Back to Journals » Infection and Drug Resistance » Volume 12
Characterization of Antibiotic-Susceptibility Patterns, Virulence Factor Profiles and Clonal Relatedness in Proteus mirabilis Isolates from Patients with Urinary Tract Infection in Iran
Authors Mirzaei A, Habibi M , Bouzari S , Asadi Karam MR
Received 15 September 2019
Accepted for publication 10 December 2019
Published 27 December 2019 Volume 2019:12 Pages 3967—3979
DOI https://doi.org/10.2147/IDR.S230303
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Arezoo Mirzaei, Mehri Habibi, Saeid Bouzari, Mohammad Reza Asadi Karam
Department of Molecular Biology, Pasteur Institute of Iran, Tehran 13164, Iran
Correspondence: Mehri Habibi; Mohammad Reza Asadi Karam
Department of Molecular Biology, Pasteur Institute of Iran, No. 69, Pasteur Ave, Tehran 1316943551, Iran
Tel +9821 66953311
Fax +9821 66492619
Email [email protected]; [email protected]
Purpose: Proteus mirabilis is one of the most important agents of urinary tract infection (UTI). As there are limited data abou the pathogenicity P. mirabilis isolated from Iran, we investigated the virulence characteristics and antibiotic resistance in the isolates. Finally, the genotypic patterns were evaluated by Pulse field gel electrophoresis (PFGE).
Methods: A total of 110 isolates of P. mirabilis causing UTIs were isolated from patients in Tehran, Iran. The virulence characteristics and antimicrobial susceptibility were assayed using phenotypic methods. Extended-spectrum β-lactamases (ESBLs) production was assayed by the combination disk diffusion test (CDDT). Presence of virulence genes and antimicrobial-resistant genes was detected by Polymerase chain reaction (PCR). Finally, thirty-three isolates were selected for PFGE.
Results: All isolates showed the ability of biofilm and hemolysin formation. Antibiotic resistance ranged from 59.1% about cotrimoxazole to 2.7% about amoxicillin-clavulanic acid. Sixteen (14.5%) of the isolates were classified as multi-drug resistant (MDR). All isolates amplified mrpH, mrpA, pmfA, ureG and hpmA genes. Furthermore, the prevalence of zapA, fliC, ptaA, and ucaA genes was 98.2%, 98.2%, 95.5%, and 95.5%, respectively. The prevalence of plasmid-mediated quinolone resistance (PMQR) genes was 4.5% and 0.9% for aac(6ʹ)-Ib-cr and qnrA, respectively. Twenty-eight pulsotypes were detected among the 33 isolates by PFGE that pulsotypes 1, 2 and 4 with two isolates and pulsotype 3 with three isolates were the most prevalent ones.
Conclusion: It was found that the P. mirabilis isolates had high frequency of virulence factors. In addition, antibiotic resistance to some antibiotics and also production of ESBLs is alarming and shows the need for hygienic procedures to prevent the dissemination of antibiotic resistance. Although PFGE showed genetic diversity among the isolates, finding of several pulsotypes among the isolates should be considered an alarm to prevent these infections in hospital environments.
Keywords: Proteus mirabilis, urinary tract infection, virulence characteristics, antibiotic resistance, MDR, PFGE
Introduction
Proteus mirabilis is among the most common agents of urinary tract infection (UTI), especially in individuals with structural abnormalities in the urinary tract or those using catheters or other urinary instruments.1,2 Cystitis and acute pyelonephritis are common UTI types caused by P. mirabilis. Moreover, P. mirabilis infections result in the formation of urinary stones, which have the risk of severe damage in the bladder and kidneys and blockage of urinary catheters.3
The severity of UTI caused by P. mirabilis depends on the expression of different virulence factors, production of biofilm and expression of resistance to different classes of antibiotics.4,5 Fimbriae and hemagglutinins as critical adherence apparatus have diversity in P. mirabilis, including mannose-resistant Proteus-like fimbriae (MR/P), mannose-resistant Klebsiella-like hemagglutinin (MR/K), and P. mirabilis fimbriae (PMF).6,7 Flagella mediate the swarming motility in P. mirabilis and increase bacterial virulence by bacterial dissemination to the upper urinary tract.1 Moreover, P. mirabilis generates urease, which catalyzes the hydrolysis of urea to ammonia and carbamate and elevates the pH of the urine to produce struvite or apatite crystals.2 The most important cytotoxins in this uropathogen are HpmA and HlyA hemolysins and Pta cytotoxic agglutinin that have been implicated in the pathogenicity of the host.8 Other virulence factors include metalloproteinase and serralysin (ZapA) that cleave serum and secretory immunoglobulin A (IgA) and IgG, thereby providing protection from the mucosal immune response.1,2 Biofilm structures of P. mirabilis like crystalline biofilms also play an important role in the development of UTIs and reinfection, especially in catheterized UTIs by protecting bacteria from antibiotics and the host defense system, and contribute to surfaces strains.5
Fluoroquinolones are used as one of the antibiotic classes for the treatment of UTIs.9 Unfortunately, extensive misuse of this class of antibiotics resulted in a significant increase in resistance to them in P. mirabilis isolates.10,11 Furthermore, resistance to other classes of antibiotics like aminoglycosides has been reported among clinical P. mirabilis strains. There are different mechanisms for resistance to quinolones and fluoroquinolones including chromosomal mutations in DNA gyrase, and DNA topoisomerase IV, as well as acquisition of plasmid-mediated quinolone resistance (PMQR) genes encoding pentapeptide repeat proteins (qnr), aac(6ʹ)-Ib-cr (modified acetyltransferase), and the qepA (efflux pump).12
Extended-spectrum β-lactamases (ESBLs), as one of the major resistance mechanisms among gram-negative bacteria, have been associated with resistance to different classes of antibiotics such as quinolones, aminoglycosides and trimethoprim–sulfamethoxazole.13 There are limited reports on the prevalence of PMQR or its correlation with ESBL production in P. mirabilis isolates or relationship of ESBL producer P. mirabilis with resistance to different antibiotic classes in Iran. Thus, it is necessary to recognize the mechanisms of antibiotic resistance, especially PMQR and ESBLs among P. mirabilis isolates in hospital environments to have programmed control for these infections.
Detection of isolates with identical or similar genotypes indicates the endurance capability of these isolates in hospital environments and possible transmission between the patients. This issue is considered an alarm for the hygiene conditions of health-care providers to prevent and eliminate infections caused by P. mirabilis, and shows the importance and requirement of epidemiological studies into P. mirabilis causing UTIs.14 Accordingly, the present study aimed to study phenotypic and genotypic characterization in P. mirabilis isolated from patients with UTI among three hospitals in Tehran, Iran. Additionally, we investigated the prevalence of antibiotic resistance, ESBLs and PMQR genes among the isolates. Finally, the genotypic patterns of a number of the isolates were evaluated by PFGE.
Materials and Methods
Sample Collection and Identification
This study was performed in Department of Molecular Biology, Pasteur Institute of Iran, Tehran, Iran. Ethical approval for this research was confirmed by the scientific and ethics committee of Pasteur Institute of Iran. A total of 1200 urine samples from patients with UTI symptoms were screened and finally 110 isolates of P. mirabilis causing UTIs were isolated. The isolates were collected from urine of outpatients (33 isolates) and inpatients (77 isolates) in several hospitals in Tehran, Iran between June and December 2017. At the sampling time, no symptom of urolithiasis or other complications in the urinary tract of the patients was monitored. Furthermore, humans who received antibiotic during sampling or 1 month after hospital admission were excluded from this study. Identification of the isolates was done based on a series of microbiological and biochemical tests that routinely are used in microbiological diagnostic laboratories. Then, identified P. mirabilis isolates were inoculated into brain–heart infusion (BHI) broth media containing 20% glycerol and stored at –20°C until further analysis.
Biofilm Formation Assay
Biofilm formation in the P. mirabilis isolates was assayed according to the previous protocol15 with some modifications. Briefly, the overnight culture of the isolates was diluted 1:100 in BHI broth medium and cultured in polystyrene microtiter 96-well plates (Greiner, Germany). Then, the plates were incubated for 2 days at 37°C. Following this, the plates were washed three times with double-distilled water (DDW) and dyed with 0.1% (w/v) crystal violet for 20 mins at room temperature (RT). Then, 200 µL of ethanol–acetic acid (90:10) solution was added to the wells to measure their absorbance with an Enzyme-linked immunosorbent assay (ELISA) reader. Each assay was done in triplicate and the mean absorbance ± standard deviation was calculated for all repetitions of the tests. E. coli K12 and Pseudomonas aeruginosa ATCC 27853 were used as negative and positive control strains, respectively.
Hemolysis Assay Using Culture and Titration Methods
At first, hemolytic ability of the P. mirabilis isolates was determined by culture of bacteria on sheep blood agar medium (Merck co, Germany). The bacteria were inoculated into the culture medium and incubated overnight at 37°C. Hemolysin production was detected by the presence of clear zone of the erythrocytes around the colonies. This test was performed in duplicate. Then, hemolytic ability of the isolates was assayed with hemolytic titer method, according to the procedure described by Mobley and Chippendale.16 Briefly, isolates were cultured overnight in BHI broth. Then, 100 µL of two folds dilution of the bacterial suspensions (OD600 nm was adjusted to 1.0 in PBS 1×) were mixed with 50 µL of 3% suspension of sheep erythrocytes in PBS 1× and incubated for 1 hr at 37°C. The highest dilution in which no visible erythrocyte was observed at the bottom of plates was defined as hemolytic titer. This test was repeated three times and means of obtained titers were compared.
Hemagglutination Assay (HA)
For HA assay, the isolates were sub-cultured twice for 48 hrs in nutrient broth at 37°C. Then, bacterial cells were harvested by centrifugation and cell pellets were resuspended in 0.5 mL of PBS 1× to prepare bacterial suspensions with absorbance (OD600 nm) equal to 1. The suspensions were mixed with an equal volume of 1% suspension of erythrocytes human blood group A, guinea pig and chicken in PBS 1× in a 96-well microtiter plate (Greiner, Germany). The results were read as visible clumping after incubation for about 10 mins in RT. For evaluation, the inhibitory effect of D-mannose on hemagglutination, solution of 1% erythrocyte suspension containing 1% D-mannose was added to each well containing bacterial suspensions, and the results were recorded as weak (+) or strong (++) hemagglutination after incubation in RT. Then, expression of MR/P and MR/K fimbriae was investigated by microplate agglutination of tannic acid-treated and non-treated erythrocytes, as described previously by Mobley and Chippendale.16 This test was repeated four times, and the mean results were compared with each other.
Urease Assay
Urease activity of the isolates was assayed according to the protocol Mobley and Chippendale.16 Briefly, bacteria were cultured overnight in Luria bertani (LB) broth medium supplemented with 0.1% urea, harvested by centrifugation, and resuspended in PBS 1× until OD600 nm=0.5. Then, the suspensions were washed twice with PBS 1×, suspended in 20 mM sodium phosphate, and lysed by passage through a French pressure cell. The lysates were centrifuged at 14,000 g for 5 mins, and the supernatant-containing soluble protein was used to determine the urease activity by a spectrophotometric method using 3 mM sodium phosphate, 120 mM urea, and phenol red. Finally, absorbance was monitored at 650 nm. This test was repeated three times and mean urease activities of the isolates were compared.
Antimicrobial Susceptibility Patterns
The P. mirabilis isolates were subjected to antibiotic susceptibility testing against nine antibiotics by disk diffusion. The antibiotics tested against the isolates were including amoxicillin (10μg), ceftazidime (30µg), ceftriaxone (30μg), cefpodoxime (30μg), aztreonam (30µg), amoxicillin-clavulanic acid (20/10µg), imipenem (10µg), meropenem (10µg), tobramycin (10µg), norfloxacin (10µg), ciprofloxacin (5 µg), and cotrimoxazole (25µg) (Mast Diagnostics, Mast Group Ltd., Merseyside, UK). Inhibition zone diameter (mm) of each antimicrobial disc was measured, and the isolates were classified as resistant, intermediate, and susceptible.
Then, Minimal Inhibition Concentration (MIC) of fluoroquinolones including ciprofloxacin and norfloxacin and also ceftazidime was evaluated by the broth dilution method. Both disk diffusion and MIC tests were done according to the guidelines of the National Committee for Clinical Laboratory Standards (NCCLS).17 According to the guidelines, MIC ≤ 1μg/mL was considered as sensitive, ≥ 4μg/mL as resistant, and equal to 2μg/mL as intermediate for ciprofloxacin. About norfloxacin and ceftazidime, MIC ≤ 4μg/mL was considered as sensitive, ≥ 16μg/mL as resistant, and equal to 8μg/mL as intermediate. The E. coli ATCC 25922 was used as control strain.
ESBL Phenotypic Confirmation Test
Those isolates with ceftazidime zone ≥22 mm and ceftriaxone zone ≥25 mm were selected to evaluate ESBL production by combination disk diffusion test (CDDT). Briefly, disks containing 30μg of ceftazidime with and without 10μg of clavulanic acid were placed at a distance of 20 mm, center to center, on an MHA plate inoculated with a bacterial suspension of 0.5 McFarland turbidity standards and incubated overnight at 37°C. An increase in the inhibition zone diameter of ≥5 mm for a combination disc versus disc alone confirmed ESBL production.17
Detection of Virulence and PMQR Genes by PCR
DNA of the isolates was extracted using the DNA extraction kit (Roche, Germany), and their purity and quality were evaluated by a NanoDrop™ spectrophotometer (OD260/OD280 nm ratio ≥1.8) and electrophoresis on a 1% agarose gel (Sigma Chemical Co.). Then, the confirmed DNA was used to amplify nine important virulence genes in P. mirabilis isolates including mrpA, mrpH, ucaA, pmfA, ptaA, zapA, fliC, ureG, and hpmA. Antimicrobial-resistant isolates were further screened by PCR for five types of PMQR genes (qnrA, qnrB, qnrS, aac(6ʹ)-Ib-cr, and qepA) against fluoroquinolones. Briefly, PCR reaction of these genes was carried out in a total volume of 25 μL with the program: an initial denaturation at 94°C for 5 mins, followed by 30 cycles, each consisting of denaturation at 94°C for 1 min; annealing at specific temperature for 1 min; extension at 72°C for 1 min, and a final extension at 72°C for 5 mins. Then, the amplified samples were electrophoresed on agarose gel containing ethidium bromide to observe the PCR products. 50-bp and 1-kb DNA ladder mix (Fermentas, Lithuania) were used as DNA size markers. Table 1 lists the used oligonucleotide primers in the present study.
 |
Table 1 Oligonucleotide Primers Used for PCR Amplification |
PFGE Analysis
According to the phenotypic and genotypic results, thirty-three isolates were selected from the 110 P. mirabilis isolates for PFGE typing. Briefly, isolates were diluted in a buffer (75 mM NaCl and 25 mM EDTA) and mixed with 2% low-melting-temperature agarose (Invitrogen, USA). Then, 100 µL of the mixture was loaded in plug molds and allowed to set at 4°C. Plugs were incubated with lysis buffer containing 0.5 M EDTA, 1% N-lauryl sarcosine, and proteinase K at 50°C. Then, plugs were washed, digested with NotI restriction enzyme (Fermentas, Lithuania), and loaded on 1% ultrapure agarose gel (Invitrogen, USA) wells. Electrophoresis was carried out with CHEF-DR III apparatus (Bio-Rad, USA), and DNA fragments were separated by electrophoresis at 6 V/cm for 24 hrs with 2 blocks, the first block switch time was 1 to 30 s for 8 hrs and the second block was 30 to 70 s for 16 hrs. Salmonella enterica serovar Branderup strain H9812 (ATCC BAA-664) was used as size marker. The band patterns were analyzed with Gel compare II software (Applied math, Belgium). Furthermore, clonal relationship between the tested isolates was evaluated by using the unweighted pair group method with arithmetic mean (UPGMA). Relatedness of isolates was defined on the basis of PFGE profiles with ≥80% similarity.21
Statistical Analysis
Statistical data analysis was performed using SPSS software version 19.0 for Windows (IBM, Chicago, USA). Frequency analysis and mean values were used for statistical analysis of the tests. Chi-square and two-tailed Fisher's exact were among the tests that were used to assess and compare the relationships between the virulence properties of the isolates. Furthermore, p value less than 0.05 was considered as significant in all comparisons.
Results
Collection of Isolates and Identification
A total of 110 P. mirabilis isolates were collected from patients with UTI. Mean patient age was 37.7 years (range 1 to 88 years). In this study, 71.8% of isolates were recovered from females and the remaining 28.2% were from males. In general, 1–20 and 41–60 age groups were the most studied population (Figure 1). In our study, 30% and 70% of isolates were related to outpatients and inpatients, respectively. Among the 77 isolates collected from inpatients, the highest prevalence belonged to internal ward with 18 (23.4%), followed by Pediatric (19.5%), Intensive care unit (ICU) (16.9%), Infectious Diseases (13%), Emergency (7.8%), Surgery (7.8%), CPR (7.8%), and Coronary care unit (CCU) (3.8%) wards.
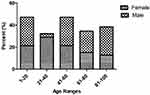 |
Figure 1 Distribution of P. mirabilis isolates according to the age and gender of UTI patients. |
Biofilm Formation Assay
In this assay, all P. mirabilis isolates showed the ability to produce biofilms. Biofilm formation capability in these isolates was classified into three groups, strong biofilm producers (71.8%) (OD590nm≥2.5), moderate biofilm producers (21.8%) (1.5≤OD590nm<2.5), and weak biofilm producers (6.4%) (0.7≤OD590nm<1.5). The weak biofilm producer isolates were collected from age groups 1–20 (3 isolates) and 21–40 (4 isolates). Furthermore, the isolates were obtained from outpatients (3 isolates) and inpatients (2 from pediatric, 1 from ICU and 1 isolate from infectious diseases ward). There was no significant difference between the strong or moderate biofilm formation in the isolates with age groups or gender (p>0.05).
Hemolysin Activity
We evaluated the ability of the isolates in hemolysin production on the blood culture medium and observed that all P. mirabilis isolates had hemolytic activity, including 97.3% β-hemolytic and 2.7% α-hemolytic. Then, hemolytic titers of the isolates were determined that all of the isolates showed hemolytic activity with titers ranging from 1:2 to 1:512. It was found that all of the isolates with the ability of α-hemolysin production had the hemolytic titers 1:2 to 1:4. There was no specific relationship between the hemolytic titers and biofilm intensity levels. In this regard, all isolates producing α-hemolysin were able to produce strong biofilm and were collected from females. We could not find any other significant relationship between the hemolytic titers of the isolates with the age groups, gender and outpatients or inpatients (p>0.05).
Hemagglutination Assay
The results of hemagglutination showed that all P. mirabilis isolates had the ability to hemagglutinate both guinea pig blood and human blood group A with a strong pattern of hemagglutination in the absence of mannose, but the isolates showed a weak severity in hemagglutination of chicken erythrocytes (Table 2). Furthermore, similar to the patterns observed in the absence of mannose, all of the tested isolates indicated the same ability in hemagglutination of guinea pig blood, human blood group A, and chicken erythrocytes in the presence of mannose (Table 2). According to the criteria of Mobley and Chippendale16 in the evaluation of expression MR/P and MR/K fimbriae, it was found that 81% of the isolates expressed both MR/P and MR/K fimbriae, whereas 19% of isolates produced MR/P alone. There were no isolates that displayed expression MR/K without MR/P fimbriae.
 |
Table 2 Hemagglutination Patterns of the P. mirabilis Isolates |
Urease Assay
In the present study, urease activity of the isolates was measured in the supernatant of the lysed cells containing soluble protein. We found that all isolates had the urease activity and the mean activity of the isolates ranged from 50 to 125 µmol of NH3/min/mg of protein. There was no significant correlation between the levels of urease activity of the isolates with biofilm intensity, hemolytic titers or hemagglutination (p>0.05). In addition, there were no significant differences between the mean activities of urease in the isolates with age groups and outpatients or inpatients (p>0.05).
Antibiotic Susceptibility Test
The bacterial isolates showed a high resistance to cotrimoxazole with 59.1%, followed by amoxicillin with 44.5%. Resistance to other antibiotics was 15.5% to ciprofloxacin, 13.6% to norfloxacin, 12.7% to tobramycin, 11.8% to imipenem, 4.5% to meropenem, and 2.7% to amoxicillin-clavulanic acid. Resistance to extended-spectrum cephalosporins was 11.8%, 10%, 10%, and 10% for ceftazidime, cefpodoxime, ceftriaxone, and aztreonam, respectively. According to Table 3, 21 different patterns of antibiotic resistance were detected among the tested isolates. Totally, 24.5% of isolates were sensitive to all of the tested antibiotics, whereas one of the isolates was resistant to all of the antibiotics. According to our results, resistance rate of fluoroquinolones among P. mirabilis isolated from male patients was significantly higher than those isolated from female patients (p<0.05). About antibiotics meropenem and amoxicillin-clavulanic acid, all resistance isolates were collected from females. Differences in distribution of antimicrobial resistance in different age groups of patients were not statistically significant (p>0.05). There was a significant difference in the antimicrobial susceptibility between isolates collected from inpatients and outpatients about the majority of tested antibiotics (p<0.05) (Table 4). Furthermore, relationship of antibiotic resistance with biofilm formation in the isolates is mentioned in Table 5. According to the table, overall resistance to the tested antibiotics was observed in P. mirabilis isolates with the ability of strong and moderate biofilm production, except for amoxicillin and cotrimoxazole. Also, resistance to amoxicillin-clavulanic acid was observed in three isolates with the ability of strong biofilm production.
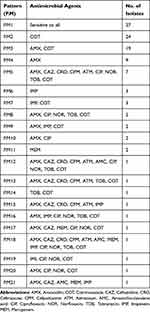 |
Table 3 Different Patterns of Antibiotic Resistance Among the Isolates |
 |
Table 4 Distribution of Antibiotic Resistance According to Inpatients and Outpatients |
 |
Table 5 Relationship Between Antimicrobial Resistance and Biofilm Formation in P. mirabilis Isolates |
In the recent study, sixteen (14.5%) of the isolates were classified as MDR, since they were resistant to at least three classes of antibiotics, and all of them belonged to isolates that collected from inpatients (Table 4).
This study showed that the MIC of ciprofloxacin ranged from 2μg/mL to ≥ 128μg/mL. It was found that of 17 (15.5%) resistant isolates to ciprofloxacin in disk diffusion, 15 (13.6%) of isolates showed MIC ≥ 4μg/mL that classified as resistant isolates and two isolates showed intermediate MIC to ciprofloxacin (MIC=2μg/mL). Furthermore, we found that fourteen (13.6%) resistant isolates to norfloxacin in disk diffusion had MIC ≥ 256μg/mL and regarded as resistant isolates in MIC. In addition, all ceftazidime-resistant isolates (11.8%) showed MIC ≥ 16μg/mL as resistant isolates.
From 13 isolates resistant to one of the antibiotics ceftazidime or ceftriaxone or both of them, 11 (10% of total isolates) were able to produce ESBLs phenotypically. The majority of the ESBL-positive isolates were isolated from women than men (73% vs 27%). Furthermore, the ESBL-positive isolates were collected from ICU (27.3%), internal (27.3%), pediatric (27.3%), and infectious diseases (18.1%) wards. The isolates were recovered from different age groups including age groups 0–20 (3 isolates), 21–40 (1 isolate), 41–60 (2 isolates), 61–80 (3 isolate), and 81–100 (2 isolate). Resistance to antibiotics among ESBL-positive isolates was higher than ESBL-negative isolates (Table 6). In the present study, it was found that all resistant isolates to ceftriaxone, cefpodoxime, and aztreonam were able to produce ESBLs. Accordingly, MDR P. mirabilis isolates were more frequently isolated from ESBL-positive than from the ESBL-negative isolates (p=0.000).
 |
Table 6 Prevalence of Antimicrobial Resistance Among ESBL-Positive and ESBL-Negative Isolates |
PCR Amplification Results
All tested isolates amplified mrpH, mrpA, pmfA, ureG, and hpmA genes. Furthermore, the prevalence of zapA, fliC, ptaA, and ucaA genes among the isolates was 98.2%, 98.2%, 95.5%, and 95.5, respectively. There was no significant difference between the prevalence of the virulence genes with age groups, gender, and hospital wards. The prevalence of PMQR genes among seventeen resistant isolates to one or both fluoroquinolones was 4.5% and 0.9% for aac(6ʹ)-Ib-cr and qnrA, respectively. The qnrB, qnrS, and qepA genes were not detected by PCR among the isolates. In one of the isolates, both aac(6ʹ)-Ib-cr and qnrA genes were detected, while 4 isolates contained only aac(6ʹ)-Ib-cr gene. In our study, isolates carried aac(6ʹ)-Ib-cr and qnrA genes were separated from inpatients in three wards of hospitals. Three isolates were related to internal ward, one isolate related to ICU and one isolate was collected from pediatric ward. In addition, the isolates containing aac(6ʹ)-Ib-cr gene were separated from patients with age ranges 1–20 (1 isolate), 41–60 (3 isolates), and 61–80 (1 isolate). The isolates were collected from both males (60%) and females (40%). The only isolate with qnrA gene was related to a 44-years-old male who was hospitalized in internal ward and the isolate had the ability to produce strong biofilm, hemolytic titer 1:128 and urease activity 75 µmol.
PFGE Typing
According to identity level 80% in PFGE, 28 pulsotypes were detected among the 33 isolates that pulsotypes (P) 1, 2, and 4 with two isolates and pulsotype 3 with three isolates were the most prevalent pulsotypes and 24 isolates were located in 24 distinct pulsotypes (Figure 2). Totally, similarity of all pulsotypes was ranged from 36.9% to 100%. It was found that all of the isolates in pulsotype 3 with identical PFGE profile were collected from infectious diseases ward. However, further analyses proved the isolates are similar in antibiotic-resistance patterns, virulence gene profiles, and biofilm production.
Discussion
Several reports from Iran show that similar to other countries, P. mirabilis isolates with the prevalence range 1% to 10% are usually among the common causative agents of UTIs, especially in catheterized patients.1,22–25 Furthermore, our study indicated that the rate of P. mirabilis was approximately 9% among the UTI agents in Iran that was in the range of other studies in Iran. There are limited data about the pathogenicity and antibiotic resistance of P. mirabilis in Iran, thus these characteristics were evaluated in a population of the isolates. The result of the current study showed that all P. mirabilis isolates had the capacity to form biofilm on polystyrene that certainly is important to establish P. mirabilis in the urinary tract, especially among catheterized patients.7 In accordance with our findings, Alves et al26 showed that all of the P. mirabilis isolates were able to produce biofilm. Aniejurengho et al27 also classified biofilm intensity of P. mirabilis isolates within moderate to strong which was near to our results. In the other study conducted in Iran, Sheikh-bardsiri et al28 reported a lower rate of biofilm formation in the isolates as compared to our study. They demonstrated that 36%, 39% and 17% of isolates produced weak, moderate and strong biofilms, respectively.28
Our results indicated high hemolysin activity for P. mirabilis isolates that could be important, especially in providing iron for bacteria and destruction of the kidney tissue.29,30 In the other studies, Zunino et al31 and Sosa et al30 showed that all P. mirabilis strains were able to produce beta-hemolysis on the blood agar medium. In accordance with several studies from other countries, P. mirabilis isolates showed hemolytic activity titers ranging from 1:2 to 1:512.16,29 In the present study, all of the isolates were able to agglutinate three kinds of erythrocytes with different severity levels, and the reactions were not inhibited by mannose. Additionally, Mobley and Chippendale16 and Sosa et al30 showed that almost all of the P. mirabilis isolates had the ability to agglutinate the erythrocytes in the presence of mannose. Furthermore, Mishra et al29 reported that 63% of the P. mirabilis isolates in India agglutinated the erythrocytes from different sources. Although we had no P. mirabilis isolated from catheter-associated or fecal sources, Mobley and Chippendale16 reported that 37.5% and 71.4% of isolates collected from pyelonephritis and catheter-associated bacteriuria expressed MR/P and MR/K simultaneously. In one study,29 the rate of simultaneous expression of MR/P and MR/K was lower than our results in Iran (44% vs 81%). In addition, our study and Mobley16 could not find MR/K alone in the isolates, whereas Mishra reported that 19% of the isolates produced MR/K alone.29
PCR amplification of fimbrial genes indicated that all isolates carried mrpH, mrpA and pmfA, whereas ucaA was not detected in 4.5% of isolates. Others have reported simultaneous presence of various types of fimbriae in P. mirabilis.6,30,32 Presence of several fimbriae in one isolate suggests that colonization of P. mirabilis is multifactorial, and presence of fimbriae together needs for colonization.6 It is also possible that expression of MR/P fimbriae containing mrpA and mrpH resulted in formation of biofilm and hemagglutination in all isolates.6,33 Our results consistent with a previous study indicated the presence of ureG in 100% of isolates, suggesting the crucial role of ureG in producing urease enzyme in P. mirabilis.34 Furthermore, in accordance with other studies, the presence of urease activity in all isolates suggests the critical role of ureG in the formation of urease enzyme.16,30 In agreement with a previous study, almost all of the isolates harbored gene encoding flagella,34 suggesting the potential of the isolates for infecting the kidneys. The results of the present study also showed that almost all isolates possessed zapA gene as a toxin gene, and this result was in agreement with other studies.34,35
In the past, the majority of P. mirabilis isolates were susceptible to common classes of antibiotics, but the recent studies in different countries have indicated that antibiotic resistance among P. mirabilis isolates is increasing.36 In our study, the highest resistance was related to cotrimoxazole followed by amoxicillin, while resistance to other antibiotics was between 2.7% and 15.5%. There are limited reports from Iran about antibiotic resistance among P. mirabilis isolates. The rate of resistance to cotrimoxazole in our study was near to that in a study conducted in Zahedan, Iran,23 whereas a higher rate of resistance to the antibiotic reported in Isfahan (78%)24 and Mazandaran (100%)25 in Iran. In one study conducted by Fallah et al22 in Iran (2019), resistance to ciprofloxacin, ceftriaxone, and imipenem was reported to be higher rate than that of our study, suggesting that the resistance to the antibiotics has increased among P. mirabilis isolates in Iran. Similar to our findings, several studies in Iran indicated that most of P. mirabilis isolates were sensitive to ciprofloxacin24 and had high resistance to amoxicillin.23 The rate of resistance in our study is also similar to those reported from the United States and European hospitals,37,38 however the rate was somewhat lower than that in a study in Ghana.39 This difference in results may result from the type of used antibiotics and the frequency of their use in countries.
Although third-generation cephalosporins are used as routine antibiotics against UTIs, the resistance against them has increased in the recent years, and it can be owing to the production of high levels of ESBL enzymes. In this study, the prevalence of ESBLs among P. mirabilis isolates was 10%, which was near to the results reported by previous studies in Korea (6.5–12.6%).40,41 The prevalence of ESBLs in P. mirabilis also is reported from 0.7% to 57% in different studies and shows that the prevalence has increased over time.40
There was a significant difference between the levels of antimicrobial resistance in inpatient and outpatient, whereas all MDR and ESBLs isolates belonged to inpatients, and this may be due to the selective pressure of excessive consumption of antibiotics in hospitals.
In the present study, susceptibility pattern of ESBL-producing isolates showed that these isolates were resistant to not only β-lactams but also to other classes of antibiotics, including cotrimoxazole, fluoroquinolones, tobramycin, and amoxicillin-clavulanic acid. In accordance with our findings, co-resistance to aminoglycosides, fluoroquinolones and cotrimoxazole has also been reported among ESBLs-positive P. mirabilis strains.11 Furthermore, in the present study, there was a direct relationship between the prevalence of MDR and ESBL production, and most of MDR isolates belonged to ESBLs-producing isolates, which was consistent with the other studies.10,11
In the current study, there was a direct relationship between the resistance to antibiotics and the intensity of biofilm formation that was shown in the other studies and it may be related to the transfer of resistance genes via genetic elements within biofilms.42,43 Previous studies also have shown increased resistance to ciprofloxacin in the strong biofilm structure of P. mirabilis strains.42,43 Nucleo et al found that ESBL-positive isolates also exhibited a higher ability to form biofilm with higher intensity, compared to ESBL (−) strains.5 However, the present study similar to the study by Kwiecinska-Piróg et al did not find such a correlation (p>0.05).44 In fact, both ESBL (+) and ESBL (−) isolates formed biofilm at comparable levels.
We examined the presence of PMQR determinants among the isolates that only qnrA and aac(6ʹ)-Ib-cr genes were detected in the isolates. The resistance mechanisms of fluoroquinolones-resistant isolates in disk diffusion that had none of the PMQR genes could be owing to other mechanisms such as the presence of other qnr genes, changes in DNA gyrase or topoisomerase IV, and efflux systems systems,45 which were not evaluated in our study. The isolates containing PMQR genes belonged to ESBLs-producing isolates and showed high-level resistance to fluoroquinolones (MIC ≥128 μg/mL for ciprofloxacin and ≥256 μg/mL for norfloxacin), demonstrating the potential of these genes in transferring the resistance to fluoroquinolones among P. mirabilis strains. Furthermore, our results showed that all of the isolates containing aac(6ʹ)-Ib-cr and qnrA genes were resistant to all of the antibiotics except amoxicillin-clavulanic acid and carbapenems. It is mentioned that aac(6ʹ)-Ib gene is also one of the most prevalent aminoglycoside-modifying enzymes, conferring resistance to antibiotics such as tobramycin, kanamycin and amikacin. However, in this study, all of the isolates harbored aac(6ʹ)-Ib-cr gene had resistance to tobramycin. In addition, all of the isolates carrying these resistance genes had the ability to produce biofilm with strong intensity. Although owing to the side effects of quinolones in children, hospitals do not recommend these drugs for children, the presence of aac(6ʹ)-Ib-cr in one child hospitalized in pediatric ward may result from the transfer of this gene from adults or other sources.
The present findings demonstrated that aac(6ʹ)-Ib-cr gene was more prevalent than qnrA gene similar to another study in Iran conducted by Majlesi et al,46 whereas Liang et al detected qnrA in 2 and qnrB in 3 P. mirabilis strains.47 In another study in Nigeria, aac(6ʹ)-Ib-cr and qnrA genes were detected in 3 (5%) and 22 (36.7%) of the MDR P. mirabilis isolates, respectively, while none was positive for qepA.48
According to the PFGE analysis, most of the isolates were clonally unrelated, and only four pulsotypes with more than one isolate were defined. Pulsotype 4 was related to 2 identical isolates (100% similarity) from different wards at the same hospital that maybe alarming for the dissemination of these isolates in different wards in a hospital. Pulsotype 1 showed that clonal dissemination was not only limited to inpatients in different hospital wards but also involved a non-hospitalized patient admitted to the same internal ward. Pulsotype 2 was related to two different hospitals with 85.7% similarity that we guess these isolates in phylogenetic evolution are separating from each other. Pulsotype 3 was associated with three identical isolates (100% similarity) collected from different patients in the same ward (infectious diseases) and from one hospital on different dates that we guess these isolates were spread from the hospital environment by personnel or nurses or a nosocomial infection might have been in this ward. This issue can be considered about alarming in terms of hygiene conditions of healthcare providers to prevent or eliminate these infections and shows the importance and requirement of periodically epidemiological studies about UTI in different countries.14
Conclusion
Our results indicated that all of P. mirabilis isolated from urine samples possessed virulence characteristics including hemolysin, hemagglutination and urease activity. Most of the isolates were able to form strong biofilm, and they produced biofilm-associated genes such as ucaA, pmfA, mrpA, mrpH, and fliC. Furthermore, the high prevalence of virulence genes along with increasing antibiotic resistance indicates that the properties can be transferred via the same plasmids and cause serious clinical problems in the treatment of UTIs. Although the overall rate of antibiotic resistance was not significant among the isolates, it is alarming and demonstrates the need for hygienic procedures to prevent the increased rate of antibiotic resistance among the isolates in the future.
Acknowledgements
The study was supported by Pasteur Institute of Iran.
Disclosure
Prof. Dr. Saeid Bouzari report grants and personal fees from Pasteur Institute of Iran, Tehran, Iran, during the conduct of the study as well as grants and personal fees from Pasteur Institute of Iran, Tehran, outside the submitted work. The authors report no other conflicts of interest in this work.
References
1. Schaffer JN, Pearson MM. Proteus mirabilis and urinary tract infections. Microbiol Spectr. 2015;3(5). doi:10.1128/microbiolspec.UTI-0017-2013
2. Armbruster CE, Mobley HL. Merging mythology and morphology: the multifaceted lifestyle of Proteus mirabilis. Nat Rev Microbiol. 2012;10(11):743–754. doi:10.1038/nrmicro2890
3. Armbruster CE, Mobley HL, Pearson MM. Pathogenesis of Proteus mirabilis infection. EcoSal Plus. 2018;8(1). doi:10.1128/ecosalplus.ESP-0009-2017
4. Luzzaro F, Mezzatesta M, Mugnaioli C, et al. Trends in production of extended-spectrum β-lactamases among enterobacteria of medical interest: report of the second Italian nationwide survey. J Clin Microbiol. 2006;44(5):1659–1664. doi:10.1128/JCM.44.5.1659-1664.2006
5. Nucleo E, Fugazza G, Migliavacca R, et al. Differences in biofilm formation and aggregative adherence between β-lactam susceptible and β-lactamases producing P. mirabilis clinical isolates. New Microbiol. 2010;33(1):37–45.
6. Rocha SP, Elias WP, Cianciarullo AM, et al. Aggregative adherence of uropathogenic Proteus mirabilis to cultured epithelial cells. FEMS Immunol Med Microbiol. 2007;51(2):319–326. doi:10.1111/j.1574-695X.2007.00308.x
7. Rocha SP, Pelayo JS, Elias WP. Fimbriae of uropathogenic Proteus mirabilis. FEMS Immunol Med Microbiol. 2007;51(1):1–7. doi:10.1111/j.1574-695X.2007.00284.x
8. Alamuri P, Eaton KA, Himpsl SD, Smith SN, Mobley HL. Vaccination with proteus toxic agglutinin, a hemolysin-independent cytotoxin in vivo, protects against Proteus mirabilis urinary tract infection. Infect Immun. 2009;77(2):632–641. doi:10.1128/IAI.01050-08
9. Zanichelli V, Huttner A, Harbarth S, Kronenberg A, Huttner B. Antimicrobial resistance trends in Escherichia coli, Klebsiella pneumoniae and Proteus mirabilis urinary isolates from Switzerland: retrospective analysis of data from a national surveillance network over an 8-year period (2009–2016). Swiss Med Wkly. 2019;149:2930.
10. Hernandez J, Martinez-Martinez L, Pascual A, Suarez A, Perea E. Trends in the susceptibilities of Proteus mirabilis isolates to quinolones. J Antimicrob Chemother. 2000;45(3):407–408. doi:10.1093/jac/45.3.407
11. Endimiani A, Luzzaro F, Brigante G, et al. Proteus mirabilis bloodstream infections: risk factors and treatment outcome related to the expression of extended-spectrum β-lactamases. Antimicrob Agents Chemother. 2005;49(7):2598–2605. doi:10.1128/AAC.49.7.2598-2605.2005
12. Martínez-Martínez L, Eliecer Cano M, Manuel Rodríguez-Martínez J, et al. Plasmid-mediated quinolone resistance. Expert Rev Anti Infec Ther. 2008;6(5):685–711. doi:10.1586/14787210.6.5.685
13. Pitout JD, Nordmann P, Laupland KB, Poirel L. Emergence of Enterobacteriaceae producing extended-spectrum β-lactamases (ESBLs) in the community. J Antimicrob Chemother. 2005;56(1):52–59. doi:10.1093/jac/dki166
14. Kanayama A, Kobayashi I, Shibuya K. Distribution and antimicrobial susceptibility profile of extended-spectrum beta-lactamase-producing Proteus mirabilis strains recently isolated in Japan. Int J Antimicrob Agents. 2015;45(2):113–118. doi:10.1016/j.ijantimicag.2014.06.005
15. O’Toole GA. Microtiter dish biofilm formation assay. J Vis Exp. 2011;47:1–2.
16. Mobley HL, Chippendale GR. Hemagglutinin, urease, and hemolysin production by Proteus mirabilis from clinical sources. J Infect Dis. 1990;161(3):525–530. doi:10.1093/infdis/161.3.525
17. Wayne P CLSI. Performance standards for antimicrobial susceptibility testing; twenty-fifth informational supplement. CLSI Document M100-S25. Clinical and Laboratory Standards Institute; 2015.
18. Cattoir V, Poirel L, Rotimi V, Soussy C-J, Nordmann P. Multiplex PCR for detection of plasmid-mediated quinolone resistance qnr genes in ESBL-producing enterobacterial isolates. J Antimicrob Chemother. 2007;60(2):394–397. doi:10.1093/jac/dkm204
19. Kim HB, Park CH, Kim CJ, Kim EC, Jacoby GA, Hooper DC. Prevalence of plasmid-mediated quinolone resistance determinants over a 9-year period. Antimicrob Agents Chemother. 2009;53(2):639–645. doi:10.1128/AAC.01051-08
20. Chen X, Zhang W, Pan W, et al. Prevalence of qnr, aac (6′)-Ib-cr, qepA, and oqxAB in Escherichia coli isolates from humans, animals, and the environment. Antimicrob Agents Chemother. 2012;56(6):3423–3427. doi:10.1128/AAC.06191-11
21. Tenover FC, Arbeit RD, Goering RV, et al. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33(9):2233–2239.
22. Fallah F, Parhiz S, Azimi L. Distribution and antibiotic resistance pattern of bacteria isolated from patients with community-acquired urinary tract infections in Iran: a cross-sectional study. IJHS. 2019;4(2):14–19.
23. Keyhan H, Sedighi S, Mashayekhi B, Fathi M, Mokhtari M. Community acquired urinary tract infections’ etiological organisms and antibiotics susceptibility patterns. Nephrourol Mon. 2017;9(5). doi:10.5812/numonthly.62146
24. Mirzarazi M, Rezatofighi SE, Pourmahdi M, Mohajeri MR. Antibiotic resistance of isolated gram negative bacteria from urinary tract infections (UTIs) in Isfahan. Jundishapur J Microbiol. 2013;6(8). doi:10.5812/jjm
25. Nasrolahei M, Poorhagibagher M, Vahedi M, Maleki I. Urinary tract infection among intellectual disability individuals” Etiology and antibiotic resistance patterns” in rehabilitation centers of Mazandaran province, Northern Iran. J Prev Med Hyg. 2013;54(3):170–175.
26. Alves MJ, Barreira J, Carvalho I, et al. Propensity for biofilm formation by clinical isolates from urinary tract infections: developing a multifactorial predictive model to improve the antibiotherapy. J Med Microbiol. 2014;63:471–477. doi:10.1099/jmm.0.071746-0
27. Aniejurengho O, Dessi M, Meikle S, Santin M, Okoli I. Biofilm formation in uropathogenic strains of Proteus mirabilis and their susceptibility to poly (epsilon-lysine) dendron. Biosci Res Today World. 2015;1(1):1–9.
28. Sheykh-Bardsiri H, Shakibaie M, Amini Kafiabad S. Plasmid pattern of biofilm producing Proteus mirabilis and Proteus vulgaris among clinical isolates in Kerman university hospitals during 2011–2012. JKMU. 2013;20(2):146–157.
29. Mishra M, Thakar Y, Pathak A. Haemagglutination, haemolysin production and serum resistance of proteus and related species isolated from clinical sources. Ind J Med Microbiol. 2001;19(2):5–11.
30. Sosa V, Schlapp G, Zunino P. Proteus mirabilis isolates of different origins do not show correlation with virulence attributes and can colonize the urinary tract of mice. Microbiol. 2006;152(7):2149–2157. doi:10.1099/mic.0.28846-0
31. Zunino P, Sosa V, Schlapp G, Allen AG, Preston A, Maskell DJ. Mannose-resistant Proteus-like and P. mirabilis fimbriae have specific and additive roles in P. mirabilis urinary tract infections. FEMS Immunol Med Microbiol. 2007;51(1):125–133. doi:10.1111/j.1574-695X.2007.00285.x
32. Mobley HL, Belas R. Swarming and pathogenicity of Proteus mirabilis in the urinary tract. Trends Microbiol. 1995;3(7):280–284. doi:10.1016/S0966-842X(00)88945-3
33. Jansen AM, Lockatell V, Johnson DE, Mobley HL. Mannose-resistant Proteus-like fimbriae are produced by most Proteus mirabilis strains infecting the urinary tract, dictate the in vivo localization of bacteria, and contribute to biofilm formation. Infect Immun. 2004;72(12):7294–7305. doi:10.1128/IAI.72.12.7294-7305.2004
34. Ali HH, Yousif MG. Detection of some virulence factors genes of Proteus mirabilis that isolated from urinary tract infection. IJAR. 2015;3(1):156–163.
35. Stankowska D, Kwinkowski M, Kaca W. Quantification of Proteus mirabilis virulence factors and modulation by acylated homoserine lactones. J Microbiol Immunol Infect. 2008;41(3):243–253.
36. Magyar A, Köves B, Nagy K, et al. Spectrum and antibiotic resistance of uropathogens between 2004 and 2015 in a tertiary care hospital in Hungary. J Med Microbiol. 2017;66(6):788–797. doi:10.1099/jmm.0.000498
37. Sader HS, Farrell DJ, Flamm RK, Jones RN. Antimicrobial susceptibility of gram-negative organisms isolated from patients hospitalized in intensive care units in United States and European hospitals (2009–2011). Diagn Microbiol Infect Dis. 2014;78(4):443–448. doi:10.1016/j.diagmicrobio.2013.11.025
38. Cernohorska L, Chvilova E. Proteus mirabilis isolated from urine, resistance to antibiotics and biofilm formation. Klin Mikrobiol Infek Lek. 2011;17(3):81–85.
39. Feglo PK, Gbedema SY, Quay SNA, Adu-Sarkodie Y, Opoku-Okrah C. Occurrence, species distribution and antibiotic resistance of Proteus isolates: a case study at the Komfo Anokye Teaching Hospital (KATH) in Ghana. Int J Pharm Sci Res. 2010;1(9):347–352.
40. Ahn JY, Ann HW, Jeon Y, et al. The impact of production of extended-spectrum β-lactamases on the 28-day mortality rate of patients with Proteus mirabilis bacteremia in Korea. BMC Infect Dis. 2017;17(1):327. doi:10.1186/s12879-017-2431-8
41. Song W, Kim J, Bae IK, et al. Chromosome-encoded AmpC and CTX-M extended-spectrum β-lactamases in clinical isolates of Proteus mirabilis from Korea. Antimicrob Agents Chemother. 2011;55(4):1414–1419. doi:10.1128/AAC.01835-09
42. Sahal G, Bilkay IS. Multidrug resistance by biofilm-forming clinical strains of Proteus mirabilis. Asian Biomed. 2015;9(4):535–541.
43. Subramanian P, Shanmugam N, Sivaraman U, Kumar S, Selvaraj S. Antibiotic resistance pattern of biofilm-forming uropathogens isolated from catheterised patients in Pondicherry, India. Australas Med J. 2012;5(7):344–348. doi:10.4066/AMJ.2012.1193
44. Kwiecińska-Piróg J, Skowron K, Zniszczol K, Gospodarek E. The assessment of Proteus mirabilis susceptibility to ceftazidime and ciprofloxacin and the impact of these antibiotics at subinhibitory concentrations on Proteus mirabilis biofilms. Biomed Res Int. 2013;2013.
45. Jacoby GA, Strahilevitz J, Hooper DC. Plasmid-mediated quinolone resistance. Microbiol Spectr; 2014;2(5):1–42.
46. Majlesi A, Kakhki RK, Nejad ASM, et al. Detection of plasmid-mediated quinolone resistance in clinical isolates of Enterobacteriaceae strains in Hamadan, West of Iran. Saudi J Biol Sci. 2018;25(3):426–430. doi:10.1016/j.sjbs.2016.11.019
47. Liang C, Kong Q, Yongmei F, Feng Y, Yang X. Mechanisms of plasmid-mediated quinolone resistance in Proteus mirabilis from urine. J Pract Med. 2017;33(24):4152–4155.
48. Alabi OS, Mendonça N, Adeleke OE, da Silva GJ. Molecular screening of antibiotic-resistant determinants among multidrug-resistant clinical isolates of Proteus mirabilis from Southwest Nigeria. Afr Health Sci. 2017;17(2):356–365. doi:10.4314/ahs.v17i2.9
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.

