | ||
Parsons LM, Salfinger M, Clobridge A, et al. Phenotypic and molecular characterization of Mycobacterium tuberculosis isolates resistant to both isoniazid and ethambutol. Antimicrob Agents Chemother. 2005;49(6):2218–2225. | ||
Wang T, Jiao WW, Shen AD. Progress on mechanism of ethambutol resistance in Mycobacterium Tuberculosis. Yi Chuan. 2016;38(10):910–917. | ||
Sugawara I, Otomo K, Yamada H, et al. The molecular epidemiology of ethambutol-resistant Mycobacterium tuberculosis in Henan Province, China. Jpn J Infect Dis. 2005;58(6):393–395. | ||
Zhao LL, Sun Q, Liu HC, et al. Analysis of embCAB mutations associated with ethambutol resistance in multidrug-resistant Mycobacterium tuberculosis isolates from China. Antimicrob Agents Chemother. 2015;59(4):2045–2050. | ||
Kent PT, Kubica GP. Public Health Mycobacteriology: A Guide for the level III Laboratory. Atlanta, GA: Centers for Disease Control, U.S. Department of Health and Human Services; 1985. | ||
Desmond E. Susceptibility testing for Mycobacteria, Nocardiae, and other Aerobic Actinomycetes; Approved Standard. CLSI Document M24-A2. 2nd ed. Wayne, PA: Clinical and Laboratory Standards Institute; 2011. | ||
Hosek J, Svastova P, Moravkova M, Pavlik I, Bartos M. Methods of mycobacterial DNA isolation from different biological material: a review. Veterinarni Medicina. 2012;51(No. 5):180–192. | ||
Xu Y, Jia H, Huang H, Sun Z, Zhang Z. Mutations found in embCAB, embR, and ubiA genes of ethambutol-sensitive and -resistant Mycobacterium tuberculosis clinical isolates from China. Biomed Res Int. 2015;2015:1–8. | ||
Tracevska T, Jansone I, Nodieva A, Marga O, Skenders G, Baumanis V. Characterisation of rpsL, rrs and embB mutations associated with streptomycin and ethambutol resistance in Mycobacterium tuberculosis. Res Microbiol. 2004;155(10):830–834. | ||
Frieden TR, Sterling TR, Munsiff SS, Watt CJ, Dye C. Tuberculosis. Lancet. 2003;362(9387):887–899. | ||
Ramaswamy SV, Dou SJ, Rendon A, Yang Z, Cave MD, Graviss EA. Genotypic analysis of multidrug-resistant Mycobacterium tuberculosis isolates from Monterrey, Mexico. J Med Microbiol. 2004;53(Pt 2):107–113. | ||
Shi D, Li L, Zhao Y, et al. Characteristics of embB mutations in multidrug-resistant Mycobacterium tuberculosis isolates in Henan, China. J Antimicrob Chemother. 2011;66(10):2240–2247. | ||
Guerrero E, Lemus D, Yzquierdo S, et al. Association between embB mutations and ethambutol resistance in Mycobacterium tuberculosis isolates from Cuba and the Dominican Republic: reproducible patterns and problems. Rev Argent Microbiol. 2013;45(1):21–26. | ||
Plinke C, Rüsch-Gerdes S, Niemann S. Significance of mutations in embB codon 306 for prediction of ethambutol resistance in clinical Mycobacterium tuberculosis isolates. Antimicrob Agents Chemother. 2006;50(5):1900–1902. | ||
Cui Z, Li Y, Cheng S, et al. Mutations in the embC-embA intergenic region contribute to Mycobacterium tuberculosis resistance to ethambutol. Antimicrob Agents Chemother. 2014;58(11):6837–6843. | ||
Srivastava S, Ayyagari A, Dhole TN, Nyati KK, Dwivedi SK. emb nucleotide polymorphisms and the role of embB306 mutations in Mycobacterium tuberculosis resistance to ethambutol. Int J Med Microbiol. 2009;299(4):269–280. | ||
Safi H, Sayers B, Hazbón MH, Alland D. Transfer of embB codon 306 mutations into clinical Mycobacterium tuberculosis strains alters susceptibility to ethambutol, isoniazid, and rifampin. Antimicrob Agents Chemother. 2008;52(6):2027–2034. | ||
Tavanaee Sani A, Shakiba A, Salehi M, Bahrami Taghanaki HR, Ayati Fard SF, Ghazvini K. Epidemiological Characterization of Drug Resistance among Mycobacterium tuberculosis Isolated from Patients in Northeast of Iran during 2012–2013. BioMed Res Int. 2015;2015(1):1–6. | ||
Seif S, Malekshahian D, Shamsi M. Prevalence of primary ethambutol resistance in new smear-positive pulmonary TB cases. Int J Mycobacteriol. 2015;4:171–172. | ||
Farazi A, Sofian M, Zarrinfar N, Katebi F, Hoseini SD, Keshavarz R. Drug resistance pattern and associated risk factors of tuberculosis patients in the central province of Iran. Caspian J Intern Med. 2013;4(4):785–789. | ||
Aung HL, Tun T, Moradigaravand D, et al. Whole-genome sequencing of multidrug-resistant Mycobacterium tuberculosis isolates from Myanmar. J Glob Antimicrob Resist. 2016;6:113–117. | ||
Casali N, Nikolayevskyy V, Balabanova Y, et al. Evolution and transmission of drug-resistant tuberculosis in a Russian population. Nat Genet. 2014;46(3):279–286. | ||
Rezaei F, Haeili M, Mohajeri P, et al. Frequency of mutational changes in the embB among the ethambutol-resistant strains of Mycobacterium tuberculosis in Iran. J Infect Dev Ctries. 2016;10(4):363–368. |
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.