Back to Journals » Infection and Drug Resistance » Volume 11
Characterization of rifampin-resistant Staphylococcus aureus nasal carriage in patients receiving rifampin-containing regimens for tuberculosis
Authors Huang YT , Liao CH , Chen SY , Yang CJ, Hsu HS, Teng LJ, Hsueh PR
Received 24 January 2018
Accepted for publication 10 April 2018
Published 14 August 2018 Volume 2018:11 Pages 1175—1182
DOI https://doi.org/10.2147/IDR.S163634
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Professor Suresh Antony
Yu-Tsung Huang,1–3 Chun-Hsing Liao,1,4 Shey-Ying Chen,5 Chia-Jui Yang,1,4 Hsin-Sui Hsu,1 Lee-Jene Teng,2,3 Po-Ren Hsueh2,6
1Department of Internal Medicine, Far Eastern Memorial Hospital, New Taipei City, Taiwan; 2Department of Laboratory Medicine, National Taiwan University Hospital, National Taiwan University College of Medicine, Taipei, Taiwan; 3Department of Clinical Laboratory Science and Medical Biotechnology College of Medicine, National Taiwan University, Taipei, Taiwan; 4Department of Internal Medicine, College of Medicine, Yang-Ming University, Taipei, Taiwan; 5Department of Emergency Medicine, National Taiwan University Hospital, National Taiwan University College of Medicine, Taipei, Taiwan; 6Department of Internal Medicine, National Taiwan University Hospital, National Taiwan University College of Medicine, Taipei, Taiwan
Objectives: This study investigated molecular characteristics of rifampin (RIF)-resistant (RIF-R) Staphylococcus aureus isolates recovered from patients receiving RIF-containing regimens for tuberculosis (TB).
Patients and methods: Patients with TB who received RIF-containing regimens from November 2009 to May 2011 at a medical center were enrolled. Nasal swabs for S. aureus culture were obtained at the time of enrollment, and then every two months until two months after RIF treatment had been completed. Genetic relatedness of the isolates was determined using pulsed-field gel electrophoresis, multilocus sequence typing, and gene typing of spa and SCCmec. The presence of RIF resistance-associated mutations in rpoB, and fusidic acid resistance genes fusB and fusC in the S. aureus isolates were analyzed.
Results: Among the 200 patients enrolled in this study, 152 completed follow-ups during treatment, and 114 completed two months of follow-up after discontinuing use of RIF. At enrollment, ten patients (5%) had nasal colonization with S. aureus, namely eight with methicillin-susceptible S. aureus (MSSA), and two with methicillin-resistant S. aureus (MRSA, ST59-SCCmecIV-RIF-susceptible). All these patients were decolonized after RIF usage. Two patients with MSSA colonization at enrollment showed recolonization with genetically unrelated MSSA strains two months after completion of RIF treatment. There were five ST45-SCCmecVT-RIF-R strains from two patients isolated during RIF exposure. Sequencing of rpoB in the RIF-R S. aureus isolates revealed different mutation sites between the MSSA and MRSA isolates.
Conclusion: RIF-R S. aureus strains are more likely to result in persistent nasal carriage in TB patients receiving RIF treatment. Monitoring of emergence and possible dissemination of the MRSA ST45 strains among TB patients treated with RIF in Taiwan is warranted.
Keywords: Staphylococcus aureus, methicillin-resistant S. aureus, rifampin resistance, tuberculosis, nasal carriage, Taiwan
Introduction
Rifampin (RIF), a semisynthetic antibiotic that inhibits DNA-dependent RNA polymerase, is commonly used as the first-line drug for the treatment of tuberculosis (TB). The drug is also effective when used in combination with vancomycin for the treatment of infections due to methicillin-resistant Staphylococcus aureus (MRSA).1 RIF has potent bactericidal activity, good tissue penetration, modest activity against non-growing cells, and has been shown to broaden the coverage of vancomycin-intermediate S. aureus (VISA) when used in combination with linezolid or daptomycin.1–3 Studies have shown that RIF may improve outcomes of patients with S. aureus bacteremia when used as an adjunct to β-lactams or glycopeptides.1,4 However, long-term use of RIF, as well as its use as a monotherapy, has been shown to be associated with the development of RIF-resistant (RIF-R) S. aureus.1,4 In 2006, Sekiguchi et al reported the emergence of RIF-R MRSA strains in a TB ward in Japan, and all of the RIF-R isolates had mutations in rpoB.5 In addition, isolates from patients receiving RIF as treatment for TB who had developed subsequent MRSA bacteremia, were shown to have higher rates of resistance to RIF, especially in patients with concurrent usage.6
Patients with persistent S. aureus colonization are more likely to develop subsequent S. aureus infections compared to intermittent carriers or patients without colonization.7,8 Moreover, patients with more MRSA colonization sites are at increased risk of developing MRSA infections.7 Decolonization of MRSA nasal carriage has been proposed as a preventative measure for patients prior to surgery, and for those admitted to intensive care units.7 However, the development of drug resistance during eradication treatment has been shown to occur in 1%–9% of patients receiving mupirocin ointment and oral antibiotics, including RIF and fusidic acid.7,8
A short course of oral RIF treatment for decolonization of S. aureus carriage has been shown to be effective in patients at risk of infection, although the rates of eradication vary from 25% to 100%, depending on the type and duration of RIF-containing regimen.9 Studies have shown that S. aureus develops resistance to RIF during or after treatment in up to 40% of cases.8,9 However, it remains unclear whether resistance to RIF among S. aureus isolated from patients with TB who are on long-term RIF therapy originates from an extrinsic source, or occurs de novo in the colonized strains.
In the present study, we prospectively screened for nasal S. aureus carriage among patients who received RIF-containing regimens for the treatment of TB. We determined antimicrobial susceptibilities, RIF-R hot-spot mutations, fusidic acid resistance mechanisms, and genetic relatedness of serial isolates using pulsed-field gel electrophoresis (PFGE), multilocus sequence typing (MLST), and spa typing. The evolution of colonized S. aureus strains in TB patients during RIF usage was also determined.
Patients and methods
Study design and population
This prospective observational cohort study was conducted at the Far Eastern Memorial Hospital (FEMH), a 1200-bed tertiary-care center in northern Taiwan. The research ethics committee of the hospital approved the study design, and informed consent was obtained from all participants (FEMH institutional review board [IRB] number: 098069-3). Written informed consent was obtained from the patients enrolled. All patients were adults (≥20 years) who received RIF-containing regimens for the treatment of TB during the period November 1, 2009, to May 31, 2011. RIF treatment included 600 mg or 10 mg/kg per day, with a mean exposure time of 207 days. Nasal swabs were taken from the anterior nares for culture of S. aureus prior to the initiation of RIF treatment, every two months during follow-up, and then two months after completion of treatment. Patients who had more than one positive surveillance culture for S. aureus with indistinguishable pulsotypes were considered persistent carriers.
Isolation and identification of S. aureus
Samples were obtained by rotating sterile swabs (BBL CultureSwab Plus; BD, Franklin Lakes, NJ, USA) over the bilateral nasal vestibule. The swabs were then inoculated onto blood agar plates and incubated for 48 hours for colony identification. Demographic data from the enrolled patients were collected.
Antimicrobial susceptibility testing
Identification and antimicrobial susceptibility testing of methicillin-susceptible S. aureus (MSSA) and MRSA isolates were performed using the Phoenix PMIC/ID panels and the Phoenix automated microbiology system (BD). In addition, susceptibility to RIF was also determined using the Etest (bioMerieux, Marcy l’Etoile, France), according to the manufacturer’s instructions. S. aureus American Type Culture Collection (ATCC) 29213 was used as the control strain for each test. Minimum inhibitory concentration (MIC) results were interpreted according to the criteria recommended by the Clinical and Laboratory Standards Institute for all agents tested, except fusidic acid, for which a clinical breakpoint (MIC of ≤ 1 mg/L) for susceptible isolates suggested by the European Committee on Antimicrobial Susceptibility Testing was adopted.10,11 Isolates were defined as resistant to one individual antimicrobial agent including those intermediate and fully resistant to that agent.
Molecular typing
Typing of spa was carried out using the public spa-type database (tools.egenomics.com) for all S. aureus isolates as reported previously.12 MLST was performed for all MRSA isolates and for persistent isolates of MSSA by analyzing seven housekeeping genes as previously reported.13 The allelic profile (allele number) was obtained from the MLST website (www.mlst.net/). The SCCmec elements (I–V) and mecA were determined for all MRSA isolates, as previously described.14 Genetic correlation of all isolates was determined by PFGE using SmaI as the restriction enzyme.13 Strains were considered part of the same cluster if their bands had indistinguishable restriction patterns. Isolates with >80% similarity were considered to be closely related.
Determination of genes conferring resistance to RIF and fusidic acid
RIF resistance-associated mutations were determined by sequencing a 460 bp fragment comprising clusters I and II of the RIF resistance-determining region of the S. aureus rpoB as previously described.16 The presence of fusidic acid resistance genes (fusB and fusC) was detected in isolates with low-level fusidic acid resistance (MICs ≤32 mg/L) by using polymerase chain reaction (PCR) as described previously.17 The size of PCR amplicons was 492 bp for fusB and 411 bp for fusC.
Statistical analysis
The demographic distribution and clinical characteristics of patients with nasal colonization at the time of entry into the study were compared using univariate analysis by Fisher exact test and Mann-Whitney U test for categorical and non-categorical data, respectively. Mean and SD were calculated for continuous variables, and percentages were calculated for categorical variables. Odds ratios (ORs) and the corresponding 95% CIs were calculated. A P-value <0.05 was considered statistically significant. All statistical analyses were performed using the statistical package SPSS for Windows (Version 15.0; SPSS Inc., Chicago, IL, USA).
Results
Colonization rate of S. aureus during RIF-containing anti-TB treatment
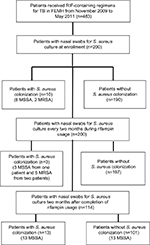
During the period November 2009 to May 2011, a total of 483 patients received RIF-containing regimens for TB. Of them, 200 provided consent to participate in the study, and had nasal swabs taken for culture of S. aureus (Figure 1). A total of 114 (57%) patients completed two months of follow-up after the discontinuation of treatment for TB. The demographic data are listed in Table 1. At enrollment, ten patients (5.0%) had S. aureus nasal carriage, including eight patients (4.0%) with MSSA and two patients (1.0%) with MRSA (Figure 1). Three of the ten patients (30.0%) with S. aureus nasal colonization had prior antibiotic usage within one month of enrollment, while 89 out of 190 (48.6%) patients without S. aureus colonization had previous exposure to antibiotics (P=0.347; Table 1). A total of 30 S. aureus isolates were obtained from surveillance cultures, including 23 MSSA and seven MRSA. Diabetes mellitus was the most common underlying disease (21%, n=42). Of the 200 participants in the study, 92 (46.0%) had been exposed to antibiotics other than RIF, isoniazid, ethambutol, or pyrazinamide within one month prior to the first sample collection. Twenty-five patients (12.5%) had been admitted to the hospital within three months prior to enrollment, of which one was colonized with S. aureus.
All ten patients with S. aureus nasal carriage at enrollment were decolonized after RIF usage and remained sterile during RIF exposure. Two of them had re-colonization with unrelated MSSA strains two months after discontinuing RIF treatment (Figure 1). Three patients free from S. aureus colonization at enrollment were colonized during RIF exposure, including one patient with MSSA (two isolates) and two with MRSA (five isolates). Follow-up cultures two months after discontinuation of RIF revealed MSSA colonization in 13 of the 114 patients who completed screening at the end of the study (11.4%).
Antimicrobial susceptibilities
The MICs of ten antimicrobial agents tested against the 23 MSSA and seven MRSA isolates are shown in Tables 2 and 3. All 30 of the S. aureus isolates were susceptible to vancomycin and linezolid. The resistance rates of MSSA to tetracycline were 52.2%, RIF 17.4%, erythromycin and gentamicin 8.7%, trimethoprim/sulfamethoxazole 4.3%, and to clindamycin and ciprofloxacin both were 0%. Of the seven MRSA isolates, all were susceptible to trimethoprim/sulfamethoxazole, five were resistant to tetracycline and ciprofloxacin, four were resistant to fusidic acid, two were resistant to erythromycin and gentamicin, and one resistant to clindamycin (Table 3). All ten of the isolates from the ten patients with nasal carriage at enrollment were susceptible to RIF according to the Etest (median MIC, 0.004 mg/L; range, 0.004–0.008 mg/L), and the Phoenix system (MICs of ≤0.5 mg/L). Two patients were considered persistent carriers, by definition, and the isolates from these patients were all resistant to RIF (>32 mg/L) including four MRSA isolates from one patient, and three MSSA isolates from the other patient. The four MRSA isolates from the persistent carrier were also resistant to ciprofloxacin, tetracycline, and fusidic acid (Table 3). Of the 11 isolates (MSSA, n=10; MRSA, n=1) from 11 patients with non-persistent colonization during RIF usage, all ten MSSA isolates were susceptible to RIF, while the MRSA isolate was resistant to RIF (>32 mg/L). Six isolates from three patients were resistant to fusidic acid, including four MRSA isolates from one of the two persistent carriers (Phoenix System MIC, 8 mg/L), and two MSSA isolates from two patients (Phoenix System MICs, 2 and 4 mg/L, respectively).
  | Table 2 Susceptibilities of the 23 methicillin-susceptible Staphylococcus aureus isolates to ten antimicrobial agents as determined by the Phoenix automation system and the Etest (for rifampin only) Notes: aMIC interpretive breakpoints are not available in the current version of CLSI.10 Clinical breakpoint (susceptible, MIC ≤1 mg/L) recommended by the EUCAST was applied.11 Abbreviations: MIC, minimum inhibitory concentration; CLSI, Clinical and Laboratory Standards Institute; EUCAST, European Committee on Antimicrobial Susceptibility Testing. |
Molecular typing of colonized isolates
The results of spa typing revealed a diverse distribution among all the S. aureus isolates and no major clone was identified. The spa types of the 23 MSSA were t084 (three patients), t091 (three patients, five isolates), t189 (two patients), t002, t012, t015, t094, t491, t1492, t2284, t4494, t5554, t6508, and three undetermined. Of the three MSSA carriers with paired isolates, one was colonized by t091 (Patient 13, three isolates), one was colonized by t1492 and then by an undetermined type, and one was colonized by t189 and then by t1508. The spa types of the seven MRSA isolates were t2365, t437, t026, t6508, and the four isolates from the persistent carriers belonged to t026. The results of PFGE of the 30 isolates revealed that MRSA isolates belonging to Patient 7 with the same spa-typing shared indistinguishable pulsotypes. No clustering of S. aureus was identified by the PFGE analysis among the S. aureus isolates belonging to the other 23 patients.
There were two sequence types (STs) identified among the seven MRSA isolates obtained from four patients (Table 3). Two patients were colonized by t2365-ST59-SCCmecIV and t437-ST59-SCCmecV at enrollment and remained free of MRSA during the follow-up surveillance. These two MRSA isolates were susceptible to RIF. The other two patients, one of whom was a persistent carrier, had five MRSA isolates cultured during follow-up surveillance. The five MRSA isolates recovered during RIF usage belonged to strains t026-ST45-SCCmecV (four isolates; persistent carrier) and t6508-ST45-SCCmecV. These isolates were all resistant to RIF. The t091-ST7 MSSA isolates from the persistent carrier were resistant to RIF, while the ST97 and t1492-ST15 isolates remained susceptible to RIF (the Etest MICs: 0.008 mg/L and 0.012 mg/L, respectively). The t091-ST7 MSSA isolate persisted two months after the patient had discontinued RIF treatment.
Genetic mutations in rpoB, fusB, and fusC
The results of rpoB mutation analysis of seven MRSA isolates are listed in Table 2, and all the mutations were located within cluster I. All ST45-SCCmecV-RIF-R isolates exhibited the same mutation sites in the regions sequenced (Table 2). However, the mutation sites in the MSSA t091-ST7 isolates differed from those in the MRSA isolates. The mutation sites included a silent mutation at amino acid position 475 and an S486L substitution. Of the six isolates with low-level fusidic acid resistance, four were strain t026-ST45-SCCmecV, and all contained fusC. One MSSA isolate (t5554) contained both fusB and fusC, and the other (t1508) contained none of these genes.
Discussion
This surveillance study revealed that RIF used during TB treatment could eradicate S. aureus nasal carriage. However, patients could be colonized with RIF resistant S. aureus isolates during treatment. Our findings indicate that the resistant strains acquired resistance extrinsically, rather than developing de novo resistance by the initial colonized strain during treatment.
In a review article on the efficacy of RIF, Falagas et al reported that oral RIF treatment for eradication of nasal S. aureus carriage is effective against the development of infection due to MRSA in 25% to 100% of patients, depending on the type and duration of the RIF-containing regimen.9 The review also provided evidence that RIF combined with trimethoprim/sulfamethoxazole and minocycline is associated with a high rate of isolation of S. aureus resistance, whereas RIF combined with other agents is associated with lower rates of resistance.8,9 However, the genetic relatedness of the resistant strains was not confirmed using molecular methods in the reviewed studies.9 In our study, we found that during long-term RIF usage, patients could be transiently colonized with RIF-susceptible strains. When persistent nasal carriage occurred in those patients, isolates were more likely to be resistant to RIF. Two persistent carriers were colonized with strains that were resistant to RIF. Their cultures at enrollment were negative for S. aureus, indicating that the resistant strains emerged after the initiation of RIF treatment.
Although Sekiguchi et al reported on the emergence of RIF-R MRSA in TB wards in two hospitals in Japan, the authors did not conduct molecular analyses of the outbreak strains, so it is not clear whether they evolved from previous susceptible strains.5 Tan et al reported that MRSA blood isolates recovered from patients on current RIF-containing anti-TB regimen are more likely to be resistant to RIF (87.5% vs. 36%) and result in a higher mortality rate than isolates from patients who were previously on a RIF-containing anti-TB regimen (79.2% vs. 40%).6 We also found that isolates obtained from patients two months after discontinuing RIF were more likely to remain susceptible to the agent. RIF resistance can rapidly emerge during therapy, with the resistant mutants being present in the inocula at a frequency of 10–6 to 10–8.1 Although we did not quantitate bacterial loads of each culture, a previous study revealed low bacterial loads of nasal carriers (1.8 to 2.9 log10 colony-forming units per nares culture).7 This may explain, at least in part, why none of the RIF-R isolates emerged from the original strains in our study.
Administration of suboptimal antibiotics, especially fluoroquinolones, can result in RIF resistance in S. aureus by increasing deletions or insertions in rpoB.18 In our study, nearly half of the patients (46%) had antibiotics exposure one month prior to the initiation of RIF treatment. However, we did not find a high rate of RIF resistance in isolates collected at enrollment, or during the follow-up surveillance period. Mutations in rpoB are not only associated with resistance to RIF, but also to other antibiotics including vancomycin and daptomycin.19,20 Watanabe et al reported that 71% (27 out of 38) of their VISA isolates had mutations in rpoB, and that 95.6% of the RIF-R MRSA mutants showed decreased vancomycin susceptibilities.19 Chen et al reported that mutations in rpoB (RpoB A477V, RpoB S529L) and fusA resulted in resistance to both agents, and were associated with the convergence of heterogeneous VISA to VISA in their isolates.21 In the report by Baek et al, the first isolate following daptomycin exposure had a single-nucleotide polymorphism in rpoB (RpoB A477D), which had decreased susceptibility to daptomycin, RIF, and vancomycin.20 The MRSA isolates in our study also carried the same rpoB mutation site, but we did not observe an increase of daptomycin and vancomycin MICs for these isolates. However, it is not clear whether our isolates would have had increased MICs after exposure to these agents. It is interesting that the MICs of linezolid gradually increased in RIF-R isolates obtained from persistent carriers. Further study is needed to understand whether cross-resistance to other antibiotics developed during prolonged RIF exposure, or whether resistance rapidly evolved during exposure to antibiotics.
The MRSA strain t026-ST45-SCCmecVT obtained from one of the two persistent carriers carried fusC, which confers low-level resistance to fusidic acid. Fusidic acid is commonly used in combination with RIF as a treatment for MRSA infections.1 The MRSA ST45 strain originated from Europe, and was rarely isolated in Taiwan before 2011.22,23 It is now the leading strain of MRSA isolated from staff and residents at nursing homes.22 In a recent study, >40% (21 out of 52) of MRSA ST45 isolates were shown to be resistant to fusidic acid.24 However, the authors of that study did not provide the rates of susceptibility of MRSA ST 45 to RIF.24 It is important to monitor whether this strain disseminated further in Taiwan, in order to better understand its multiple drug-resistant characteristics.
Limitations
There are several limitations in this study. First, the sample size was small, and the rate of nasal S. aureus carriage was low, especially compared to the rates reported in other studies from Taiwan. Second, we used culture-based methods to identify the isolates to the species level, the results of which can be influenced by collection techniques and culture conditions. Third, some patients were lost to follow-up after discontinuing RIF treatment. We, therefore, were not able to accurately determine the reinfection and RIF resistance rates after RIF exposure. Fourth, antibiotics usage at enrollment may be one of the reasons for the low nasal carriage rate, although the culture rates did not differ significantly. Fifth, detailed medical information, including patients’ immunosuppression and exposure to invasive devices, especially central venous catheters, was not available for assessing risk factors for S. aureus colonization. Finally, we did not fully sequence rpoB. The full-length sequence of the gene should be determined to understand whether mutations in rpoB accumulated during prolonged RIF exposure.
Conclusion
S. aureus nasal carriage of TB patients could be eradicated after initiation of RIF-containing regimens. RIF-R S. aureus strains, which are acquired during RIF treatment for TB patients, are more likely to result in persistent nasal carriage. Re-colonization with RIF-susceptible S. aureus strains after discontinuation of the agent is also common. Monitoring of emergence and possible dissemination of the MRSA ST45 strains among TB patients on RIF treatment in Taiwan is warranted.
Acknowledgment
This study was supported by internal funding (FEMH-99- C-039).
Disclosure
The authors report no conflicts of interest in this work.
References
Forrest GN, Tamura K. Rifampin combination therapy for nonmycobacterial infections. Clin Microbiol Rev. 2010;23(1):14–34. | ||
Deresinski S. Vancomycin in combination with other antibiotics for the treatment of serious methicillin-resistant Staphylococcus aureus infections. Clin Infect Dis. 2009;49(7):1072–1079. | ||
Tang HJ, Lai CC, Hsueh PR, et al. RNA polymerase B subunit gene mutations in biofilm-embedded methicillin-resistant Staphylococcus aureus following rifampin treatment. J Microbiol Immunol Infect. 2016;49(3):394–401. | ||
Russell CD, Lawson McLean A, Saunders C, Laurenson IF. Adjunctive rifampicin may improve outcomes in Staphylococcus aureus bacteraemia: a systematic review. J Med Microbiol. 2014;63(Pt 6):841–848. | ||
Sekiguchi J, Fujino T, Araake M, et al. Emergence of rifampicin resistance in methicillin-resistant Staphylococcus aureus in TB wards. J Infect Chemother. 2006;12(1):47–50. | ||
Tan CK, Lai CC, Liao CH, et al. Increased rifampicin resistance in blood isolates of meticillin-resistant Staphylococcus aureus (MRSA) amongst patients exposed to rifampicin-containing antituberculous treatment. Int J Antimicrob Agents. 2011;37(6):550–553. | ||
Septimus EJ, Schweizer ML. Decolonization in prevention of health care-associated infections. Clin Microbiol Rev. 2016;29(4):201–222. | ||
Ammerlaan HS, Kluytmans JA, Wertheim HF, Nouwen JL, Bonten MJ. Eradication of methicillin-resistant Staphylococcus aureus carriage: a systematic review. Clin Infect Dis. 2009;48(4):922–930. | ||
Falagas ME, Bliziotis IA, Fragoulis KN. Oral rifampin for eradication of Staphylococcus aureus carriage from healthy and sick populations: a systematic review of the evidence from comparative trials. Am J Infect Control. 2007;35(3):106–114. | ||
Clinical and Laboratory Standards Institute. Performance Standards for Antimicrobial Susceptibility Testing: 27th Informational Supplement M100-S27. Wayne, PA: CLSI; 2018. | ||
European Committee on Antimicrobial Susceptibility Testing. Clinical Breakpoints (version 8.0). EUCAST; 2018. Available from: http://www.eucast.orgclinical_breakpoints/. Accessed May 31, 2018. | ||
Shopsin B, Gomez M, Montgomery SO, et al. Evaluation of protein A gene polymorphic region DNA sequencing for typing of Staphylococcus aureus strains. J Clin Microbiol. 1999;37(11):3556–3563. | ||
Enright MC, Day NP, Davies CE, Peacock SJ, Spratt BG. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-susceptible clones of Staphylococcus aureus. J Clin Microbiol. 2000;38(3):1008–1015. | ||
International Working Group on the Classification of Staphylococcal Cassette Chromosome Element (IWG-SCC). Classification of staphylococcal cassette chromosome mec (SCCmec): guidelines for reporting novel SCCmec elements. Antimicrob Agents Chemother. 2009;53(12):4961–4967. | ||
Huang YH, Tseng SP, Hu JM, et al. Clonal spread of SCCmec type IV methicillin-resistant Staphylococcus aureus between community and hospital. Clin Microbiol Infect. 2007;13(7):717–724. | ||
Wichelhaus TA, Schäfer V, Brade V, Böddinghaus B. Molecular characterization of rpoB mutations conferring cross-resistance to rifamycins on methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 1999;43(11):2813–2816. | ||
Chen HJ, Hung WC, Tseng SP, et al. Fusidic acid resistance determinants in Staphylococcus aureus clinical isolates. Antimicrob Agents Chemother. 2010;54(12):4985–4991. | ||
Didier JP, Villet R, Huggler E, et al. Impact of ciprofloxacin exposure on Staphylococcus aureus genomic alterations linked with emergence of rifampin resistance. Antimicrob Agents Chemother. 2011;55(5):1946–1952. | ||
Watanabe Y, Cui L, Katayama Y, Kozue K, Hiramatsu K. Impact of rpoB mutations on reduced vancomycin susceptibility in Staphylococcus aureus. J Clin Microbiol. 2011;49(7):2680–2684. | ||
Baek KT, Thogersen L, Mogenssen RG, et al. Stepwise decrease in daptomycin susceptibility in clinical Staphylococcus aureus isolates associated with an initial mutation in rpoB and a compensatory inactivation of the clpX gene. Antimicrob Agents Chemother. 2015;59(11):6983–6991. | ||
Chen CJ, Lin MH, Shu JC, Lu JJ. Reduced susceptibility to vancomycin in isogenic Staphylococcus aureus strains of sequence type 59: tracking evolution and identifying mutations by whole-genome sequencing. J Antimicrob Chemother. 2014;69(2):349–354. | ||
Lee YT, Lin DB, Wang WY, et al. First identification of methicillin-resistant Staphylococcus aureus MLST types ST5 and ST45 and SCCmec types IV and Vt by multiplex PCR during an outbreak in a respiratory care ward in central Taiwan. Diagn Microbiol Infect Dis. 2011;70(2):175–182. | ||
Ho CM, Lin CY, Ho MW, et al. Concomitant genotyping revealed diverse spreading between methicillin-resistant Staphylococcus aureus and methicillin-susceptible Staphylococcus aureus in central Taiwan. J Microbiol Immunol Infect. 2016;49(6):363–370. | ||
Tsao FY, Kou HW, Huang YC. Dissemination of methicillin-resistant Staphylococcus aureus sequence type 45 among nursing home residents and staff in Taiwan. Clin Microbiol Infect. 2015;21(5):451–458. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.