Back to Journals » Infection and Drug Resistance » Volume 13
Characterization of Phenotypic and Genotypic Traits of Klebsiella pneumoniae from Lung Cancer Patients with Respiratory Infection
Authors Ding L, Yang Z , Lu J, Ma L, Liu Y, Wu X, Yao W, Zhang X, Zhu K
Received 30 August 2019
Accepted for publication 17 December 2019
Published 29 January 2020 Volume 2020:13 Pages 237—245
DOI https://doi.org/10.2147/IDR.S229085
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Lingchi Ding, 1,* Zhiqiang Yang, 1, 2,* Junguo Lu, 1 Lichao Ma, 2 Ying Liu, 2 Xiaoyan Wu, 3 Weidong Yao, 1 Xiaodong Zhang, 1 Kui Zhu 2
1Oncology Department, Nantong Tumor Hospital, Nantong 226361, People’s Republic of China; 2Beijing Advanced Innovation Center for Food Nutrition and Human Health, College of Veterinary Medicine, China Agricultural University, Beijing 100193, People’s Republic of China; 3Clinical Laboratory, Nantong Tumor Hospital, Nantong 226361, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Xiaodong Zhang; Weidong Yao Email [email protected]; [email protected]
Background: Klebsiella pneumoniae has been a leading healthcare-acquired infection (HAI) agent worldwide for decades. However, the epidemiological characteristics of K. pneumoniae in lung cancer patients with respiratory infection are unclear. Here, we characterized the frequency of K. pneumoniae in lung cancer patients with respiratory infection in a cancer hospital in China and determined the antibiotic resistance profile, virulence phenotype and clonal relationships among these K. pneumoniae strains.
Methods: The clinical data of lung cancer patients with respiratory infection from September 2017 to October 2018 were retrospectively evaluated. Microbiological methods, antimicrobial susceptibility tests, pulsed-field gel electrophoresis (PFGE), polymerase chain reaction (PCR) assays, Sanger sequencing and Galleria mellonella larvae infection model were used in this study.
Results: During the study period, a total of 47 lung cancer patients with respiratory infection caused by bacteria were identified, among 27 patients were identified as positive for K. pneumoniae and the positive rate was 57.45%. Among 37 nonduplicate K. pneumoniae strains from these 27 patients, 19 isolates (51.4%) were classified as multidrug resistant (MDR) with high-level resistance to, at least one agent in three or more antibiotic categories, including polymyxin B and tigecycline. Sixteen of the 37 strains (43.2%) were hypermucoviscous isolates. Extended spectrum β-lactamases-producing K. pneumoniae strains consisted of two dominant PFGE types. Furthermore, the assessment of virulence potential using a G. mellonella larvae infection model showed that K. pneumoniae isolated from these patients exhibited a high virulence level.
Conclusion: Our data showed that K. pneumoniae is the most critical cause of lung infection in patients with lung cancer in this hospital. The various drug resistance and virulence backgrounds of K. pneumoniae may make this clinical center a breeding ground for superbugs. It is paramount to enhance surveillance of K. pneumoniae strains and take control measures.
Keywords: K. pneumoniae, lung cancer, multidrug-resistant, virulence
Background
Lung cancer is the dominant cause of cancer-related death worldwide.1 Infection is the most common cause of death in lung cancer patients since tumor bleeding, bronchial obstruction, radiation, chemotherapy, and transplantation could lead to immune deficiency. The most common types of infection in these patients' population are pneumonia and septicemia, and gram-negative bacilli are the most common pathogens.2 Klebsiella spp cause a large number of these infections. Klebsiella pneumoniae remains a significant cause of bloodstream infections (BSIs) and directly threatens the health and life of cancer patients.3
K. pneumoniae is an important opportunistic pathogen, and it has been a leading healthcare-acquired infection (HAI) agent worldwide for decades. Since the mid-1980s, reports from the Asian Pacific Rim have described a unique clinical syndrome of community-acquired, tissue-invasive K. pneumoniae infection in healthy individuals.4 These strains often present in multiple sites or metastatically spread but are sensitive to antibiotics. Currently, K. pneumoniae shows high resistance to a broad spectrum of drugs, including β-lactam antibiotics, aminoglycosides and fluoroquinolones.5,6 Notably, carbapenem-resistant and hypervirulent K. pneumoniae strains have emerged recently.7 These strains are both hypervirulent and multidrug-resistant, and they may also be highly transmissible and able to cause severe infections in both the hospital and the community. A group found that the cancer patients with long hospital stay and previous antibiotics use were more easily infected with ESBL- K. pneumoniae.8 It increased the difficulty to combat K. pneumoniae infection for the currently available antibiotics. The limited treatment options of antibiotics impose high mortality rates, prolonged hospitalization and costs on public healthcare.9
The aim of this study was to investigate the molecular epidemiology characteristics of K. pneumoniae collected from lung cancer patients with respiratory infection during an outbreak in a cancer hospital in China and to provide insights into the prevention and control of this nosocomial infection.
Methods
Identification of Patients
This retrospective study was conducted in the Nantong Tumor Hospital, a 1200-bed tertiary hospital with about 9000 new cases per year in Nantong, East China. In this hospital, about 1400 lung cancer patients were admitted to medical oncology per year. About 10% of the 1400 lung cancer patients were accompanied with respiratory infection. From September 2017 to October 2018, patients who satisfied the symptoms of respiratory infection were screened for bacterial isolates by culturing sputum, blood, pleural cavity or excretion.10 The inclusion and exclusion criteria of samples are in the Supplementary information. The relevant clinical and microbiological data were extracted from electronic or paper medical records and microbiological databases. This study was reviewed and approved by the Ethics Committee of Nantong Tumor Hospital following the Declaration of Helsinki (project No. 2019002), and the patients or their family members were informed and their written consents were obtained.
Bacterial Isolates and Antimicrobial Susceptibility Testing
All samples were collected at the bedside and then transported immediately to the microbiology laboratory for inoculation on media and preliminary analysis. For sputum sampling, we collected the first deep sputum samples after gargle in the morning. Briefly, subculture of the samples were performed on sheep blood agar. Two single colonies of similar morphology in each plate were picked for further identification. All the isolates were identified by the Vitek 2 system (bioMérieux, Marcyl’E´ toile, France), and the species identification of all isolates was confirmed via matrix-assisted laser desorption/ionization time of flight mass spectrometry (MALDI-TOF/MS) (Shimadzu AXIMA PerformanceTM, Manchester, UK). Minimal inhibitory concentrations (MICs) for colistin and tigecycline were determined using the broth dilution method. The 2014 European Committee on Antimicrobial Susceptibility Testing (EUCAST) breakpoints were used for colistin and tigecycline. The MICs for other antibiotics, as shown in Supplementary Table 1, were determined using the agar dilution method according to the Clinical & Laboratory Standards Institute (CLSI) criteria of 2018–28.
Pulsed-Field Gel Electrophoresis (PFGE) and Multilocus Sequence Typing (MLST)
Pulsed-field gel electrophoresis (CHEF-MAP-PER System, Bio-Rad Laboratories, Hercules, CA, USA) was used to assess the genetic relatedness between the test isolates, as previously described.7 Briefly, K. pneumoniae isolates from the patients were embedded in SeaKem® Gold Agarose gels, digested with XbaI, and subjected to PFGE. Salmonella enterica serovar Braenderup H9812 digested with XbaI was used as a reference marker. Chromosome DNA of different strains was separated at 6 V/cm for 19.5 hrs at 14°C, with pulse times of 2.16–63.8 s and an included angle of 120°. PFGE profiles were analyzed by InfoQuest software version 4.5 (Bio-Rad Laboratories, Hercules, CA, USA). MLST was performed on all isolates to investigate the scale of clonal dissemination of the test strains by multiplex polymerase chain reaction (PCR) assay.11
DNA Extraction and Antibiotic Resistance Gene Detection
DNA templates were extracted using the boiling lysis method described previously.12 The detection of resistance genes was performed by PCR, and their identities were confirmed by sequencing. Isolates were screened by PCR amplification using specific primers for the detection of extended-spectrum β-lactamase (ESBL)-encoding genes (blaTEM, blaSHV, blaOXA-1, blaOXA-4, blaOXA-30 and blaCTX-M),13 carbapenemase genes (blaKPC, blaVIM, blaIMP, blaNDM, and blaOXA-48),14 a tigecycline resistance determinant (tet (A)),15 and colistin resistance genes (mcr-1, 2, 3, 4, and 5).16 The specific primers and the length of the expected PCR products are presented in Supplementary Table 3. Amplicons were analyzed by gel electrophoresis in 2% agarose and visualized under ultraviolet (UV) light. The forward primers were used for DNA sequencing.
String Test and Infection of G. mellonella Larvae
The string test was considered indicative of a hypermucoviscous K. pneumoniae isolate if the inoculation loop was able to generate a viscous string 5 mm in length while stretching a single colony away from an overnight culture agar plate containing 5% sheep blood.17 The virulence of K. pneumoniae isolates was evaluated in vivo using a G. mellonella larvae infection model performed as previously described.18 Briefly, K. pneumoniae isolates were grown in 1 mL LB broth, harvested during exponential phase, and washed once with 10 mM phosphate-buffered saline (PBS; pH 7.4). Isolates were diluted in PBS to a McFarland turbidity of 0.5, which corresponds to approximately 1×108 CFU/mL. After surface disinfection, larvae were injected with 10 μL of bacterial suspension, containing approximately 1×107 CFU/mL, into the last right proleg by use of a Gas Chromatogram syringe with a 30-gauge needle. A group of 10 larvae were injected with 10 μL of PBS in parallel to ensure that death was not due to injection trauma. Larvae were placed in 9-cm petri-dishes with food and kept at 37°C in the dark. Insects were considered dead when they did not respond to physical stimuli. Time of death was recorded. Assays were allowed to proceed for only 72 hrs. At least two independent experiments were performed.
Serotype and Virulence-Associated Gene Detection
PCR was used to detect the presence of capsule serotypes (K1 and K2) and virulence-associated genes. These virulence-associated genes included those encoding regulators of mucoid phenotype A (rmpA), K1 serotype (magA), enterobactin (entB), yersiniabactin (ybtS), phenolate-type siderophore (kfu), the aerobactin siderophore system (iutA), type 3 adhesins (mrkD), allantoin metabolism-associated genes (allS), peg-344, iroB, iucA, and the K2 capsular serotype-specifying wzi gene (Supplementary Table 2). Isolated DNA samples were screened using specific primers for the detection of virulence genes.11,19,20 The forward primers were used for DNA sequencing.
Statistical Analysis
Data were analyzed by SAS (V9.01). Continuous variables were described as mean ± SD and categorical variables as No (percentage). T-test process was conducted for normal distribution continuous data, while Chi-square or Fisher’s exact analysis was for categorical data. P-value <0.05 was considered as statistical significance.
Results
Frequency of K. pneumoniae in Lung Cancer Patients with Respiratory Infection
From September 2017 to October 2018, 260 lung cancer cases with cough and fever were collected. After excluding 213 non-respiratory infections cases, there were 47 respiratory infection cases to be processed. As a result, a total of 60 clinical samples were collected from the 47 cases. The 60 clinical samples were composed of 47 sputum samples, 8 blood samples, 3 hydrothorax samples, 1 ascites sample and 1 secreta sample. One representative sample was taken from one-sampled isolates sharing the same PFGE patterns and MIC phenotypes. Thus, 37 nonduplicate K. pneumoniae strains were collected. These 37 K. pneumoniae strains were isolated from 27 of the 47 (57.45%) patients tested (Figure 1). Among all the patients, 13 patients were coinfected with K. pneumoniae and other bacteria or fungi, such as Staphylococcus aureus, Staphylococcus epidermidis, Acinetobacter baumannii or Candida albicans. No significance was demonstrated for age and sex between K. pneumoniae infection and non-infection.
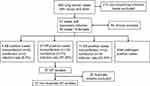 |
Figure 1 Flowchart diagram of samples collection. Abbreviations: KP, Klebsiella pneumoniae; SA, Staphylococcus aureus; AB, Acinetobacter baumannii. |
Antibiotic Susceptibility and Potential Mechanisms of Resistance to Important Antibiotics in K. pneumoniae Strains
Among the 37 K. pneumoniae isolates, 19 isolates (51.4%) were classified as MDR with high-level resistance to at least one agent in three or more antibiotic categories. Of the 19 isolates, 18 isolates were aztreonam-resistant K. pneumoniae and embodied resistance to amoxicillin/clavulanate and more than two non-β-lactam antimicrobial agents, including ciprofloxacin and tetracycline. Two of the ESBL-producing K. pneumoniae isolates were resistant to carbapenems, including ertapenem, imipenem and meropenem. Overall, the rates of resistance to tetracycline, ceftazidime, aztreonam, ceftazidime/avibactam, ciprofloxacin and gentamicin were 64.9%, 35.1%, 48.6%, 48.6%, 51.4% and 21.6%, respectively (Table 1). Twelve isolates (31.1%) were found to be resistant to polymyxin B; however, the mcr-1, 2, 3, 4, and 5 genes were not detected in any of the isolated strains. In addition, tigecycline susceptibility was reduced in some strains, with an MIC ≥ 0.5 µg/mL (64.9%).
 |
Table 1 Antibiotic Susceptibility Profiles of K. pneumoniae |
Acquired β-lactamase genes were detected in most isolates with a ceftazidime MIC ≥ 4 µg/mL. blaCTX-M-type and blaSHV genes were found to be dominantly carried in all these isolates. Fifteen of the 18 aztreonam-resistant K. pneumoniae isolates carried blaTEM. Two isolates of carbapenem-resistant K. pneumoniae (CRKP) carried the blaKPC-2 gene. Among the 37 isolates tested, we found that 20 strains carried the tet(A) variant, accounting for 54.1% of the test isolates. These 20 strains were all resistant to tetracycline with high MIC values. The tigecycline MIC values of strains harboring the tet(A) variant were mostly ≥0.5 µg/mL (18/20; 90.0%). Interestingly, all pulsotype I and J isolates harboring blaCTX-M were found to harbor the tet(A) gene (Figure 2).
Virulence-Associated Features
All K. pneumoniae isolates were subjected to string tests to determine their virulence level, with results showing that 16 of the 37 strains (43.2%) were positive for the test. The virulence genes peg-344, iroB, iucA, and rmpA were detected as markers for identifying hypervirulent K. pneumoniae (HvKP) strains.20 Seven of the 16 hypermucoviscous K. pneumoniae isolates harbored all the 4 hypervirulent marker genes. Two virulence-encoding genes were detected (entB and mrkD) in all isolates. Additionally, the rmpA gene was found in 27.0% (10/37), the iutA gene in 27.0% (10/37), the allS gene in 13.5% (5/37), the kfu gene in 18.9% (7/37), the ybtS gene in 43.2% (16/37), and the iucA gene in 94.6% (35/37).
KP10.1, KP14.2, KP28.1, KP37.1, KP49, KP54.1, and KP63.1 were selected as representative isolates according to PFGE patterns and MIC phenotypes and then subjected to further assessment of virulence potential using a G. mellonella larvae infection model, using an inoculum of 1×105 CFU per larva. The hypermucoviscous strain of K. pneumonia ATCC43816 was used as a hypervirulent-reference strain. As shown in Figure 3, it was clear that the death rate caused by strains isolated from this hospital was higher than that of ATCC43816 by 24 hrs, 48 hrs, and 72 hrs. Notably, not all strains from different patients showed consistent virulence.
Clonal Relationship of K. pneumoniae Strains
To investigate whether K. pneumoniae strains were clonally transmitted within this hospital and identify the prevalent clones, all 37 isolates were subjected to XbaI-PFGE and MLST analyses. With 80% genetic similarity as the cutoff, a total of 17 different pulsotypes were identified (Figure 2), while strain KP57.1 was untypable and did not give any bands. The most predominant pulsotype was pulsotype I (27.0%), followed by pulsotypes E (13.5%) and H (10.8%). In addition, there were seven PFGE patterns represented by a single strain. Strains isolated from different patients often showed diverse PFGE profiles, suggesting that K. pneumoniae isolates had diverse genetic backgrounds. The predominant pulsotype I consisted of 10 strains containing blaCTX and blaSHV genes obtained from June 12, 2018 to July 12, 2018. The pulsotype E strain also harbored 5 β-lactam-resistant MDR strains isolated from September 13, 2018 to October 12, 2018. The pulsotype H consisted of four hypervirulent ST23 K. pneumoniae isolates. The two ST11 CRKP isolates belonged to pulsotype K. The similarity of PFGE patterns of K. pneumoniae from different patients suggested that clonal transmission was observed in the hospital (Figure 3). We also found that one patient could carry various K. pneumoniae isolates harboring different phenotypes and genetic diversity at the same time, exhibiting the coexistence of hypervirulence and low virulence or coexistence of MDR strains and sensitive strains.
Discussion
Globally, lung cancer is the most common cancer, contributing to one in five cancer deaths.21 Although the operative mortality after lung resection has decreased over the past decades, the mortality rate from postoperative pneumonia following lung cancer surgery remains significant.22 Patients with cancer are more likely to suffer from infection due to pathogenic bacteria than benign patients.23 K. pneumoniae is a leading cause of lung infection in hospitalized patients.24 High mortality is closely related to healthcare-associated acute infection due to KPC-K. pneumoniae in cancer patients.25 However, the epidemiology characteristics of K. pneumoniae in lung cancer patients with respiratory infection are unclear.
The present study showed important information regarding the current status of the frequency and transmission kinetics of respiratory infection due to K. pneumoniae in lung cancer patients in a cancer hospital. Our data showed that K. pneumoniae were the dominant strain causing lung infection in lung cancer patients, with an isolation rate of 57.45%, exhibiting a wide distribution of genetic diversity among the 37 K. pneumoniae strains. ESBL-encoding genes, carbapenemase genes, a tigecycline-resistance determinant and colistin resistance genes which covered the grave-concerned genes for the treatment of K. pneumoniae infections were screened here.26 A total of 19 (51.4%) nonduplicate MDR isolates were recovered from sputum, blood or deep body fluid samples.Eighteen of the 19 MDRKP isolates were ESBL-carrying K. pneumoniae isolates; there seems to be an intimate connection between the occurrence of ESBL-carrying and MDR K. pneumoniae. Carriage of ESBL genes contributes to high mortality, prolonged length of hospital stay and high cost in hospitals. Globally, 14% of healthy individuals are colonized with ESBL Enterobacteriaceae.27 The frequency of ESBL- K. pneumoniae in our study was higher in lung cancer patients than in the healthy population.28 The most common determinants were the blaCTX-M, blaSHV and blaTEM genes in two dominant pulsotypes, which have been reported to be the most frequent ESBLs in K. pneumoniae isolates.26 Epidemiological studies suggest that the increasingly widespread occurrence of CRKP has become a major public health crisis.29 In our study, ST11 CRKP strains were isolated from two patients, and these strains could bring difficulties for infection control if the hospital does not prevent the transmission of CRKP. The high rates of resistance to polymyxin B and the prevalence rate of tet(A) variant genes found in this study deserve particular attention because these antibiotic categories have typically been used as the drugs of last resort for the treatment of severe infections caused by CRKP. High rates of resistance to polymyxin B and tet(A) variant genes mediating reduced tigecycline susceptibility of CRKP or Salmonella enterica were reported in other previous studies.15,30
A hypervirulent variant of K. pneumoniae has emerged worldwide.17 Since hypervirulent K. pneumoniae often confers the hypermucoviscous (HM) phenotype, we conducted a string test of 37 isolates. The positive rate of hypermucoviscous K. pneumoniae was 43.2%. The virulent serotype K1 and K2 HM strains were isolated. Then, representative strains of different genotypes and phenotypes (including hypermucoviscous and nonhypermucoviscous strains) were subjected to a G. mellonella infection model. Notably, all the K. pneumoniae strains were demonstrated to be highly virulent in the G. mellonella infection model. A review has suggested that HM and hypervirulence are two distinct phenotypes of K. pneumoniae that should not be used synonymously.31 Our results may be caused by the high prevalence rate of the iucA gene, which is one of the biomarkers for hvKP.20 In addition, diverse drug resistance and virulent phenotype K. pneumoniae strains were isolated, including typical ST23 hypervirulent K. pneumoniae, ESBL-producing HMKP, MDR classical K. pneumoniae, polymyxin B-resistant HMKP, tigecycline-resistant K. pneumoniae and ST11 CRKP.
In order to control MDR K. pneumoniae infection, we conduct measures as followed. Firstly, the pre-screening of MDR isolates and active surveillance for lung cancer patients will be protocolled. Secondly, we implement stringent isolation procedures for MDR isolates-bearing patients, and early-targeted therapies were used to kill the MDR isolates according to the MIC results. Thirdly, it would be essential to separate the MDR-isolates patients in isolation wards.
Conclusion
Although the sample size of this study is limited, this is the first report of the molecular epidemiology of K. pneumoniae in lung cancer patients with respiratory infection in Nantong, eastern China. Our study highlights the urgent need to enhance the clinical prevention and management of K. pneumoniae, especially the MDR hypervirulent isolate-causing respiratory infection in cancer hospitals. Careful and continued surveillance of K. pneumoniae strains and enhanced control measures are needed during the hospitalization of cancer patients.
Acknowledgments
We thank Jin Tao at the College of Veterinary Medicine, China Agricultural University for his help in the pulsed-field gel electrophoresis experiment, we also thank Huahua Yi for statistics, who works in Shanghai Jiao Tong University. This work was supported by the National Key Research and Development Program of China (2016YFD0501304).
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.v68.6
2. Bodey GP. Infection in cancer patients: a continuing association. Am J Med. 1986;81(1A):11–26. doi:10.1016/0002-9343(86)90510-3
3. Bodey GP, Elting LS, Rodriquez S, Hernandez M. Klebsiella bacteremia. A 10-year review in a cancer institution. Cancer. 1989;64(11):2368.
4. Liu Y-C, Cheng D-L, Lin C-L. Klebsiella pneumoniae liver abscess associated with septic endophthalmitis. Arch Intern Med. 1986;146(10):1913–1916. doi:10.1001/archinte.1986.00360220057011
5. Dsouza R, Pinto NA, Hwang I, et al. Panel strain of Klebsiella pneumoniae for beta-lactam antibiotic evaluation: their phenotypic and genotypic characterization. Peer J. 2017;5:e2896. doi:10.7717/peerj.2896
6. Fair RJ, Tor Y. Antibiotics and bacterial resistance in the 21st century. Perspect Med Chem. 2014;6:25–64. doi:10.4137/PMC.S14459
7. Gu D, Dong N, Zheng Z, et al. A fatal outbreak of ST11 carbapenem-resistant hypervirulent Klebsiella pneumoniae in a Chinese hospital: a molecular epidemiological study. Lancet Infect Dis. 2018;18(1):37–46. doi:10.1016/S1473-3099(17)30489-9
8. Li D, Chen Y, Zhang W, et al. Risk factors for hospital-acquired bloodstream infections caused by extended-spectrum beta-lactamase Klebsiella pneumoniae among cancer patients. Ir J Med Sci. 2014;183(3):463–469. doi:10.1007/s11845-013-1043-6
9. Gandra S, Barter DM, Laxminarayan R. Economic burden of antibiotic resistance: how much do we really know? Clin Microbiol Infect. 2014;20(10):973–980. doi:10.1111/1469-0691.12798
10. Garner JS, Jarvis WR, Emori TG, Horan TC, Hughes JM. CDC definitions for nosocomial infections, 1988. Am J Infect Control. 1988;16(3):128–140. doi:10.1016/0196-6553(88)90053-3
11. Yu F, Lv J, Niu S, et al. Multiplex PCR analysis for rapid detection of Klebsiella pneumoniae carbapenem-resistant (Sequence Type 258 [ST258] and ST11) and hypervirulent (ST23, ST65, ST86, and ST375) strains. J Clin Microbiol. 2018;56:9. doi:10.1128/JCM.00731-18
12. Chen L, Chavda KD, Findlay J, et al. Multiplex PCR for identification of two capsular types in epidemic KPC-producing Klebsiella pneumoniae sequence type 258 strains. Antimicrob Agents Chemother. 2014;58(7):4196–4199. doi:10.1128/AAC.02673-14
13. Dallenne C, Da Costa A, Decre D, Favier C, Arlet G. Development of a set of multiplex PCR assays for the detection of genes encoding important beta-lactamases in Enterobacteriaceae. J Antimicrob Chemother. 2010;65(3):490–495. doi:10.1093/jac/dkp498
14. Development of a set of multiplex, Poirel L, Walsh TR, Cuvillier V, Nordmann P. Multiplex PCR for detection of acquired carbapenemase genes. Diagn Microbiol Infect Dis. 2011;70(1):119–123. doi:10.1016/j.diagmicrobio.2010.12.002
15. Shu LB, Lu Q, Sun RH, et al. Prevalence and phenotypic characterization of carbapenem-resistant Klebsiella pneumoniae strains recovered from sputum and fecal samples of ICU patients in Zhejiang Province, China. Infect Drug Resist. 2019;12:11–18. doi:10.2147/IDR.S175823
16. Rebelo AR, Bortolaia V, Kjeldgaard JS, et al. Multiplex PCR for detection of plasmid-mediated colistin resistance determinants, mcr-1, mcr-2, mcr-3, mcr-4 and mcr-5 for surveillance purposes. Euro Surveill. 2018;23:6. doi:10.2807/1560-7917.ES.2018.23.6.17-00672
17. Shon AS, Bajwa RP, Russo TA. Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: a new and dangerous breed. Virulence. 2013;4(2):107–118. doi:10.4161/viru.22718
18. Insua JL, Llobet E, Moranta D, et al. Modeling Klebsiella pneumoniae pathogenesis by infection of the Wax Moth Galleria mellonella. Infect Immun. 2013;81(10):3552–3565. doi:10.1128/IAI.00391-13
19. Multiplex PCR for Detection of Seven Virulence Factors and K1/K2 Capsular Serotypes of Klebsiella pneumoniae.
20. Russo TA, Barker JH, Ricardo JM, La Hoz K. Identification of biomarkers for differentiation of hypervirulent Klebsiella pneumoniae from classical K. pneumoniae. J Clin Microbiol. 2018;56(9). doi:10.1128/JCM.00776-18
21. Dunn J, Garvey G, Valery PC, et al. Barriers to lung cancer care: health professionals’ perspectives. Support Care Cancer. 2017;25(2):497–504. doi:10.1007/s00520-016-3428-3
22. Lee JY, Jin SM, Lee CH, et al. Risk factors of postoperative pneumonia after lung cancer surgery. J Korean Med Sci. 2011;26(8):979–984. doi:10.3346/jkms.2011.26.8.979
23. Lin L, Jia L, Fu Y, et al. A comparative analysis of infection in patients with malignant cancer: a clinical pharmacist consultation study. J Infect Public Health. 2019;12:789–793. doi:10.1016/j.jiph.2019.03.021
24. Bachman MA, Breen P, Deornellas V, et al. Genome-wide identification of Klebsiella pneumoniae fitness genes during lung infection. MBio. 2015;6(3):e00775. doi:10.1128/mBio.00775-15
25. Freire MP, Pierrotti LC, Filho HH, et al. Infection with Klebsiella pneumoniae carbapenemase (KPC)-producing Klebsiella pneumoniae in cancer patients. Eur J Clin Microbiol Infect Dis. 2015;34(2):277–286. doi:10.1007/s10096-014-2233-5
26. Navon-Venezia S, Kondratyeva K, Carattoli A. Klebsiella pneumoniae: a major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol Rev. 2017;41(3):252–275. doi:10.1093/femsre/fux013
27. Karanika S, Karantanos T, Arvanitis M, Grigoras C, Mylonakis E. Fecal colonization with extended-spectrum beta-lactamase-producing enterobacteriaceae and risk factors among healthy individuals: a systematic review and metaanalysis. Clin Infect Dis. 2016;63(3):310–318. doi:10.1093/cid/ciw283
28. Ulstad CR, Solheim M, Berg S, Lindbaek M, Dahle UR, Wester AL. Carriage of ESBL/AmpC-producing or ciprofloxacin non-susceptible Escherichia coli and Klebsiella spp. in healthy people in Norway. Antimicrob Resist Infect Control. 2016;5:57. doi:10.1186/s13756-016-0156-x
29. Wyres KL, Holt KE. Klebsiella pneumoniae population genomics and antimicrobial-resistant clones. Trends Microbiol. 2016;24(12):944–956. doi:10.1016/j.tim.2016.09.007
30. Akiyama T, Presedo J, Khan AA. The tetA gene decreases tigecycline sensitivity of Salmonella enterica isolates. Int J Antimicrob Agents. 2013;42(2):133–140. doi:10.1016/j.ijantimicag.2013.04.017
31. Catalan-Najera JC, Garza-Ramos U, Barrios-Camacho H. Hypervirulence and hypermucoviscosity: two different but complementary Klebsiella spp. phenotypes? Virulence. 2017;8(7):1111–1123. doi:10.1080/21505594.2017.1317412
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.


