Back to Journals » Infection and Drug Resistance » Volume 12
Characterization of a novel blaNDM-5-harboring IncFII plasmid and an mcr-1-bearing IncI2 plasmid in a single Escherichia coli ST167 clinical isolate
Authors Xu L, Wang P, Cheng J, Qin S, Xie W
Received 31 October 2018
Accepted for publication 29 January 2019
Published 1 March 2019 Volume 2019:12 Pages 511—519
DOI https://doi.org/10.2147/IDR.S192998
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Sahil Khanna
Lijuan Xu,1,* Ping Wang,2,* Jing Cheng,2 Shangshang Qin,2,3 Weihong Xie1
1The First Affiliated Hospital of Zhengzhou University, Zhengzhou, Henan 450001, China; 2School of Pharmaceutical Sciences, Zhengzhou University, Zhengzhou, Henan 450001, China; 3Key Laboratory of Advanced Drug Preparation Technologies, Ministry of Education, Zhengzhou University, Zhengzhou, Henan 450001, China
*These authors contributed equally to this work
Background and purpose: The spread of the plasmid-mediated, colistin-resistance gene mcr-1 into New Delhi metallo-β-lactamase (NDM)-producing bacterial isolates can cause untreatable infections. In this study, we conducted a molecular characterization of a novel, conjugative, bla NDM-5-positive IncFII plasmid (pNDM-EC16-50) together with an mcr-1-bearing IncI2 plasmid in a single Escherichia coli ST167 clinical isolate EC16-50.
Methods and results: EC16-50, which belongs to the E. coli strain ST167 and phylogroup A, was identified to co-produce NDM-5 and MCR-1. S1-PFGE and Southern blotting showed that blaNDM-5 and mcr-1 genes were located on ~95 kb and ~65 kb plasmids, respectively. A conjugation assay revealed that both blaNDM-5- and mcr-1-bearing plasmids were self-transmissible. Comparative plasmid analysis suggested that blaNDM-5-harboring F2:A-:B-plasmid might have evolved from the well-reported NDM-carrying pMC-NDM-like plasmid via recombination with a locally emerged pSJ_94 plasmid, whereas the mcr-1-carrying IncI2 plasmid was similar to previously reported mcr-1-bearing plasmids in China.
Conclusion and impact: This study represents the first report of the NDM-5 carrying IncFII- but not IncX3-type plasmid in an MCR-1-producing E. coli isolate. More striking was the dissemination of mcr-1 in a successful epidemic NDM-5-producing E. coli clone ST167, which could facilitate the spread of colistin resistance in carbapenemase-producing E. coli isolates.
Keywords: Escherichia coli, ST167, MCR-1, IncFII plasmid, NDM-5
Introduction
Colistin is considered to be the last resort for the treatment of serious infections caused by carbapenem-resistant Enterobacteriaceae (CRE). However, the emergence and dissemination of the plasmid-mediated, colistin-resistance gene mcr-1 have seriously compromised current treatment options for CRE infections.1 The spread of the mcr-1 gene into CRE, such as New Delhi metallo-β-lactamase (NDM)-producing isolates, is of great clinical concern as this results in the emergence of true pan-drug-resistant pathogens. NDM-5, which differs from NDM-1 by two amino-acid substitutions (Val88Leu and Met154Leu) and possesses increased resistance to carbapenems and extended-spectrum cephalosporins, has become a predominant variant expressed by NDM-producing Escherichia coli since they were first reported in 2014 in China.2,3 To date, several cases of infection caused by NDM-5 and MCR-1 co-producing E. coli have been reported in the United States and China.4–10 In these bacterial isolates, the mcr-1 gene was carried by plasmids of different replicon types, including IncX4, HI2, and I2, whereas the blaNDM-5 gene in each isolate was located on IncX3 plasmids. Nearly all of the published studies from China revealed that the blaNDM-5 gene was carried by IncX3-type plasmids in E. coli isolates, and E. coli ST167 was regarded as an epidemic clone involved in the transmission of the blaNDM-5 gene.11–14 More recently, two NDM-5-carrying IncF plasmids were identified in E. coli ST167 strains from China (pNDM5_020007, CP025626) and Italy (pNDM-5-IT, MG649062),15,16 but there have been no reports of NDM-5-encoding IncF plasmids in MCR-1-producing E. coli.
In this study, we conducted a molecular characterization of a novel NDM-5-carrying F2:A-:B-plasmid and an IncI2 plasmid co-harboring mcr-1 and blaCTX-M-55 in a single E. coli ST167 clinical isolate. Our results revealed new molecular features of E. coli that co-produce NDM-5 and MCR-1.
Materials and methods
Bacterial isolates and antimicrobial susceptibility testing
A male patient without a history of foreign travel was admitted to the department of hepatobiliary surgery in a teaching hospital of Zhengzhou University in July 2016. He was diagnosed with bile-duct stones complicated by cholangitis. During the surgery, a bile sample was collected from this patient for microbiological analysis. A multidrug-resistant E. coli isolate named EC16-50 was recovered from this bile culture; the isolate was identified using the VITEK®2 system (bioMérieux, Marcy-l’Étoile, France) and further confirmed by 16S-rRNA sequencing. Antimicrobial susceptibility testing was conducted using several different assays: the broth microdilution method for ampicillin, piperacillin-tazobactam, cefazolin, ceftazidime, aztreonam, imipenem, meropenem, ciprofloxacin, gentamicin, amikacin, chloroamphenicol, doxycycline, tigecycline, and colistin; the agar dilution method for fosfomycin; and the Etest method (AB bioMérieux, Sweden) for trimethoprim/sulfamethoxazole. The results were analyzed according to the Clinical and Laboratory Standards Institute (CLSI) guidelines.17 Breakpoints for colistin and tigecycline were interpreted according to European Committee on Antimicrobial Susceptibility testing (EUCAST) guidelines (http://www.eucast.org/clinical_breakpoints/). E. coli ATCC 25922 was used as a quality control strain.
Detection of resistance determinants and bacterial genotyping
The modified Hodge test and the imipenem-EDTA double-disk synergy test were performed according to CLSI guidelines to detect carbapenemase activity.17 PCR and nucleotide sequencing were employed to screen for the presence of carbapenemase-encoding genes (blaOXA-48-like, blaIMP, blaVIM, blaKPC, and blaNDM) and the mcr-1 gene as described previously.1,18 Multi-locus sequence typing and phylogenetic grouping were performed according to published protocols (http://mlst.warwick.ac.uk/mlst/dbs/Ecoli/) and a previously described method, respectively.19
S1 nuclease-pulsed-field gel electrophoresis (S1-PFGE) and Southern blot analysis
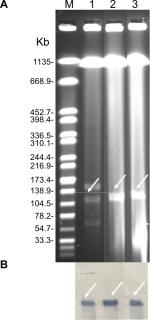
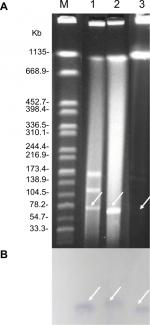
To evaluate whether the blaNDM and mcr-1 genes were present in plasmids, genomic DNA of E. coli EC16-50 was used for S1-PFGE and Southern blotting with blaNDM- and mcr-1-specific probes as previously described.18 The Salmonella enterica serotype Braenderup H9812 was used as the size marker.
Conjugation assay
A conjugation assay was conducted as previously described using E. coli J53 (sodium-azide-resistant strain) as the recipient. Transconjugants were selected on Mueller-Hinton (MH) agar supplemented with sodium azide (100 µg/mL) and imipenem (1 µg/mL) or colistin (2 µg/mL).18 PCR sequencing and antimicrobial susceptibility testing of the transconjugants were subsequently performed to confirm whether the plasmid was successfully transferred to the recipient.
Plasmid analysis
For plasmid analysis, each plasmid was transferred to E. coli DH5α by electroporation. Transformant colonies were selected on Luria-Bertani agar plates containing imipenem (1 µg/mL) or colistin (2 µg/mL) and were examined for the presence of the blaNDM and mcr-1 genes by PCR and sequencing. Plasmid DNA was extracted from a confirmed transformant using a QIAGEN Midi Kit (QIAGEN, Hilden, Germany) and subjected to complete plasmid sequencing using an Illumina MiSeq platform (Shanghai Majorbio Company, Shanghai, China). Sequencing reads were assembled de novo using SOAPdenovo v.2.04 (http://soap.genomics.org.cn/). The gaps between different contigs were closed by PCR and sequencing. Prediction and annotation of open reading frames (ORFs) were done with Glimmer 3.02 (http://cbcb.umd.edu/software/glimmer/) and Basic Local Alignment Search Tool (BLAST) of the National Center for Biotechnology Information (NCBI) (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
Nucleotide sequence accession numbers
The complete nucleotide sequences of plasmids pEC16-50-MCR and pNDM-EC16-50 have been deposited in GenBank under accession nos. MG515249 and MG515250, respectively.
Ethics statement
The bile sample and clinical isolate of E. coli EC16-50 were generated as part of routine hospital laboratory procedures. The patient’s next of kin provided written, informed consent for details that were included in the manuscript.
Results and discussion
Isolate characteristics
The antimicrobial susceptibility testing results showed that strain EC16-50 was resistant to all of the β-lactams including carbapenems, fluoroquinolones, gentamicin, amikacin, doxycycline, and colistin, but susceptible to fosfomycin, tigecycline, and trimethoprim/sulfamethoxazole (Table 1). The positive result for the imipenem-EDTA double-disk synergy test revealed that EC16-50 was a metallo-β-lactamase producer. Finally, PCR and sequencing showed that EC16-50 co-harbored both blaNDM-5 and mcr-1 genes.
Strain EC16-50 was assigned to the E. coli ST167 strain and phylogroup A. To date, E. coli ST167 is classified as an internationally disseminated clonal lineage associated with the global spread of CTX-M-15 extended-spectrum-β-lactamase and NDM metallo-β-lactamases in humans and other animals.20,21 Recently, reports from different parts of China indicated that E. coli ST167 exhibited close linkages with the blaNDM-5 gene,13,14 and dissemination of mcr-1 within the successful epidemic clone may facilitate further spread of colistin resistance in carbapenemase-producing E. coli isolates. This led to the emergence of extensively drug-resistant pathogens. To prevent such pathogens from developing further, the spread of E. coli ST167 that co-produces MCR-1 and NDM-5 should be closely monitored in China.
Plasmid location
S1-PFGE followed by Southern blotting was able to identify that the blaNDM-5 and mcr-1 genes in EC16-50 were on ~95 kb and ~65 kb plasmids, respectively (Figures S1 and S2). Then we performed conjugative assays to confirm their transferability, and both mcr-1- and blaNDM-5-carrying plasmids in EC16-50 were successfully transferred into E. coli J53.
Characterization of the blaNDM-5-carrying plasmid
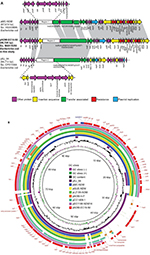
We found that the blaNDM-5-carrying plasmid, named pNDM-EC16-50, was 95,724 base pairs (bp) in size. It consisted of an average guanine and cytosine (GC) content of 51.2% and 84 predicted ORFs. The replication region of pNDM-EC16-50 comprised repA2, repA1, and repA4 and was completely identical to other F2:A-:B-plasmids, such as pMC-NDM (HG003695) and pGUE-NDM (JQ364967). BLASTn results for pNDM-EC16-50 against the NCBI non-redundant database showed that the plasmid shared 99% identity with several IncFII-type plasmids carrying blaNDM (including pMC-NDM, pGUE-NDM, pEC1188-NDM16 [MH213345], pLZ135-NDM [MF353156], and pCC1409 [KT725789]), with query coverages ranging from 70% to 81%. The blaNDM-5-containing multidrug resistance mosaic region (MRR) in pNDM-EC16-50 was almost identical to that of pMC-NDM, except for 1) two point mutations in blaNDM-1 leading to the occurrence of the blaNDM-5 allele, 2) a 160-bp sequence insertion between the IS26 and ΔISAba125 elements, and 3) an intact ISCR1 element without IS26 inserted. The extensive backbone region upstream of the conjugative transfer operon tra-trb in pNDM-EC16-50 showed new features compared to typical blaNDM-harboring IncFII plasmids, such as pGUE-NDM (blaNDM-1) and pMC-NDM (blaNDM-1) reported in Europe and pCC1409 (blaNDM-5) reported in Korea (Figure 1B).22–24 A 13,592 bp fragment in pNDM-EC16-50’s backbone region included scaffold genes responsible for plasmid partition (parM) and stability (stbB) and a group II intron that was virtually identical (100% query coverage, 99.6% identity by BLASTn analysis in NCBI) to the plasmid pGUE-NDM/pMC-NDM (the group II intron is missing from pMC-NDM). As shown in Figure 1A, Region 1 (ydaAB-ydbA-ycdA-ssb-parB-psiAB-mok-hok-yubOP-X-polypeptide), which is present in both pGUE-NDM and pMC-NDM, was replaced by a 16,182 bp pSJ_94 (CP011064)-derived accessory module (100% query coverage and 100% nucleotide identity; designated as Region 2) composed of mobile genetic elements and a number of functional genes, such as those encoding putative alpha-galactosidase, lactose permease, and sucrose-6-phosphate hydrolase (Figure 1A). In addition, we observed that the entire tra-trb region, excluding traX gene in pNDM-EC16-50, was more similar to its counterpart on pSJ_94, except for 1) a 50-nucleotide difference in the traI gene, 2) a 1-nucleotide difference in the traT and traP genes, and 3) the number of CAACAGCCA tandem repeats in the traD gene (61 and 11 repeats in pNDM-EC16-50 and pSJ_94, respectively). Thus, the overall structure of pNDM-EC16-50 can be divided into two distinct modules (Figure 1A): an ~47.9 kb pMC-NDM-derived module containing the IncFII replication region, partial backbone region, and the entire MRR and an ~47.8 kb pSJ_94-like module encompassing multiple functional genes and the tra-trb gene clusters. These findings suggested that pNDM-EC16-50 might have evolved from a recombination of the blaNDM-carrying F2:A-:B- type pMC-NDM-like plasmid and the pSJ_94-like plasmid belonging to F36:A4:A20:B- type. Moreover, we speculated that pNDM-EC16-50 may have been created by Region 2 of a pSJ_94-like plasmid displacing Region 1 on a pMC-NDM-like plasmid.
Previous studies from China revealed that the IncX3-type pNDM-MGR194-like plasmid contributed to the dissemination of the blaNDM-5 gene among Enterobacteriaceae, and blaNDM-5 gene is mostly carried on this type of plasmid in E. coli ST167.12,13 Recently, two novel blaNDM-5-positive IncF plasmids harboring two or more IncF replicons belonging to F36:A4:B- (pNDM5_020007, China) and F36:F31:A4:B1 (pNDM-5-IT, Italy) plasmid types were identified in E. coli ST167 strains.15,16 In addition, three blaNDM-5-carrying IncFII-type plasmids, whose sequences were submitted to the GenBank database in 2017, were detected in clinical E. coli (pGZ3_NDM5, CP017981 and pLZ135-NDM, MF353156) isolates and the S. enterica serovar Typhimurium strain (pST41-NDM, CP019444) in China.8,25 These observations, together with our findings, suggested that conjugative IncFII-type plasmids might have emerged as a potential vehicle (other than the IncX3-type plasmid) to mediate the spread of blaNDM-5 in China. In this study, the transfer frequency of pNDM-EC16-50 determined by conjugation assay (2.5×10−7 transconjugants per donor cell) was much lower than that of the representative blaNDM-bearing IncFII plasmids pGUE-NDM (2.5×10−4 transconjugants per donor cell) and pMC-NDM (2×10−3 transconjugants per donor cell)22,23 and blaNDM-1-bearing IncX3 plasmids (10−1 to 10−5 transconjugants per donor cell).26 In addition, Shin et al reported a prominent instability profile of the blaNDM-5-bearing IncFII plasmid in a Klebsiella pneumoniae isolate.27 In light of such evidence, we speculated that the low transfer frequency and instability in the host of the IncFII-type plasmid compared to the IncX3-type plasmid may be responsible for the scarce reporting of blaNDM-5-bearing IncFII plasmids in NDM-producing E. coli isolates. Clearly, these elusive plasmids need closer investigation.
Characterization of the plasmid bearing mcr-1
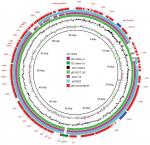
We found that the mcr-1-carrying plasmid in EC16-50, named pEC16-50-MCR, was 65,978 bp in size and belonged to the IncI2 incompatibility group. It contained 80 ORFs and had an average GC content of 42.6%. We further revealed that pEC16-50-MCR consisted of 1) a typical IncI2 plasmid backbone region that is responsible for plasmid replication, maintenance, and transfer, 2) a 3,090 bp ISEcp1-blaCTX-M-55 transposition unit, and 3) a 2,471 bp mcr-1 cassette composed of the mcr-1 gene and a PAP2-like ORF (Figure 2). Similar IncI2 plasmids co-harboring mcr-1 and blaCTX-M-55 genes have been described, such as pA31-12 (KX034083) and pSCS23 (KU934209) in E. coli and S. enterica isolates, respectively.28,29 It is noteworthy that an ISApl1 mobile element was present upstream of the mcr-1 cassette in both pA31-12 and pSCS23, whereas it was absent in pEC16-50-MCR. Snesrud et al hypothesized that the loss of ISApl1 during the course of evolution increased the stability of mcr-1 in plasmid vectors due to ISApl1’s pivotal role in the mobilization of mcr-1.30 Therefore, the mcr-1 cassette lacking ISApl1 and co-occurring with blaCTX-M genes on pEC16-50-MCR-like conjugative plasmids may facilitate widespread dissemination of mcr-1. A BLASTn comparison revealed that the sequence of pEC16-50-MCR was very similar to that of the plasmid pE15017_00 (KX772778), with 99% query coverage and 99% nucleotide identity. We found that the major difference between pEC16-50-MCR and pE15017_00 lay in the shufflon region, a DNA segment that consists of multiple DNA inversions and is involved in site-specific recombination in IncI2-type plasmids.31 Both segments C and BD of IncI2 shufflons were found to be present in the shufflon region of pEC16-50-MCR, while segment C was found to be missing from pE15017_00, indicating that rearrangement events had taken place in the shufflon region of pE15017_00 mediated by the recombinase (Figure 2).
Conclusion
In summary, we characterized a novel, conjugative, blaNDM-5-carrying IncFII plasmid that coexists with a self-transmissible IncI2 plasmid harboring both mcr-1 and blaCTX-M-55 genes in a successful epidemic E. coli clone ST167. Unlike IncX3-type plasmids, which are common vehicles for dissemination of blaNDM-5, the IncF plasmid is currently only sporadically reported to carry blaNDM-5 in E. coli from China. Therefore, the role of this type of plasmid in the dissemination of blaNDM-5 in E. coli needs further investigation. In addition, the acquisition of mcr-1-bearing plasmids in widespread NDM-5-producing E. coli ST167 clones could contribute to the spread of polymyxin resistance within CRE strains. Overall, our study highlights the urgent need for enhanced surveillance of the spread of E. coli ST167 strains co-producing MCR-1 and NDM-5.
Acknowledgments
This work was supported by grant 81501782 from the National Natural Science Foundation of China and the Outstanding Young Teacher Research Fund of Zhengzhou University (32210464).
Author contributions
All authors contributed toward data analysis, drafting and critically revising the paper and agree to be accountable for all aspects of the work. LX, SQ, and WX designed and supervised the study. PW conducted the primary and confirmatory diagnostics for carbapenemase gene presence, species identification, and bacterial genotyping. PW and JC prepared the samples for Illumina MiSeq, performed the data analyses, and conducted most of the molecular experiments. SQ wrote the manuscript. All of the authors reviewed and approved the final manuscript.
Disclosure
The authors report no conflicts of interest in this work.
References
Liu Y-Y, Wang Y, Walsh TR, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16(2):161–168. | ||
Yang P, Xie Y, Feng P, et al. blaNDM-5 carried by an IncX3 plasmid in Escherichia coli sequence type 167. Antimicrob Agents Chemother. 2014;58:7548–7452. | ||
Zhang Y, Wang Q, Yin Y, et al. Epidemiology of Carbapenem-Resistant Enterobacteriaceae Infections: report from the China CRE Network. Antimicrob Agents Chemother. 2018;62:e01882–17. | ||
Mediavilla JR, Patrawalla A, Chen L, et al. Colistin- and carbapenem-resistant Escherichia coli Harboring mcr-1 and blaNDM-5, causing a complicated urinary tract infection in a patient from the United States: TABLE 1 . mBio. 2016;7(4):e01191–16. | ||
Yu H, Qu F, Shan B, et al. Detection of the mcr-1 Colistin resistance gene in Carbapenem-resistant Enterobacteriaceae from different hospitals in China. Antimicrob Agents Chemother. 2016;60(8):5033–5035. | ||
Mao J, Liu W, Wang W, et al. Antibiotic exposure elicits the emergence of colistin- and carbapenem-resistant Escherichia coli coharboring MCR-1 and NDM-5 in a patient. Virulence. 2018;9(1):1001–1007. | ||
Feng S, Shen C, Chen H, et al. Co-production of MCR-1 and NDM-5 in Escherichia coli isolated from a colonization case of inpatient. Infect Drug Resist. 2018;11:1157–1161. | ||
Zhang Y, Liao K, Gao H, et al. Decreased Fitness and Virulence in ST10 Escherichia coli Harboring blaNDM-5 and mcr-1 against a ST4981 Strain with blaNDM-5. Front Cell Infect. Microbiol.. 2017;7:242. | ||
Zheng B, Lv T, Xu H, et al. Discovery and characterisation of an Escherichia coli ST206 strain producing NDM-5 and MCR-1 from a patient with acute diarrhoea in China. Int J Antimicrob Agents. 2018;51(2):273–275. | ||
Liu L, Feng Y, Zhang X, Mcnally A, Zong Z. New Variant of mcr-3 in an Extensively Drug-Resistant Escherichia coli Clinical Isolate Carrying mcr-1 and bla NDM-5. Antimicrob. Agents Chemother. 2017;61(12):01757–17. | ||
Chen D, Gong L, Walsh TR, et al. Infection by and dissemination of NDM-5-producing Escherichia coli in China: Table 1. J. Antimicrob. Chemother.. 2016;71(2):563–565. | ||
Zhang F, Xie L, Wang X, et al. Further Spread of bla NDM-5 in Enterobacteriaceae via IncX3 Plasmids in Shanghai, China. Front Microbiol. 2016;7:424. | ||
Li X, Fu Y, Shen M, et al. Dissemination of blaNDM-5 gene via an IncX3-type plasmid among non-clonal Escherichia coli in China. Antimicrob Resist Infect Control. 2018;7(1):59. | ||
Huang Y, Yu X, Xie M, et al. Widespread Dissemination of Carbapenem-Resistant Escherichia coli Sequence Type 167 Strains Harboring blaNDM-5 in Clinical Settings in China. Antimicrob. Agents Chemother.. 2016;60(7):4364–4368. | ||
Feng Y, Liu L, Mcnally A, Zong Z. Coexistence of Two bla NDM-5 Genes on an IncF Plasmid as Revealed by Nanopore Sequencing. Antimicrob Agents Chemother. 2018;62(5):00110–00118. | ||
Giufrè M, Errico G, Accogli M, et al. Emergence of NDM-5-producing Escherichia coli sequence type 167 clone in Italy. Int J Antimicrob Agents. 2018;52(1):76–81. | ||
Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing. CLSI Supplement M100. 27th ed. Wayne, PA: Clinical and Laboratory Standards Institute; 2017. | ||
Qin S, Fu Y, Zhang Q, et al. High incidence and endemic spread of NDM-1-positive Enterobacteriaceae in Henan Province, China. Antimicrob Agents Chemother. 2014;58(8):4275–4282. | ||
Qin S, Zhou M, Zhang Q, et al. First identification of NDM-4-producing Escherichia coli ST410 in China. Emerg Microbes Infect. 2016;5(1):e118:1–3. | ||
Irrgang A, Falgenhauer L, Fischer J, et al. CTX-M-15-producing E. coli isolates from food products in germany are mainly associated with an IncF-Type plasmid and belong to two predominant clonal E. coli lineages. Front. Microbiol.. 2017;8:2318. | ||
Grönthal T, Österblad M, Eklund M, et al. Sharing more than friendship – transmission of NDM-5 ST167 and CTX-M-9 ST69 Escherichia coli between dogs and humans in a family, Finland, 2015. Euro Surveill. 2018;23(27):1560–7917. | ||
Bonnin RA, Poirel L, Carattoli A, Nordmann P. Characterization of an IncFII plasmid encoding NDM-1 from Escherichia coli ST131. PLoS One. 2012;7(4):e34752. | ||
Fiett J, Baraniak A, Izdebski R, et al. The first NDM metallo-β-lactamase-producing Enterobacteriaceae isolate in Poland: evolution of IncFII-type plasmids carrying the bla(NDM-1) gene. Antimicrob Agents Chemother. 2014;58(2):1203–1207. | ||
Shin J, Baek JY, Cho SY, et al. bla NDM-5 -Bearing IncFII-type plasmids of Klebsiella pneumoniae sequence type 147 transmitted by cross-border transfer of a patient. Antimicrob. Agents Chemother. 2016;60(3):1932–1934. | ||
Li X, Jiang Y, Wu K, et al. Whole-genome sequencing identification of a multidrug-resistant Salmonella enterica serovar Typhimurium strain carrying bla NDM-5 from Guangdong, China. Infect Genet Evol. 2017;55:195–198. | ||
Ho P-L, Li Z, Lo W-U, et al. Identification and characterization of a novel incompatibility group X3 plasmid carrying bla NDM-1 in Enterobacteriaceae isolates with epidemiological links to multiple geographical areas in China. Emerg Microbes Infect. 2012;1(1):e39:1–6. | ||
Shin J, Baek JY, Chung DR, Ko KS. Instability of the IncFII-type plasmid carrying blaNDM-5 in a Klebsiella pneumoniae Isolate. J Microbiol Biotechnol. 2017;27(9):1711–1715. | ||
Sun J, Li XP, Yang RS, et al. Complete nucleotide sequence of an IncI2 plasmid Coharboring blaCTX-M-55 and mcr-1. Antimicrob Agents Chemother. 2016;60(8):5014–5017. | ||
Yang Y-Q, Zhang A-Y, Ma S-Z, et al. Co-occurrence of mcr-1 and ESBL on a single plasmid in Salmonella enterica. J Antimicrob Chemother. 2016;71(8):2336–2338. | ||
Snesrud E, He S, Chandler M, et al. A Model for Transposition of the Colistin Resistance Gene mcr-1 by IS Apl1. Antimicrob. Agents Chemother. 2016;60(11):6973–6976. | ||
Sekizuka T, Kawanishi M, Ohnishi M, et al. Elucidation of quantitative structural diversity of remarkable rearrangement regions, shufflons, in IncI2 plasmids. Sci Rep. 2017;7(1):928. |
Supplementary materials
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.