Back to Journals » Cancer Management and Research » Volume 12
CFTR Regulates the Proliferation, Migration and Invasion of Cervical Cancer Cells by Inhibiting the NF-κB Signalling Pathway
Authors Wu Z, Li J, Zhang Y, Hu L, Peng X
Received 3 March 2020
Accepted for publication 7 May 2020
Published 18 June 2020 Volume 2020:12 Pages 4685—4697
DOI https://doi.org/10.2147/CMAR.S252296
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Seema Singh
Zhao Wu, 1 Jinke Li, 2 Yi Zhang, 2 Lina Hu, 3 Xue Peng 2
1Department of Obstetrics and Gynecology, Sichuan Academy of Medical Sciences and Sichuan Provincial People’s Hospital, Chengdu, Sichuan, People’s Republic of China; 2Department of Obstetrics and Gynecology, West China Second University Hospital, Sichuan University; Key Laboratory of Birth Defects and Related Diseases of Women and Children (Sichuan University), Ministry of Education, Chengdu, Chengdu, Sichuan, People’s Republic of China; 3Department of Obstetrics and Gynecology, Second Affiliated Hospital, Chongqing Medical University, Chongqing, People’s Republic of China
Correspondence: Xue Peng
Department of Obstetrics and Gynecology, West China Second University Hospital and Key Laboratory of Birth Defects and Related Diseases of Women and Children (Sichuan University), Ministry of Education, No. 20 3rd Section, South Renmin Road, Chengdu 610041, Sichuan, People’s Republic of China
Tel +86-13880487168
Email [email protected]
Background: The cystic fibrosis transmembrane conductance regulator (CFTR) and nuclear factor κB (NF-κB) signalling pathways are currently regarded as co-regulators of the occurrence of cervical cancer. However, the detailed mechanism of CFTR- and NF-κB-mediated effects in cervical cancer remains to be elucidated. This study aimed to investigate the mechanism by which CFTR and NF-κB influence the development of cervical cancer.
Patients and Methods: CFTR ΔF508 mutation and CFTR promoter methylation were detected in cervical tissue samples. NF-κB p65 and IκBα protein levels were tested in HeLa cells with CFTR overexpression and knockdown by Western blotting. The effects of CFTR on cell proliferation, migration, and invasion were examined in HeLa cells by WST-1 and soft agar assays, cell wound scratch assay, and Matrigel invasion assays, respectively. The protein–protein interaction (PPI) network was constructed by using GeneMANIA. GeneCoDis3 was used to perform Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) functional enrichment analysis on genes in the PPI network.
Results: CFTR mutation and CFTR promoter methylation were not associated with the occurrence of cervical cancer. NF-κB p65 protein levels were decreased in CFTR overexpression lines and increased in CFTR knockdown lines, and IκBα levels were affected in the opposite manner, indicating that CFTR inhibited the NF-κB signalling pathway. CFTR also regulated the cell proliferation, migration, and invasion ability of cervical cancer cells. When CFTR was overexpressed, cell proliferation, migration, and invasion ability were decreased. There were 20 genes that interacted with CFTR. KEGG pathway analysis showed enrichment in the gastric acid secretion, chemokine signalling, bile secretion and apoptosis pathways.
Conclusion: CFTR plays an important role in cancer cell proliferation, migration and invasion by inhibiting the NF-κB signalling pathway in cervical cancer.
Keywords: cervical cancer, cystic fibrosis transmembrane conductance regulator, nuclear factor κB, signalling pathway
Introduction
Cervical cancer is one of the major malignant tumours, seriously threatening the life and health of women all over the world and bringing heavy burdens to each affected country, society and family.1 Human papillomavirus (HPV) infection has been extensively investigated and proven to be a pathogenic mechanism of both cervical cancer and its precursor lesions.2 Recently, some studies have reported the existence of non-viral molecular pathways contributing to the disease, among which long-term chronic inflammation plays an important role .3,4 However, the molecular mechanism of non-viral molecular pathways in cervical cancer is not well understood. Therefore, elucidation of the involved molecular mechanism can not only fully explain the aetiology and pathogenesis but can also prevent and treat the disease.
Cystic fibrosis transmembrane conductance regulator (CFTR) is a cAMP-dependent chloride channel protein encoded by the cystic fibrosis (CF) gene.5 It is present in a number of human tissues, including the uterine cervix.6 There are more than 1500 mutation forms of the CFTR gene, and one of the most common forms CFTR ΔF508 can lead to a high incidence of chromosome recessive genetic disease CF in white people.7 CFTR gene mutation, protein expression or dysfunction may induce a series of reproductive health-related diseases.8
A variety of human malignancies are associated with chronic inflammation, including cervical cancer and chronic cervicitis, and it is estimated that approximately 15% of human cancers are associated with chronic inflammation. Many inflammatory cytokines, such as tumour necrosis factor α (TNF-α) and interleukin-1 (IL-1), play important biological roles mainly by activating NF-κB inflammatory signalling pathways.9 NF-κB is a group of transcription factors closely related to cell apoptosis, growth and proliferation, and the inflammatory response. Studies have shown that abnormal activation of NF-κB is involved in the development of cervical cancer, which may be related to the sensitivity of cervical cancer cells to chemoradiotherapy. Constitutive activation of NF-κB may play a crucial part in the transformation of chronic inflammation into cancer.10
Our previous work suggested that the expression levels of CFTR and NF-κB were correlated and that abnormal expression was closely related to cervical cancer. In addition, the coexpression of the two was related to the malignant behaviour of cervical cancer cells and a poor prognosis in cervical cancer patients.11,12 Therefore, in this study, we aimed to further clarify whether CFTR is involved in the occurrence of cervical cancer and to determine the specific mechanism involved.
Patients and Methods
Patient Samples
Subjects were included from the Department of Gynaecology and Obstetrics, West China Second Hospital of Sichuan University. The patient samples were collected as previously described.12 A total of 135 cervical tissue samples were collected, including 20 samples of normal cervical tissue, 35 samples of cervical intraepithelial neoplasia, and 80 samples of squamous cell carcinoma of the uterine cervix. All study procedures were approved by the ethics committee of West China Second Hospital of Sichuan University and performed with the informed consent of the patients.
Exome Sequencing of CFTR
Genomic DNA was isolated from the tissue samples obtained from the available patients according to the standard procedures. The qualified genomic DNA was utilized to execute the exome target enrichment by Agilent SureSelect Human All Exon 50Mb Exon kit (Agilent Technologies, CA, USA). The captured library was sequenced on the Illumina HiSeq 2500 sequencer with paired-end 125-bp and mean coverage of 100X.
Cell Line and Cell Culture
The human cervical cancer cell lines HeLa, SiHa, Caski and H8 were obtained from the China Center for Type Culture Collection (CCTCC, Wuhan, China). HeLa cells originated from cervical adenocarcinoma, and all cell lines were cultured in RPMI 1640 (Gibco, Paisley, UK) supplemented with 100 μg/mL streptomycin, 100 U/mL penicillin, and 10% foetal bovine serum (Gibco) at 37°C under humidified incubator conditions with 5% CO2.
Western Blot Analysis
Nuclear and cytosolic fractions from cell lines were collected by using a Nuclear and Cytoplasmic Protein Extraction Kit (Shanghai Sangon Biotechnology Co, Ltd, Shanghai, China) according to the manufacturer’s instructions. Equal amounts of protein (50 μg/lane) were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to a polyvinylidene difluoride membrane (Millipore Corporation, Bedford, MA). Subsequently, the transferred membranes were blocked in defatted milk dissolved in TBST buffer at room temperature for 1 h. Then, the membranes were incubated with mouse anti-human CFTR monoclonal antibody (diluted 1:500; R&D Systems), rabbit anti-human IκBα monoclonal antibody (diluted 1:1000; Cell Signaling Technology) or rabbit anti-human NF-κB monoclonal antibody (diluted 1:1000; Cell Signaling Technology) at 4°C overnight. On the second day, after being washed three times in TBST, the membranes were incubated with horseradish peroxidase-conjugated secondary antibody for 1 h at room temperature. The immunoreactive bands interacted with an enhanced chemiluminescence (ECL) reagent (Millipore Corporation) and were quantified by densitometry with Bio-Rad Universal Hood and Quantity One software (Bio-Rad Laboratories, Hercules, CA). Nuclear or cytosolic protein levels in each lane were normalized to A-actin or histone H1 levels, respectively.
Methylation-Specific PCR (MSP)
Total DNA was extracted by Chelex-100, and then the genomic DNA was modified by an EZ DNA Methylation kit (Zymo Research, USA) according to the manufacturer’s instructions. The sequences of the primers were as follows: methylated CFTR forward primer 5ʹ-AGAGGTCGCGATTGTCGTT-3ʹ and reverse primer 5ʹ-CGACTTTCTCCACCCACTACG-3ʹ; unmethylated CFTR forward primer 5ʹ-TTAAAGAGAGGTTGTGATTGTTGTT-3ʹ and reverse primer 5ʹ-TCCTTCACTCCCTCACCA-3ʹ, which were designed by Primer Premier 6.0 software. The following protocol was used: 10 s at 98°C and 30 s at 55°C (30–40 cycles) and 30 s at 72°C.
WST-1 Cell Proliferation Assay
Briefly, the cells in the logarithmic phase were cultured in 96-well plates. After subsequent subculture overnight at 37°C with 5% CO2, the medium was refreshed every 24 h, and WST-1 reagent solution (Roche Diagnostics, Switzerland) was added to the experimental group. Each sample was analysed using Thermo Scientific Varioskan® Flash (Thermo Fisher Scientific, USA) at A440 nm after 4 h. As blank controls, the wells were treated with 100 μL medium and 10 μL WST-1. All counts were performed using three technical replicates of each sample. The absorbance value was positively correlated with the number of living cells, and the growth curve was plotted for 7 consecutive days.
Soft Agar Assay
Agar (Amresco Agarose, USA) was prepared as a 1.2% solution of normal saline and autoclaved for sterilization. A 0.6% agar/medium base layer added to each well prevented the cells from attaching and forming a monolayer on the plastic substrate. Cells at the logarithmic growth stage were harvested, and the cell concentration was adjusted to 1×104/mL after digestion with trypsin. The upper layer of 0.3% AGAR was prepared by mixing 0.6% low-melting agarose with 2 × cell culture medium at a 1:1 volume. A total of 1 mL of the upper layer of agarose and 100 mL of the single-cell suspension (approximately 1000 cells) were added to each well and solidified at room temperature. After incubation at 37°C with 5% CO2 for 1–2 weeks, colonies were scored under an inverted microscope, and the number of colonies containing more than 50 cells was counted.
Cell Wound Scratch Assay
The cells in logarithmic growth phase digested by pancreatin were inoculated in 6-well plates in a cell incubator with 37°C and 5% CO2 overnight. On the second day, a wound was made by scratching the monolayer with a pipette tip in a straight line. The cells were washed with PBS 3 times, and 2 mL serum-free medium was added to resuspend the cells. The cells were kept in the incubator, viewed and photographed under an inverted microscope at 0 h and 72 h, respectively.
Matrigel Invasion Assay
Transwell invasion assays were performed to determine cell invasion ability. The upper chamber was precoated with 5 mg/mL Matrigel (BD Biosciences, USA) at 37°C for at least 4 h. The cells were digested and washed 3 times with RPMI 1640 and then suspended in serum-free medium. Then, 200 μL (approximately 1×104 cells) of the suspension was added to the upper chamber, while RPMI-1640 medium containing 15% FBS was added to the lower chamber. After incubation for 24 h at 37°C with 5% CO2, the Transwell was inverted and dried at room temperature. Then, 0.1% crystal violet solution (Beyotime, China) was added to the dye for 30 min. Stained cells were counted under a light microscope (Nikon, Japan).
GeneMANIA Analysis
GeneMANIA (http://www.genemania.org) is a flexible, user-friendly web interface for establishing a protein–protein interaction (PPI) network, generating hypotheses about gene function, analysing gene lists and prioritizing genes for functional assays. The website can set the source of the edge of the network, and it features several bioinformatics methods to assess physical interactions, gene coexpression, gene co-expression, and gene enrichment and to predict gene functions. We used GeneMANIA to visualize the gene networks and predict the function of genes.
Functional Annotation
GeneCoDis3 (http://genecodis.cnb.csic.es/analysis) was used to perform Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) function enrichment analysis on genes in the PPI network. The selection criterion was P <0.05, and the top 15 GO enrichments and KEGG enrichments are graphically displayed.
Statistical Analysis
All of the assays were performed at least three times independently. All data were analysed by SPSS 17.0 statistical software. Fisher’s exact test was used. The Kruskal–Wallis H-test was used for measurement data. Pearson’s correlation coefficient was used for correlation analysis. P<0.05 was considered statistically significant.
Results
CFTR Mutation and Methylation of the CFTR Promoter Did Not Play a Role in the Occurrence of Cervical Cancer
To investigate whether CFTR was the wild type in cervical cancer tissue, the 10th exon of CFTR in normal and cervical cancer tissues was sequenced to detect the ΔF508 mutation. The results showed that there was no ΔF508 locus mutation in any inspection samples, indicating that the occurrence of cervical cancer was not associated with the CFTR mutation (Figure 1A).
According to a previous study, promoter methylation promotes high expression of the CFTR gene.13 Therefore, we detected the methylation of the CFTR promoter in normal cervical tissue, cervical cancer tissue and all kinds of cervical cell lines (H8, HeLa, SiHa, Caski). As shown in Figure 1B, methylation stripe amplification was not found in either the normal cervix or the cervical cancer tissues. This indicated that methylation of the CFTR promoter causing high CFTR expression was not related to cervical cancer.
CFTR Inhibited NF-κB P65 Expression and Promoted IκBα Expression in HeLa Cells
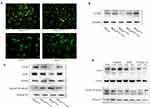
To investigate the interaction of CFTR and the NF-κB signalling pathway in cervical cancer, we constructed CFTR overexpression and knockdown HeLa cell lines using the CFTR overexpression plasmid peGFP-C3-CFTR and the control plasmid peGFP-C3, the CFTR knockdown plasmid pEF6/V5-His-CFTR and the control plasmid pEF6/V5-His. Green fluorescence was observed in all four cell lines (Figure 2A). Western blotting results revealed that CFTR expression was almost the same in the two control cell lines HeLapeGFP-control and HeLapEF/His-control and was significantly increased in HeLaCFTR cells and dramatically decreased in HeLaCFTR-kd cells (Figure 2B).
Furthermore, we examined the expression of NF-κB p65 and IκBα in HeLa cells. Compared with control cells, CFTR overexpression significantly inhibited the protein level of NF-κB p65 and increased the protein level of IκBα, and the results were reversed in CFTR knockdown cells (Figure 2C). Therefore, we predicted that CFTR was likely to act in cervical cancer by inhibiting the NF-κB signalling pathway.
To further test our conjecture, HeLa cells were treated with the CFTR activator forskolin and IBMX and the CFTR inhibitor CFTR inh-172 and the expression of NF-κB p65 was detected. The results showed that NF-κB p65 expression decreased when cells were treated with forskolin, especially at a concentration of 20 μM. During IBMX stimulation, the change in NF-κB p65 expression was similar to the change in CFTR expression, which might be because IBMX affects other signalling pathways related to NF-κB p65 in addition to CFTR. In addition, the NF-κB p65 protein level increased with the increase in CFTR inh-172 concentration (Figure 2D). The results of plasmid and drug studies suggested that CFTR may have a feedback inhibitory effect on the NF-κB signalling pathway.
CFTR Involved in HeLa Cell Proliferation
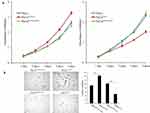
WST-1 experiment assay results indicated that HeLa cell proliferation was enhanced when CFTR was knocked out but decreased when CFTR was overexpressed (P <0.05) (Figure 3A). This may be related to the effect of CFTR on the expression and activation of genes related to cell proliferation downstream through NF-κB.
Then, the effect of the CFTR gene on cell proliferation was further verified by soft Agar clonal formation experiments. Compared to that in control cells, the number of colonies formed by HeLa cells with CFTR knockdown was significantly increased (P <0.05), while the number of colonies formed by HeLa cells with CFTR overexpression was significantly decreased (P<0.05) (Figure 3B). These results combined with those of the cell proliferation experiments demonstrate that CFTR inhibits HeLa cell proliferation.
CFTR Affected Cell Migration Ability
The effect of CFTR on HeLa cell migration was detected by a cell wound scratch assay. The migration ability of HeLa cells with CFTR knockdown was significantly improved, while it was reduced in CFTR overexpression lines compared with control cell lines (Figure 4).
CFTR Regulated the Ability of Cells Invasion
Through a Matrigel invasion assay, we clarified the effect of CFTR on the invasion ability of HeLa cervical cancer cells. The results showed that the invasion ability of HeLa cells was decreased when CFTR was overexpressed, and the number of cells passing through the Transwell chamber was significantly lower than that in the control group (P<0.05), while the invasion ability of HeLa cells was markedly increased when CFTR was knocked out (P<0.05), and the number of cells passing through the Transwell chamber was increased (Figure 5). This indicated that the change in CFTR expression was closely related to tumour invasion ability.
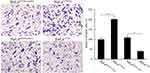 |
Figure 5 Effects of CFTR on cell invasion. Cell invasion of HeLa was measured through matrigel invasion assay. *P < 0.05. |
Genes Interacting with CFTR in Cervical Cancer
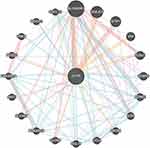
The PPI network analysis of the genes interacting with CFTR was performed using GeneMANIA (http://genemania.org/) to determine the relationship of the interactions between the genes and the main functions involved. As shown in Figure 6, the genes interacting with CFTR included SLC9A3R1, DNAJC5, STIP1, VIM, KCNJ1, CEBPD, ADRB2, EZR, ABCB1, CCL4, CCL11, CLCN3, PRKAR1B, SLC4A7, ANO1, PRKAR1A, PDZD3, PRKX, NKX3-2 and CXCL12.
GO and KEGG Pathway Analyses of Genes Correlated with CFTR in Cervical Cancer
GO term analysis of significant CFTR-related genes by GeneCoDis3 showed that genes correlated with CFTR in the PPI network were mainly related to water transport, transmembrane transport, the apical plasma membrane, chemokine activity, protein binding and PDZ domain binding. KEGG pathway analysis showed enrichment in the gastric acid secretion, chemokine signalling, bile secretion and apoptosis pathways (Figure 7).
 |
Figure 7 (Continued). |
 |
Figure 7 (Continued). |
 |
Figure 7 (Continued). |
Discussion
Several studies have shown that CFTR is involved in the occurrence and development of tumours. It has been demonstrated that the CFTR gene may be associated with certain tumours; for example, mutation of the CFTR gene could increase the risk of pancreatic cancer,14 and the occurrence of lung cancer and prostate cancer is also related to CFTR.15,16 This suggests that CFTR and its mutations play an important role in tumorigenesis. The CFTR gene has more than 1000 identified mutant forms, of which ΔF508 is the most common; this mutation is a 3-base-pair deletion (CTT) 1653–1655 in exon 10, which results in the loss of phenylalanine at 508 and accounts for approximately 70% of all mutations17,18 and is associated with various diseases, including certain cancers.14–16 CFTR ΔF508 may inhibit breast cancer cell proliferation; nonetheless, the results are not supported by clinical studies. However, the findings suggest that CFTR ΔF508 may have an effect on tumour biological behaviour, with unclear specific molecular mechanisms.19,20 Therefore, we detected the pathogenic mutation CFTR ΔF508 by gene sequencing in cervical cell lines and tissues and found that there was no ΔF508 mutation of CFTR in all samples, which might show that cervical cancer is not related to CFTR gene mutations but could be related to high expression of the normal CFTR gene.
Methylation is an epigenetic modification of genomic DNA and an important means of regulating genomic functions. Abnormal DNA methylation is widespread in tumours and closely related to the occurrence of tumours.21 It has been reported that CFTR gene expression may be concerned with promoter methylation.22 Therefore, to explore the relationship between the methylation status of the CFTR gene and its high expression in cervical cancer tissues, we applied methylation-specific PCR to detect the methylation status of the CpG is land of the CFTR gene. The results showed that the unmethylated CFTR gene promoter was found in both normal cervical cell lines and normal cervical tissues and in cervical cancer cell lines and cervical cancer tissues. This indicated that gene methylation was not correlated with the expression of CFTR in cervical cancer and that the function of the CFTR gene was tissue specific. Of course, due to the limited sample size, larger trials are needed to obtain more accurate results.
As an important transcription factor, NF-κB is involved in a variety of cellular functions and is abnormally activated in human tumours, but its exact mechanism is unclear.23 NF-κB downstream target genes are strongly linked to the inflammatory response and mediate a variety of physiological functions, such as cell proliferation, apoptosis, adhesion and cell microenvironment maintenance.24,25 It has been confirmed that NF-κB is interrelated with the occurrence and development of multiple tumours, such as breast cancer, pancreatic cancer, cervical cancer, and ovarian cancer.26–29 Abnormal activation of NF-κB can induce anti-apoptotic gene expression; regulate cyclin and oncogene expression; promote cell proliferation; regulate the expression of matrix metalloproteinases and cell adhesion genes; affect the metastasis of tumour cells; and indirectly inhibit p65, Rb, PTEN and other tumour suppressor genes.23
We constructed HeLa cell lines with CFTR knockdown and overexpression through plasmid transfection to investigate the effect of CFTR on the NF-κB signalling pathway and the biological behaviour of tumours. It was found that CFTR overexpression may inhibit NF-κB activity, while NF-κB transcription activity increased with CFTR knockdown. The results were further validated by CFTR activators and inhibitors. In addition, we examined the effect of CFTR on cell proliferation and invasion and found that CFTR overexpression reduced cell proliferation, migration and invasion abilities. This was consistent with existing research results, which indicate that CFTR overexpression can downregulate TWIST1, FGFR2, ITGA1, THBS1 and other genes related to tumour cell invasion and migration.30 However, the specific mechanism by which CFTR interacts with these genes remains unclear. It has been reported that defects in CFTR can lead to endogenous activation of NF-κB.31,32 Therefore, CFTR overexpression may affect the function of tumour cells by inhibiting the NF-κB signalling pathway.
Chemokines belong to a superfamily of small molecules. These small molecules were originally discovered because they interact with chemokine receptors and were found to regulate the transfer of leukocytes to sites of inflammation and the recirculation of secondary lymphatic vessels.33,34 Increasing evidence suggests that chemokines regulate many aspects of tumour cell biology, including survival, proliferation, migration, and angiogenesis.35,36 CXCL12/CXCR4, one of the most studied chemokine signalling axes, has been demonstrated to play a critical role in various solid tumours.37 In our study, the chemokine signalling pathway was the top 2 significantly enriched pathways in the KEGG analysis. CFTR was correlated with CXCL12 and related to the chemokine signalling pathway.
Conclusion
In conclusion, our study demonstrated that CFTR plays a role in the proliferation and invasion of cervical cancer cells through the NF-κB signalling pathway. NF-κB can activate transcription and increase CFTR expression, and in turn, CFTR has a feedback inhibitory effect on NF-κB. Although the role of this mutual regulatory function in tumours has been preliminarily studied, the relationship between CFTR and NF-κB in tumorigenesis, especially the tumorigenesis of cervical cancer, still needs further study. In addition, the mechanism by which CFTR mediates the cervical inflammatory response and microenvironment through the NF-κB inflammatory pathway and its effect on cervical lesions and cervical cancer are worth exploring. The NF-κB signalling pathway is also involved in the occurrence of cervical cancer. We hypothesize that CFTR may play a potentially important role in the treatment and prevention of cervical cancer.
Abbreviations
CFTR, cystic fibrosis transmembrane conductance regulator; NF-κB, nuclear factor κB; HPV, human papillomavirus; CF, cystic fibrosis; TNF-α, tumor necrosis factor α; IL-1, interleukin-1; SDS-PAGE, sodium dodecyl sulfate polyacrylamide gel electrophoresis; ECL, enhanced chemiluminescence; MSP, methylation-specific PCR; PPI, protein–protein interaction; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Acknowledgments
The plasmids were kindly provided by Pro. Wenming Xu, West China Second University Hospital, Sichuan University, Chengdu 610041, China.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
2. Bodily J, Laimins LA. Persistence of human papillomavirus infection: keys to malignant progression. Trends Microbiol. 2011;19(1):33–39. doi:10.1016/j.tim.2010.10.002
3. Castle PE, Rodriguez AC, Burk RD, et al. Short term persistence of human papillomavirus and risk of cervical precancer and cancer: population based cohort study. BMJ. 2009;339:b2569. doi:10.1136/bmj.b2569
4. Saslow D, Solomon D, Lawson HW, et al. American Cancer Society, American Society for Colposcopy and Cervical Pathology, and American Society for Clinical Pathology screening guidelines for the prevention and early detection of cervical cancer. CA Cancer J Clin. 2012;62(3):147–172. doi:10.3322/caac.21139
5. Moran O. The gating of the CFTR channel. Cell Mol Life Sci. 2017;74(1):85–92. doi:10.1007/s00018-016-2390-z
6. Girardet A, Viart V, Plaza S, et al. The improvement of the best practice guidelines for preimplantation genetic diagnosis of cystic fibrosis: toward an international consensus. Eur J Med Genet. 2016;24(4):469–478.
7. Pankow S, Bamberger C, Calzolari D, et al. F508 CFTR interactome remodelling promotes rescue of cystic fibrosis. Nature. 2015;528(7583):510–516. doi:10.1038/nature15729
8. He Q, Tsang LL, Ajonuma LC, Chan HC. Abnormally up-regulated cystic fibrosis transmembrane conductance regulator expression and uterine fluid accumulation contribute to Chlamydia trachomatis-induced female infertility. Fertil Steril. 2010;93(8):2608–2614. doi:10.1016/j.fertnstert.2010.01.040
9. Colotta F, Allavena P, Sica A, Garlanda C, Mantovani A. Cancer-related inflammation, the seventh hallmark of cancer: links to genetic instability. Carcinogenesis. 2009;30(7):1073–1081. doi:10.1093/carcin/bgp127
10. Ren Z, Wang L, Cui J, et al. Resveratrol inhibits NF-kB signaling through suppression of p65 and IkappaB kinase activities. Die Pharmazie. 2013;68(8):689–694.
11. Peng X, Wu Z, Yu L, et al. Overexpression of cystic fibrosis transmembrane conductance regulator (CFTR) is associated with human cervical cancer malignancy, progression and prognosis. Gynecol Oncol. 2012;125(2):470–476. doi:10.1016/j.ygyno.2012.02.015
12. Zhao W, Xue P, Jinke L, Yi Z, Lina H. Constitutive activation of nuclear factor κB contributes to cystic fibrosis transmembrane conductance regulator expression and promotes human cervical cancer progression and poor prognosis. Int J Gynecol Cancer. 2013;23(5):906–915. doi:10.1097/IGC.0b013e318292da82
13. Bergougnoux A, Rivals I, Liquori A, et al. A balance between activating and repressive histone modifications regulates cystic fibrosis transmembrane conductance regulator (CFTR) expression in vivo. Epigenetics. 2014;9(7):1007–1017. doi:10.4161/epi.28967
14. McWilliams RR, Petersen GM, Rabe KG, et al. Cystic fibrosis transmembrane conductance regulator (CFTR) gene mutations and risk for pancreatic adenocarcinoma. Cancer. 2010;116(1):203–209. doi:10.1002/cncr.24697
15. Li Y, Sun Z, Wu Y, et al. Cystic fibrosis transmembrane conductance regulator gene mutation and lung cancer risk. Lung Cancer. 2010;70(1):14–21. doi:10.1016/j.lungcan.2010.01.005
16. Qiao D, Yi L, Hua L, et al. Cystic fibrosis transmembrane conductance regulator (CFTR) gene 5T allele may protect against prostate cancer: a case-control study in Chinese Han population. J Cyst Fibros. 2008;7(3):210–214. doi:10.1016/j.jcf.2007.07.011
17. Wine JJ, Dean M, Glavac D. Natural animal models of human genetic diseases. Methods Mol Med. 2002;70:31–46. doi:10.1385/1-59259-187-6:31
18. Callebaut I, Chong PA, Forman-Kay JD. CFTR structure. J Cyst Fibros. 2018;17(2s):S5–s8. doi:10.1016/j.jcf.2017.08.008
19. Truninger K, Malik N, Ammann RW, et al. Mutations of the cystic fibrosis gene in patients with chronic pancreatitis. Am J Gastroenterol. 2001;96(9):2657–2661. doi:10.1111/j.1572-0241.2001.04047.x
20. Ciofu O, Riis B, Pressler T, Poulsen HE, Hoiby N. Occurrence of hypermutable Pseudomonas aeruginosa in cystic fibrosis patients is associated with the oxidative stress caused by chronic lung inflammation. Antimicrob Agents Chemother. 2005;49(6):2276–2282. doi:10.1128/AAC.49.6.2276-2282.2005
21. Vidal E, Sayols S. A DNA methylation map of human cancer at single base-pair resolution. Oncogene. 2017;36(40):5648–5657.
22. Yang A, Sun Y, Mao C, et al. Folate protects hepatocytes of hyperhomocysteinemia mice from apoptosis via cystic fibrosis transmembrane conductance regulator (CFTR)-activated endoplasmic reticulum stress. J Cell Biochem. 2017;118(9):2921–2932.
23. Perkins ND. The diverse and complex roles of NF-kappaB subunits in cancer. Nat Rev Cancer. 2012;12(2):121–132. doi:10.1038/nrc3204
24. Iliopoulos D, Hirsch HA, Struhl K. An epigenetic switch involving NF-kappaB, Lin28, Let-7 MicroRNA, and IL6 links inflammation to cell transformation. Cell. 2009;139(4):693–706. doi:10.1016/j.cell.2009.10.014
25. Zhang YC, Huo FC, Wei LL, et al. PAK5-mediated phosphorylation and nuclear translocation of NF-kappaB-p65 promotes breast cancer cell proliferation in vitro and in vivo. J Exp Clin Cancer Res. 2017;36(1):146. doi:10.1186/s13046-017-0610-5
26. Cho U, Kim B, Kim S, Han Y, Song YS. Pro-inflammatory M1 macrophage enhances metastatic potential of ovarian cancer cells through NF-kappaB activation. Mol Carcinog. 2018;57(2):235–242.
27. Fusella F, Secli L, Busso E, Krepelova A, Moiso E. The IKK/NF-kappaB signaling pathway requires Morgana to drive breast cancer metastasis. Nat Commun. 2017;8(1):1636.
28. Yuan P, He XH, Rong YF, et al. KRAS/NF-kappaB/YY1/miR-489 Signaling Axis Controls Pancreatic Cancer Metastasis. Cancer Res. 2017;77(1):100–111. doi:10.1158/0008-5472.CAN-16-1898
29. Ali A, Kim SH, Kim MJ, et al. O-GlcNAcylation of NF-kappaB Promotes Lung Metastasis of Cervical Cancer Cells via Upregulation of CXCR4 Expression. Mol Cells. 2017;40(7):476–484. doi:10.14348/molcells.2017.2309
30. Xie C, Jiang XH, Zhang JT, et al. CFTR suppresses tumor progression through miR-193b targeting urokinase plasminogen activator (uPA) in prostate cancer. Oncogene. 2013;32(18):
31. Vij N, Mazur S, Zeitlin PL, Hartl D. CFTR is a negative regulator of NFκB mediated innate immune response. PLoS One. 2009;4(2):e4664. doi:10.1371/journal.pone.0004664
32. Boncoeur E, Roque T, Bonvin E, et al. Cystic fibrosis transmembrane conductance regulator controls lung proteasomal degradation and nuclear factor-kappaB activity in conditions of oxidative stress. Am J Pathol. 2008;172(5):1184–1194. doi:10.2353/ajpath.2008.070310
33. Murdoch C, Finn A. Chemokine receptors and their role in inflammation and infectious diseases. Blood. 2000;95(10):3032–3043. doi:10.1182/blood.V95.10.3032
34. Mantovani A, Savino B, Locati M, Zammataro L, Allavena P, Bonecchi R. The chemokine system in cancer biology and therapy. Cytokine Growth Factor Rev. 2010;21(1):27–39. doi:10.1016/j.cytogfr.2009.11.007
35. Vicari AP, Caux C. Chemokines in cancer. Cytokine Growth Factor Rev. 2002;13(2):143–154. doi:10.1016/S1359-6101(01)00033-8
36. Nickel R, Beck LA, Stellato C, Schleimer RP. Chemokines and allergic disease. J Allergy Clin Immunol. 1999;104(4 Pt 1):723–742. doi:10.1016/S0091-6749(99)70281-2
37. Domanska UM, Kruizinga RC, Nagengast WB, et al. A review on CXCR4/CXCL12 axis in oncology: no place to hide. Eur J Cancer. 2013;49(1):219–230. doi:10.1016/j.ejca.2012.05.005
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.