Back to Journals » Infection and Drug Resistance » Volume 13
Characterization of a Novel mcr-8.2-Bearing Plasmid in ST395 Klebsiella pneumoniae of Chicken Origin
Authors Yang X, Peng K, Zhang Y , Liu L, Li R
Received 3 April 2020
Accepted for publication 16 May 2020
Published 16 June 2020 Volume 2020:13 Pages 1781—1784
DOI https://doi.org/10.2147/IDR.S256544
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Suresh Antony
Xiaorong Yang,1,* Kai Peng,2,3,* Yuxia Zhang,4 Li Liu,1 Ruichao Li2,3
1Center for Disease Control and Prevention of Sichuan Province, Chengdu, People’s Republic of China; 2Jiangsu Co-Innovation Center for Prevention and Control of Important Animal Infectious Diseases and Zoonoses, College of Veterinary Medicine, Yangzhou University, Yangzhou, People’s Republic of China; 3Institute of Comparative Medicine, Yangzhou University, Yangzhou, People’s Republic of China; 4Institute of Qinghai-Tibet Plateau, Southwest Minzu University, Chengdu, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Ruichao Li
College of Veterinary Medicine, Yangzhou University, Yangzhou, Jiangsu Province, People’s Republic of China
Email [email protected]
Abstract: The emergence of mobile colistin resistance mcr genes undermines the efficacy of colistin as the last-resort drug for multi-drug resistance infections and constitutes a great public health concern. Plasmids play a critical role in the transmission of mcr genes among bacteria. One colistin-resistant Klebsiella pneumoniae strain of chicken origin was collected and analyzed by antimicrobial susceptibility testing, PCR, conjugation assay and S1-PFGE. Whole-genome sequencing (WGS) approach combining Illumina and MinION platforms was utilized to decipher the underlying colistin resistance mechanism and genetic context. A novel mcr-8.2-bearing plasmid p2019036D-mcr8-345kb with 345 655 bp in size encoding various resistance genes including floR, sul1, aadA16, aadA2, blaCTX-M-27, blaDHA-1, tet(D), dfrA12 and qnrB4 was identified responsible for the colistin resistance phenotype. Plasmid comparison has shown that the mcr-8.2-bearing plasmid differed from other reported plasmids positive for mcr-8.2 but shared the same core mcr-8.2-bearing conserved region. This study demonstrates the emergence of mcr-8.2-bearing K. pneumoniae of animal origin is a potential risk to humans.
Keywords: mcr-8.2, Klebsiella pneumoniae, plasmids, animal origin
Antimicrobial resistance is posing a great public health concern worldwide. Since the first report of plasmid-mediated colistin resistance gene mcr-1 in 2015,1 a variety of mcr genes up to mcr-10 have been detected.2,3 These different mcr genes and the mcr-bearing plasmids are wildly distributed in Enterobacterales from humans, animals and environments.1–4 Klebsiella pneumoniae is ubiquitous in environments and is a major cause of nosocomial infections worldwide.5 The emergence of mcr genes in K. pneumoniae is a challenge to clinical treatments. To date, several mcr genes and their variants have been detected in K. pneumoniae of both human and animal origins in different countries.6–9 The first identified mcr-8 was found in a transferrable IncFII plasmid pKP91 in K. pneumoniae of swine origin.6 Then, another novel mcr-8.2 variant was reported in K. quasipneumoniae, phylogenetically similar to K. pneumoniae, isolated from a pig farm during our surveillance study in 2018.10 Recently, a cluster of Klebsiella pneumoniae carrying both blaNDM-1 and mcr-8.2 was also reported.11 In this study, we characterized a novel mcr-8.2-bearing plasmid harbored by a multi-drug resistance (MDR) K. pneumoniae strain of chicken origin, which extended the understanding of large plasmids co-harboring mcr-8.2 and other important resistance genes.
A colistin-resistant strain 2019036D, isolated through MacConkey agar plates supplemented with colistin (4 μg/mL), was recovered from a caecal microbiota sample at a broiler chicken slaughterhouse in Sichuan, China in June 2019. The genomic DNA of the purified bacteria was extracted using the TIANamp Bacteria DNA Kit (Tiangen, China) according to the manufacturer’s instruction, and PCR targeting at mcr genes from mcr-1 to mcr-8 were performed using primers as previously described.6 The PCR product was sequenced and confirmed positive for mcr-8.2 after BLASTn analysis. 16S rRNA gene sequencing identified 2019036D isolate as K. pneumoniae. Minimum inhibitory concentrations (MICs) of different antimicrobials were measured with the broth microdilution method according to the CLSI standards with E. coli ATCC 25,922 as the control. The strain 2019036D was resistant to colistin, cefazolin, cefoxitin, cefotaxime, ceftazidime, tetracycline, nalidixic acid, erythromycin, trimethoprim/sulfamethoxazole, azithromycin, ciprofloxacin and chloramphenicol, but still susceptible to imipenem (Table S1). Conjugation assay was performed to verify the transferability of the colistin resistance gene with the E. coli C600 as the recipient strain but failed after three repeats. Subsequently, S1-PFGE showed that 2019036D harbored three plasmids of ca. 350kb, 50kb and 35kb in length. To investigate the genetic structure of mcr-8.2, the complete genome sequence of 2019036D was sequenced by short-read Illumina (150bp×2) Hiseq 2500 and long-read MinION with the rapid sequencing kit simultaneously, and de novo assembled with a hybrid strategy utilizing Unicycler.12,13 The complete genome sequences were annotated using the online RAST tool (http://rast.nmpdr.org/) and were modified manually. Plasmid replicon typing and resistance genes identification were performed using the online tools (https://cge.cbs.dtu.dk/services/). Insertion sequences were identified based on the ISfinder database.14 Circular plasmids comparison was performed by BRIG.15 Phylogenetic analysis was based on the core genome analysis of Roary and FastTree,16,17 and visualized by iTOL.18
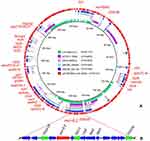
One 5369 757 bp chromosome (CP047336) and three plasmids including p2019036D-mcr8-345kb (CP047337), p2019036D-50kb (CP047338) and p2019036D-35kb (CP047339) were obtained, which was consistent with the plasmid profile observed by S1-PFGE. MLST analysis indicated that 2019036D belonged to ST395, a clinical KPC-producing and NDM-producing K. pneumoniae ST lineage.19,20 Kleborate analysis (https://github.com/katholt/Kleborate) identified no virulence genes indicating this strain was not a Hypervirulent Klebsiella pneumoniae (HvKP). Whole-genome analysis showed that a mcr-8 variant showing 100% identity to mcr-8.2 was detected in the plasmid p2019036D-mcr8-345kb belonging to IncFIB(K) replicon type. Other mcr-8.2-bearing plasmids in NCBI databases were found to harbor the backbone of IncF-type plasmid. But they showed limited homologous region to p2019036D-mcr8-345kb (Figure 1A). Among them, pD120-1_83kb belonging to IncFIB(K) showed most identity (84%) to p2019036D-mcr8-345kb but differed in most plasmid backbone, highlighting this plasmid was a novel mcr-8.2-bearing plasmid. Meanwhile, all plasmids co-harboring mcr genes and IncFIB(K) replicon in NCBI databases were retrieved and they shared few common regions to p2019036D-mcr8-345kb (Figure S1), which implied that the structure of the mcr-8.2-bearing plasmid was novel among all mcr-bearing plasmids. In addition to the mcr-8.2 gene, two multi-drug resistance regions (MRRs) were detected in p2019036D-mcr8-345kb but lacked in pD120-1_83kb, these MRRs contained floR, sul1, aac(6ʹ)-Ib-cr, aadA16, aadA2, aph(3ʹ)-Ia, blaCTX-M-27, blaDHA-1, mph(A), tet(D), dfrA12, dfrA27, aac(6ʹ)-Ib-cr and qnrB4 (Figure 1A). They were also absent in other mcr-negative IncFIB(K) plasmids (pIncFIBK and p1_020143) sharing similar backbone to p2019036D-mcr8-345kb in NCBI databases (Figure S2). The core genetic structure of mcr-8.2 with IS903B-ORF1-4-dgkA-baeS-copR-ISEcl1-ORF5-mcr-8.2-ORF6-ISKpn26-ORF7-8 in p2019036D-mcr8-345kb was identified nondistinctive to other five available mcr-8.2-bearing plasmids in nr databases (Figure 1B), demonstrating that the mcr-8.2 containing region might have a common ancestor and translocate among different plasmids. ISEcl1 was inserted in the intergenic region of mcr-8.2 and copR, reconfirming the assumption that ISEcl1 insertion occurred before mcr-8.2 mobilization and has no association with the translocation of mcr-8.2.10 Comparatively, IS903B and ISKpn26 located in the boundary regions and may play roles in the dissemination of mcr-8.2, but no circular intermediate harboring mcr-8.2 was detected. Until now, all strains positive for mcr-8.2-bearing plasmids are K. pneumoniae besides strain D120-1 identified as K. quasipneumoniae, both of which were derived from the same clade.10 Although all mcr-8.2-positive strains from different sources were Klebsiella spp., they belonged to different sequence types (STs) (Figure S3), implying that the dissemination of mcr-8.2 and its corresponding plasmids were likely limited by genus and widely spread in different clones. Thus, the prevalence of mcr-8.2 gene among Klebsiella spp. should be monitored consistently.
In addition to p2019036D-mcr8-345kb, another two resistance plasmids were identified. A multireplicon (IncR/IncN) plasmid p2019036D-50kb with 50,845 bp in length showed 100% identity at 84% coverage to plasmid sequence tig00000003 (CP021547) (Figure S4a). Another multireplicon (IncX1/IncN) plasmid p2019036D-35kb with 35,955 bp in length shared 99.67% identity at 71% coverage with p16EC-IncN (MN086778) (Figure S4b). The resistome analysis of the two plasmids indicated the presence of genes encoding resistance for beta-lactams (blaCTX-M-55, blaTEM-141), aminoglycosides (aac(3)-IV, aadA1, aadA2b, aph(3ʹ)-IIa, aph(3ʹ)-Ia, aph(4)-Ia) and sulphonamides (sul3). Together, the three MDR plasmids rendered the strain resistant to multiple antimicrobials.
In conclusion, a ST395 K. pneumoniae strain of chicken origin was found positive for a novel mcr-8.2-bearing MDR plasmid. Plasmids and core mcr-8.2-bearing structure are the genetic basis underlying the transmission of mcr-8.2 in K. pneumoniae. Continuous surveillance of mcr-8.2 in Klebsiella spp. and other bacterial pathogens of different origins is necessary to understand its potential dissemination and risk.
Acknowledgments
This work was supported by the Key Research and Development of Food Safety of the Ministry of Science and Technology of China (no. SQ2017YFC1601501), the National Key Research and Development Program of China (no. 2019YFC1605205), the Natural Science Foundation of Jiangsu Province (no. BK20180900) and the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Liu YY, Wang Y, Walsh TR, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16(2):161–168. doi:10.1016/S1473-3099(15)00424-7
2. Wang C, Feng Y, Liu L, Wei L, Kang M, Zong Z. Identification of novel mobile colistin resistance gene mcr-10. Emerg Microbes Infect. 2020;9(1):508–516. doi:10.1080/22221751.2020.1732231
3. Carroll LM, Gaballa A, Guldimann C, Sullivan G, Henderson LO, Wiedmann M. Identification of novel mobilized colistin resistance gene mcr-9 in a multidrug-resistant, colistin-susceptible Salmonella enterica serotype Typhimurium isolate. MBio. 2019;10(3). doi:10.1128/mBio.00853-19
4. Yin W, Li H, Shen Y, et al. Novel plasmid-mediated colistin resistance gene mcr-3 in Escherichia coli. mBio. 2017;8(3):e00543–17.
5. Holt KE, Wertheim H, Zadoks RN, et al. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc Natl Acad Sci U S A. 2015;112(27):E3574–3581. doi:10.1073/pnas.1501049112
6. Wang X, Wang Y, Zhou Y, et al. Emergence of a novel mobile colistin resistance gene, mcr-8, in NDM-producing Klebsiella pneumoniae. Emerg Microbes Infect. 2018;7(1):122. doi:10.1038/s41426-018-0124-z
7. Yang YQ, Li YX, Lei CW, Zhang AY, Wang HN. Novel plasmid-mediated colistin resistance gene mcr-7.1 in Klebsiella pneumoniae. J Antimicrob Chemother. 2018;73(7):1791–1795. doi:10.1093/jac/dky111
8. Hadjadj L, Baron SA, Olaitan AO, Morand S, Rolain JM. Co-occurrence of variants of mcr-3 and mcr-8 genes in a Klebsiella pneumoniae isolate from Laos. Front Microbiol. 2019;10:2720. doi:10.3389/fmicb.2019.02720
9. Farzana R, Jones LS, Barratt A, et al. Emergence of mobile colistin resistance (mcr-8) in a highly successful Klebsiella pneumoniae sequence type 15 clone from clinical infections in Bangladesh. mSphere. 2020;5(2). doi:10.1128/mSphere.00023-20.
10. Yang X, Liu L, Wang Z, Bai L, Li R. Emergence of mcr-8.2-bearing Klebsiella quasipneumoniae of animal origin. J Antimicrob Chemother. 2019;74(9):2814–2817. doi:10.1093/jac/dkz213
11. Ma K, Feng Y, Liu L, Yao Z, Zong Z. A cluster of colistin- and carbapenem-resistant Klebsiella pneumoniae carrying blaNDM-1 and mcr-8.2. J Infect Dis. 2019;221:S237–S242.
12. Wick RR, Judd LM, Gorrie CL, Holt KE. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLOS Comput Biol. 2017;13(6):e1005595. doi:10.1371/journal.pcbi.1005595
13. Li R, Xie M, Dong N, et al. Efficient generation of complete sequences of MDR-encoding plasmids by rapid assembly of MinION barcoding sequencing data. Gigascience. 2018;7(3):1–9. doi:10.1093/gigascience/gix132
14. Kichenaradja P, Siguier P, Perochon J, Chandler M. ISbrowser: an extension of ISfinder for visualizing insertion sequences in prokaryotic genomes. Nucleic Acids Res. 2010;38(suppl_1):D62–68. doi:10.1093/nar/gkp947
15. Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics. 2011;12(1):402. doi:10.1186/1471-2164-12-402
16. Page AJ, Cummins CA, Hunt M, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31(22):3691–3693. doi:10.1093/bioinformatics/btv421
17. Price MN, Dehal PS, Arkin AP. FastTree: computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol. 2009;26(7):1641–1650. doi:10.1093/molbev/msp077
18. Letunic I, Bork P. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 2016;44(W1):W242–245. doi:10.1093/nar/gkw290
19. Yang J, Ye L, Guo L, et al. A nosocomial outbreak of KPC-2-producing Klebsiella pneumoniae in a Chinese hospital: dissemination of ST11 and emergence of ST37, ST392 and ST395. Clin Microbiol Infect. 2013;19(11):E509–515. doi:10.1111/1469-0691.12275
20. Fursova NK, Astashkin EI, Gabrielyan NI, et al. Emergence of five genetic lines ST395(NDM-1), ST13(OXA-48), ST3346(OXA-48), ST39(CTX-M-14), and novel ST3551(OXA-48) of multidrug-resistant clinical Klebsiella pneumoniae in Russia. Microb Drug Resist. 2020. doi:10.1089/mdr.2019.0289.
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.