Back to Journals » Cancer Management and Research » Volume 11
Cervical Cancer Cells-Secreted Exosomal microRNA-221-3p Promotes Invasion, Migration and Angiogenesis of Microvascular Endothelial Cells in Cervical Cancer by Down-Regulating MAPK10 Expression
Authors Zhang L, Li H, Yuan M, Li M, Zhang S
Received 1 July 2019
Accepted for publication 4 November 2019
Published 9 December 2019 Volume 2019:11 Pages 10307—10319
DOI https://doi.org/10.2147/CMAR.S221527
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Antonella D'Anneo
Lu Zhang,* Huihui Li,* Ming Yuan, Mingbao Li, Shuquan Zhang
Department of Obstetrics and Gynecology, Qilu Hospital of Shandong University, Jinan 250012, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Shuquan Zhang
Department of Obstetrics and Gynecology, Qilu Hospital of Shandong University, No.107 Wenhua West Road, Jinan, Shandong 250012, People’s Republic of China
Tel +86 185 6008 1932
Email [email protected]
Purpose: Cervical cancer (CC) is recognized as a common cancer with a high risk worldwide. Exosomal microRNAs (miRNAs) have received attention for their increasing potentials in CC therapy. In this study, we identify the involvement of miR-221-3p in CC progression by affecting angiogenesis of microvascular endothelial cells (MVECs).
Methods: Microarray-based gene expression profiling was conducted to retrieve the differentially expressed genes in CC. The expression patterns of miR-221-3p were measured by RT-qPCR, while Western blot analysis and RT-qPCR were performed to determine the expression of MAPK10 in the CC tissues and cells, followed by verification of the interaction between miR-221-3p and MAPK10 using dual luciferase reporter gene assay. Then the effects of miR-221-3p and MAPK10 on cell activities were assessed through gain- and loss-of-function experiments in CC. Subsequently, the impact of exosomal miR-221-3p on MVEC proliferation, migration, invasion and angiogenesis was examined after exosomal isolation from CC cells and co-cultured with MVECs.
Results: Gene expression profile showed that MAPK10 might participate in CC with a low expression. Moreover, miR-221-3p was highly expressed and MAPK10 was poorly expressed in CC tissues and cells. It was observed that miR-221-3p targeted MAPK10. Depletion of miR-221-3p blocked the cell proliferation, invasion and migration in CC by up-regulating MAPK10. Moreover, CC cells-derived exosomes carrying miR-221-3p accelerated MVEC proliferation, invasion, migration and angiogenesis in CC by regulating MAPK10.
Conclusion: CC cells-derived exosomes harboring miR-221-3p enhanced MVEC angiogenesis in CC by decreasing MAPK10.
Keywords: cervical cancer, microvascular endothelial cells, exosomes, microRNA-221-3p, mitogen-activated protein kinase 10, angiogenesis
Introduction
Cervical cancer (CC) is among the most common female malignancies worldwide, and its incidence can be hindered by undergoing a vaccination as well as human papillomavirus (HPV) screening.1 HPV is a major causative factor of CC, and continuous exposure to high-risk HPV and sexual behavior alteration account for increased incidence of CC.2 CC predominantly presents in two histotypes as squamous cell carcinomas and adenocarcinoma.3 Besides, the reversion of refractory metastatic CC after chemotherapy can be facilitated by immunotherapy.4 Microvascular endothelial cells (MVEC) arise from a lineage of endothelial cells of the large vessels.5 MVECs have gained focus due to its function in the anti-angiogenic therapy of cancer.6 Exosomal microRNAs (miRNAs) have been recognized as therapeutic targets and markers for the development of novel therapeutic interventions for cancer.7
Exosomes originate from endosomes, short lipid bilayer-enclosed structure particles varying in length from 30–140 nm, which are a subset of extracellular vesicles derived from most cells.8 Besides, exosomes function as mediators of tumor cell interaction and long-range signaling effectors responsible for coordinating a variety of hallmarks implications in cancer, including tumor formation, development, metastasis, angiogenesis, tumor immunity and treatment resistance.9 Moreover, exosomes are secretory products from various types of cells under specific physiological or pathological conditions, which possess multiple functional proteins including miRNAs.10 Additionally, evidence has acknowledged miRNAs as diagnostic biomarkers for CC and precancerous lesions.11 For instance, the intimate relations between miR-221-3p and lymph node metastasis and peritumoral lymphangiogenesis have been detected in cervical squamous cell carcinoma.12 The biological prediction website predicted the presence of binding site between miR-221-3p and mitogen-activated protein kinase (MAPK10). MAPK10 is a highly conserved C-Jun N-terminal kinase gene in mammals.13 A prior study has reported the proapoptotic functionality of MAPK10 in addition to its role as an anti-oncogene in chromophobe renal cell carcinoma.14 Therefore, we speculated that miR-221-3p may affect CC progression by regulating MAPK10. Hence, this study was designed with an aim of verifying whether CC cells-secreted exosomal miR-221-3p could influence CC progression by targeting MAPK10.
Materials and Methods
Ethics Statement
The experiment procedures were performed in line with the Helsinki Declaration, with approval of the Ethics Committee of the Medical Ethics Committee of Qilu Hospital of Shandong University. All participants were informed of the specific experiment content, and signed informed written consent documents prior to participation. All cell lines were purchased commercially.
Microarray-Based Gene Expression Profiling
The microarray data relevant to CC were downloaded from the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo). With |logFC| > 1 and p value < 0.05 as thresholds, the R language limma package was employed for screening the differentially expressed genes (DEGs) of CC, from which the heat map of genes was plotted. “Clusterprofiler” package was adopted for Kyoto Encyclopedia of Genes and Genomes (KEGG) functional enrichment analysis of DEGs, and the “pathview” package was employed to mark the position and expression change of DEGs in the metabolic pathway. The potential upstream miRNAs capable of regulating MAPK10 were predicted using the miRDB database (http://mirdb.org/miRDB/index.html), mirDIP database (http://ophid.utoronto.ca/mirDIP/index.jsp#r), TargetScan database (http://www.targetscan.org/vert_71/) and microRNA database (http://www.microrna.org/microrna/home.do?tdsourcetag=s_pcqq_aiomsg).
Patient Enrollment
CC tissues were amassed from 52 patients (43.15 ± 3.82 years) with clinically diagnosed CC in the Qilu Hospital of Shandong University from May 2016 to December 2017. All patient diagnoses were confirmed by cervical biopsy, and none of them was administered chemoradiotherapy before the operation. Meanwhile, 28 normal cervical tissues were collected from matched patients who were diagnosed with myoma of the uterus in the Qilu Hospital of Shandong University as controls.15
Immunohistochemistry (IHC)
The paraffin sections of the tissues were deparaffinized, dehydrated using gradient ethanol, and washed under running water for 2 min. Next, the sections were immersed in 3% H2O2 for 20 min, and rinsed for 3 min using 0.1 M phosphate buffer saline (PBS). Then, the sections were subjected to antigen retrieval in a water bath and cooled down under running water. Afterwards, the section blockade was performed using normal goat serum blocking solution (C-0005, Shanghai Haoran Bio Technologies Co., Ltd., Shanghai, China) at room temperature for 20 min. The sections were subsequently incubated with the primary rabbit anti-human antibody against MAPK10 (1: 100, ab51248, Abcam Inc., Cambridge, MA, UK) at 4ºC overnight. The sections were further incubated with the secondary goat anti-rabbit antibody against immunoglobulin G (IgG) (1: 100, ab6758, Abcam Inc., Cambridge, MA, UK) at 37ºC for 20 min. Next, the sections were subjected to incubation with horse radish peroxidase (HRP)-labeled streptavidin ovalbumin solution, visualized using diaminobenzidine (DAB), and then counterstained by hematoxylin (PT001, Shanghai Bogoo Biotechnology Co., Ltd., Shanghai, China) for 1 min. Further, ammonia water was added to the sections for attaining a corresponding blue color, dehydrated using gradient ethanol, cleared by xylene, and finally mounted using a neutral gum and observed under a microscope.
Cell Culture and Transfection
CC cell lines [Caski ((ATCC® CRM-CRL-1550), Hela (ATCC® CCL-2), SiHa (ATCC® HTB-35) and SW756 (ATCC® CRL-10302)] and the normal cervical epithelial cell line End1/E6E7 (ATCC® CRL-2615) [all from American Type Culture Collection (ATCC)] were cultured together in a 37ºC incubator supplemented with 5% CO2. Upon attaining 80% cell confluence, the cells were treated with 0.25% trypsin to adjust the concentration to 1 × 106 cells/mL by the addition of dulbecco’s modified eagles medium (DMEM) containing 10% fetal bovine serum (FBS). Afterwards, the cells were quantitatively inoculated in culture plates and dishes for further experimentation. Then, following the provided instructions of the Lipofectamine® 2000 reagent (Invitrogen, Carlsbad, CA, USA), cell transfection was performed with the plasmids of miR-221-3p mimic, miR-221-3p inhibitor and overexpressed (oe)-MAPK10 alone or in combination.
Isolation and Identification of CC-Derived Exosomes
The supernatant of CC cells was collected to remove any dead cells and cell debris by differential centrifugation. The cells were successively centrifuged and the supernatant was collected. Subsequently, the supernatant was centrifuged at 200,000 × g at 4ºC for 2 h in an ultra-speed centrifuge tube in order to collect the exosome sediment. The exosome sediment was rinsed and resuspended using sterile PBS buffer after removal of the supernatant, which was then centrifuged at 100,000 × g at 4ºC for 2 h. After removal of the supernatant, the pure exosome sediment was collected and resuspended using 100 µL PBS and stored at −80ºC for further practice. The expression of the specific surface biomarkers (HSP70, CD63 and CD9) was detected by means of Western blot analysis for identification of the characteristics of exosomes. Under the transmission electron microscope (TEM), the exosome was placed on the Forvar and carbon-coated copper grids, and then the grids were placed on 2% phosphotungstic acid for 1 min and rinsed using PBS. The morphology of exosomes was observed under the TEM (Tecnai Spirit, Thermo Fisher Scientific, Hillsboro, Oregon, USA).16
Co-Culture of Exosomes and MVECs
Exosomes dissolved in PBS were combined with Exo-Red (EXOR100A-1, Byxbio, Changzhou, Jiangsu, China) at the ratio of 10: 1 and then cultured in a 37ºC incubator for 10 min. After termination of the reaction using 100 μL stop buffer, the exosomes were further incubated at 4ºC for 30 min, and centrifuged at 35,068 × g for 3 min. The fluorescence-labeled exosomes were resuspended using 200 μL PBS. Afterwards, the fluorescence-labeled exosomes were co-cultured with inoculation of the supernatant of MVEC culture medium in a 24-well plate for 48 h after confluence reached 50–60%. Briefly, MVEC was incubated with PBS (as control), exosomes from SiHa transfected with miR-221-3p mimic (Exo-miR-221-3p mimic), and the Exo-miR-221-3p inhibitor. MVEC was rinsed 3 times under PBS after incubation and then observed under an inverted fluorescence microscope.
Angiogenesis Assay
MVECs (5 × 103 cell/mL) were cultured in 96-well plates at 37ºC for 18 h in accordance with the provided instructions of the In Vitro Angiogenesis Assay Kit (K905-50, Chemicon International Inc, BioVision, Milpitas, CA, USA). Angiogenesis was observed under an inverted microscope (IX73, OLYMPUS, Olympus Corporation, Tokyo, Japan). Moreover, 10 visual fields (40 ×) were randomly selected to count the number of angiogenesis nodes, followed by calculation of the average value. Each group was set with three parallel controls.17
5-Ethynyl-2ʹ-Deoxyuridine (EdU) Assay
The logarithmical cells were seeded into the 24-well plates. Each group comprised of three duplicate wells. Subsequently, the culture medium was supplemented with EdU (C10341-1, Guangzhou RiboBio Co., Ltd., Guangzhou, China) to attain a density of 10 μmol/L, and the cells were cultured for 2 h. The cells were fixed using PBS containing 4% polyformaldehyde for 15 min after the culture medium was removed, followed by a rinse with PBS-3% bovine serum albumin (BSA) and subsequent incubation with PBS containing 0.5% Triton-100 for 20 min at room temperature. Next, the cells were cultured in Apollo® 567 (Guangzhou RiboBio Co., Ltd., Guangzhou, China) at a density of 100 µL/well at room temperature for 30 min after two rinses with PBS containing 3% BSA. After the rinses under PBS containing 3% BSA, the cells were stained with 1 × Hoechst 33342 for 30 min. Finally, a fluorescence microscope (FM-600, Shanghai Pudan Optical Instrument Co., Ltd., Shanghai, China) was used to observe and document the number of positive cells in each field.
Transwell Assay
The Transwell chamber with the 24-well plate (Corning Incorporated, Corning, NY, USA; an aperture of 8 μm) was used in the migration test. A total of 200 μL cells were inoculated to the apical Transwell chamber, while 600 μL of complete medium (Invitrogen, Carlsbad, CA, USA) containing 10% FBS was added to the basolateral chamber. After incubation at 37ºC with 5% CO2 for 48 h, the cells were fixated using 4% paraformaldehyde for 30 min. Subsequently, after treatment with 0.2% Triton X-100 (Sigma-Aldrich Chemical Company, St Louis, MO, USA) for 15 min, the Transwell chamber was stained using 0.05% crystal violet for 5 min. Cell migration was also detected.
Prior to the invasion test, the filter membrane was covered with 50 μL Matrigel (Sigma-Aldrich Chemical Company, St Louis, MO, USA). Next, 200 μL cells were inoculated to the apical Transwell chamber, while 600 μL complete medium (Invitrogen, Carlsbad, CA, USA) containing 10% FBS was added to the basolateral chamber. After incubation at 37ºC with 5% CO2 for 48 h, the aforementioned procedures were repeated for cell fixation and staining. Under an inverted microscope, five visual fields were randomly selected to count the number of stained cells (XDS-800D, Shanghai Caikon Optical Instrument Co., Ltd., Shanghai, China). The migration and invasion ability of tumor cells was assessed after calculating the average number of cells.
Dual Luciferase Reporter Gene Assay
The biological prediction website (http://www.microrna.org/microrna/home.do) was used to validate whether MAPK10 was the target gene of miR-221-3p, which was further verified by the dual luciferase reporter gene essay. The dual luciferase reporter gene vector of MAPK10 and the mutant type (MUT) on the binding site between miR-221-3p and MAPK10, namely PGLO-MAPK10 wild type (WT) and PGLO-MAPK10 MUT, were constructed respectively. Subsequently, the aforementioned plasmids were co-transfected with miR-221-3p mimic and negative control (NC) plasmids into HEK-293T cells. After 24 h, the cells were lysed and centrifuged at 25,764 × g for 1 min to collect the supernatant. The luciferase activity was detected using the dual luciferase® Reporter Assay System (E1910, Promega Corp., Madison, Wisconsin, USA). Then 100 μL of Renilla luciferase working solution and Firefly luciferase working solution were added to detect the activity of Renilla luciferase and Firefly luciferase, which served as a marker for relative luciferase activities.
Western Blot Analysis
The cells were trypsinized and lysed using an enhanced Radio Immunoprecipitation Assay (RIPA) lysis buffer (Wuhan Boster Biological Technology Co., Ltd., Wuhan, China) supplemented with the protease inhibitor. Next, the protein concentration was determined using the BCA Protein Quantitative Kit (Wuhan Boster Biological Technology Co., Ltd., Wuhan, China). The proteins were transferred onto a polyvinylidene fluoride (PVDF) membrane after separation by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). The membrane was immersed in the sealing solution for 1 h and incubated with the primary rabbit anti-human antibodies against MAPK10 (1: 2000, ab208035), c-Fos proto-oncogene (FOS) (1: 100, ab209794), c-Jun proto-oncogene (JUN) (1: 5000, ab40766), JunB proto-oncogene (JUNB) (1: 1000, ab128878), vascular endothelial growth factor (VEGF) (1: 10,000, ab32152) and glyceraldehyde-3-phosphate dehydrogenase (GAPDH; internal reference) (1: 5000, ab181602) at 4°C overnight. Following that, the membrane was incubated with the secondary goat anti-rabbit antibody against IgG (1: 10,000, ab205718) at 37°C for 1 h. The aforementioned antibodies were provided by Abcam Inc (Cambridge, MA, USA). After 3 rinses with PBS, the membrane was visualized using the enhanced chemiluminescence solution, and photographed using SmartView Pro 2000 (UVCI-2100, Major Science, Saratoga, CA, USA). Eventually, the gray value of the protein bands was analyzed using the Quanity One software.
RNA Isolation and Quantitation
The total RNA was extracted using a RNeasy Mini Kit (Qiagen, Valencia, CA, USA). Next, the total RNA extracted from the mRNAs was reversely transcribed into complementary DNA (cDNA) following the provided instructions of the First Strand cDNA Synthesis Kit (RR047A, Takara Bio Inc., Tokyo, Japan). The total RNA of miRNA was reversely transcribed into cDNA in strict accordance with the provided instructions of the miRNA First Strand cDNA Synthesis (Tailing Reaction) Kit (B532451-0020, Shanghai Sangon Biotechnology Co. Ltd., Shanghai, China). SYBR® Premix Ex TaqTM II (Perfect Real Time) Kit (DRR081, Takara Bio Inc., Tokyo, Japan) was used to load the samples. Reverse transcription quantitative polymerase chain reaction (RT-qPCR) was performed on the ABI 7500 RT-qPCR system (ABI, Foster City, CA, USA). Three duplicate wells were set for each sample. The general negative primers of miRNA and the upstream primers of U6 were provided by the miRNA First Strand cDNA Synthesis Kit. Other primers were synthesized by Shanghai Sangon Biotechnology Co. Ltd. (Shanghai, China) (Table 1). With GAPDH and U6 as internal control, the relative expression of genes was calculated based on the 2−△△Ct method.
 |
Table 1 Primer Sequences for RT-qPCR |
Statistical Analysis
Statistical analyses were conducted using the SPSS 21.0 statistical software (IBM Corp., Armonk, New York, USA). Measurement data were presented as mean ± standard deviation. Comparisons between two groups were analyzed by non-paired t-test if the data conformed to the homogeneity of variance and normal distribution. Comparisons among multiple groups were analyzed by one-way analysis of variance (ANOVA) with Tukey’s post hoc test. The comparison was significant when p < 0.05.
Results
MAPK10 Is Down-Regulated in CC Tissues and Cells
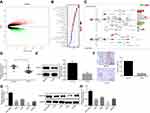
The CC GSE9750 microarray data concerning CC was retrieved from the GEO database, which comprises of 24 normal samples and 33 tumor samples. Next, the differential analysis on these samples identified 1327 potential DEGs (Figure 1A). In order to further examine the functions of these DEGs, the KEGG pathway enrichment analysis was performed, which revealed that these genes were enriched in signaling pathways such as the “cell cycle” signaling pathway (Figure 1B). Among these signaling pathways, some genes appeared to be evidently enriched in the tumor necrosis factor (TNF) signaling pathway that was indicated in various tumor regulatory mechanisms, including angiogenesis.18,19 Further analysis of the DEGs in TNF signaling pathway revealed the existence of both up-regulated genes and down-regulated genes in CC (Figure 1C). Besides, results demonstrated the predominant localization of MAPK10 gene at the core of TNF signaling pathway with a significantly low expression in CC. Thus, the expression of MAPK10 in normal cervical tissues and CC tissues was detected by regimens of RT-qPCR and Western blot analysis. The results illustrated lower mRNA and protein expression of MAPK10 in the CC tissues compared to the normal cervical tissues (p < 0.05; Figure 1D and E).
Next, IHC revealed that the CC tissues had exhibited a lower MAPK10 positive rate relative to the normal cervical tissues (p < 0.05; Figure 1F). To further investigate the correlation between MAPK10 and clinicopathological characteristics, we divided the cervical tumor samples into low and high MAPK10 expression parts. As shown in Table 2, in contrast to the high expression of MAPK10, low expression of MAPK10 was indicative of increased incidence of large tumor size, late FIGO stages and preoperative metastasis (p < 0.05). However, the expression of MAPK10 shared little correlation with parameters such as age, squamous cell associated antigen (SCC-Ag) and tumor histology (p > 0.05). Meanwhile, the expression of MAPK10 was lower in CC cell lines in respect to the normal cervical epithelial cell line with the lowest MAPK10 expression in SiHa (p < 0.05; Figure 1G and H). Therefore, the SiHa cells were selected for subsequent experimentation.
 |
Table 2 Clinical Correlations Between MAPK10 Expression and Cervical Carcinoma |
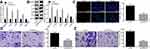
Up-Regulated MAPK10 Represses CC Cell Activities
To further investigate the downstream regulatory mechanism of MAPK10 and its effects on CC cells, the SiHa cells were transfected with oe-MAPK10. The AP-1 family was the downstream target of MAPK10 and was mediated by its transcription. The expression of the AP-1 family members (c-FOS, c-JUN and JUNB) and angiogenesis-related factor VEGF was detected. The upregulated c-FOS, MAPK10, c-JUN, and JUNB and downregulated VEGF were detected upon SiHa cell transfection with oe-MAPK10 (Figure 2A and B). Cell activities were restrained after transfection with oe-MAPK10 (Figure 2C–E). Conjointly, MAPK10 elevation repressed CC cell proliferation, migration and invasion.
miR-221-3p Silencing Induces Inhibition of CC Cell Proliferation, Migration and Invasion by Upregulating MAPK10
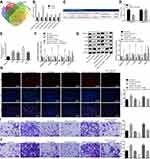
To further understand the down-regulatory mechanism of MAPK10 in CC, several databases were included to predict the upstream regulatory miRNAs of MAPK10, which detected 6 miRNAs in the intersection of predicted results (Figure 3A). Furthermore, a quantitative analysis on the expression of these 6 miRNAs in CC tissue samples was conducted, which recognized the most significant change of the expression of miR-221-3p with a higher expression of miR-221-3p in the CC tissue samples compared to the adjacent normal tissues (Figure 3B). Bioinformatics database (http://www.microrna.org/microrna/home.do) predicted the existence of a specific binding site between MAPK10 and miR-221-3p (Figure 3C). The luciferase activity of PGLO-MAPK10 WT was reduced (p < 0.05), while that of PGLO-MAPK10 MUT did not differ significantly following the transfection of miR-221-3p mimic (p > 0.05) (Figure 3D). The miR-221-3p expression was higher in the Hela, Caski, SiHa and SW756 cell lines than the normal cervical epithelial cell line End1/E6E7, with the highest expression evident in the SiHa cell line (p > 0.05) (Figure 3E). The expression of c-FOS, MAPK10, c-JUN and JUNB was reduced and that of VEGF was elevated after treatment with miR-221-3p mimic, which was conflicting to the results after transfection with the miR-221-3p inhibitor or the combination of miR-221-3p mimic with oe-MAPK10 in comparison with their matched controls (p < 0.05; Figure 3F and G). Subsequently, the cell proliferation, invasion and migration abilities were increased upon transfection of miR-221-3p mimic and lowered upon transfection of miR-221-3p inhibitor or the combination of miR-221-3p mimic with oe-MAPK10 compared to the relevant controls (p < 0.05) (Figure 3H–J). Altogether, inhibition of miR-221-3p could extensively suppress cell proliferation, invasion and migration by targeting MAPK10 in CC.
CC Cell-Derived Exosomes Carry miR-221-3p
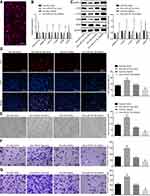
Then, we isolated exosomes from the CC cells for further identification. TEM results evidently revealed a group of circular or elliptical membranous vesicles with similar morphology, obvious heterogeneity in size and a diameter of 50–180 nm. The membranous structures were observed in the peripheral regions of vesicles, and low electron density was critical in the central region (Figure 4A). The increased expression of HSP70, CD9 and CD63 detected in exosomes isolated from CC cells was indicative of successful exosome isolation (Figure 4B). Further, the expression of miR-221-3p was detected in exosomes derived from CC, and the results revealed that exosomes derived from CC could carry miR-221-3p (Figure 4C). These findings suggested high probability of exosomes derived from CC cells to carry miR-221-3p.
Exosomes Secreted by CC Cells Carrying miR-221-3p Enters MVECs to Promote Their Proliferation, Migration and Angiogenesis by Down-Regulating MAPK10 in CC
Subsequently, a fluorescence microscope was used to observe whether the MVECs could uptake exosomes secreted by CC cells. Next, 48 h after the co-transfection, the uptake of exosomes by MVECs was evidently increased (Figure 5A). Thereafter, the expression of miR-221-3p was determined using RT-qPCR, while the mRNA expression and protein levels of MAPK10, c-FOS, c-JUN, JUNB and VEGF in MVECs after co-culture with exosomes were detected by means of RT-qPCR and Western blot analysis. Subsequently, it was unraveled that the expression of miR-221-3p and VEGF was up-regulated and the expression of MAPK10, c-FOS, c-JUN, and JUNB was lowered upon co-culture with Exo-miR-221-3p mimic, while contradictory results were evident after co-culture with Exo-miR-221-3p inhibitor (Figure 5B and C). Next, the MVEC proliferation, migration, invasion and angiogenesis abilities were evaluated. Results demonstrated enhanced MVEC proliferation, migration, invasion and angiogenesis abilities after co-culture with Exo-221-3p mimic, and weakened after co-culture with the Exo-miR-221-3p inhibitor (Figure 5D–G). In all, miR-221-3p carried by CC cell-secreted exosomes promoted MVEC proliferation, invasion migration and angiogenesis by lowering the expression of MAPK10 in CC.
Discussion
CC remains to be a global public health problem.20 Exosomes have exhibited vital importance in regulating intercellular communication and function as a potential carrier for gene therapy and drug delivery.21 Exosomal miRNAs have established the functionality as therapeutic markers and targets for their significant roles in cancer therapy.7 Thus we delineated the role of exosomal miR-221-3p derived from CC cells in CC. Conjointly, our study found that CC cell-derived exosomal miR-221-3p stimulated MVEC proliferation, migration, invasion and angiogenesis in CC by down-regulating MAPK10.
Our findings illustrated that miR-221-3p was highly expressed while MAPK10 was poorly expressed in CC. Moreover, miR-221-3p has been revealed to be expressed at a high level in pancreatic cancer.22 Similarly, notable increases in miR-221-3p have been observed in lung adenocarcinoma patients’ plasma.23 An elevated expression of miR-221-3p has been evident in clear cell renal cell carcinoma tumor tissues.24 Meanwhile, miR-221-3p is discernibly upregulated in metastatic CC tissues compared to the non-metastatic CC tissues.25 MAPK10 has been revealed to be down-regulated or depleted in Hodgkin’s lymphoma, non-Hodgkin’s lymphoma, gastric, breast, and hepatocellular cancer cell lines.26 Low expression of MAPK10 has been previously documented in nasopharyngeal carcinoma.27 Besides, dual luciferase reporter gene assay validated MAPK10 as the target gene of miR-221-3p. In consistency with our results, Lihua Li et al have elucidated the functionality of MAPK10 as a promising target of miR-27a-3p.27 Additionally, miR-221-3p could enhance the desensitization of pancreatic cancer cells to 5-fluorouracil through targeting RB1.28
Our study also speculated silencing of miR-221-3p to inhibit cell proliferation, migration, invasion and angiogenesis in CC by increasing the expression of MAPK10, in response to elevated expression of AP-1 family members and downregulated VEGF. Functioning as a heterogeneous dimer of a cluster, AP-1 is an association complex formed by FOS protein and JUN proteins that are functionally and structurally fundamental to CC progression.29 VEGF is a vital mediator of abnormal and normal growth of blood vessels.30 Links have been established between VEGF elevation and severe CC precursor lesion and invasive disease.31 In addition, miR-221-3p down-regulation functions as a blockade for the initiation and development of hepatocellular carcinoma.32 Meanwhile, lower expression of miR-221-3p could potentially inhibit cell invasion, migration and lymphatic metastasis in CC.25 MAPK signaling is capable of affecting various biological activities including cell invasion, migration, proliferation, and angiogenesis.33 As a tumor inhibitor, MAPK kinase 4 could extensively suppress the metastasis of ovarian and prostate cancers.34 Furthermore, activated p38 MAPK signaling pathway could accelerate apoptosis and impede proliferation in CC in conformity with TMPyP4.35
In addition, our findings demonstrated the ability of CC cells-secreted exosomes to carry miR-221-3p in order to enter MVEC. Exosomes, nanovesicles derivatives of cells, carry an extensive variety of molecules including DNA/RNA, lipid and protein.7 Exosomes derived from cancer cells are associated with miRNA biogenesis independent of cells and tumorigenesis.36 Also, the possibility of exosomal miRNA has been suggested to serve as therapeutic targets and biological markers for cancer therapy.7 For instance, exosomal miR-105 secreted from active cancer cells accelerates tumor growth.37 Besides, exosomes derived from glioma stem cells could enhance the angiogenic ability of endothelial cells through the miR-21/VEGF signal.38 Moreover, our results provide evidence elucidating that CC cells-secreted exosomes containing miR-221-3p enhanced MVEC proliferation, migration, invasion and angiogenesis. An existing study revealed that exosomes secreted by cervical squamous cell carcinoma carrying miR-221-3p facilitate lymphatic metastasis and lymphangiogenesis by regulating vasohibin-1.12 Furthermore, a previous report on cervical squamous cell carcinoma demonstrated the ability of exosomal miR-221-3p derived from cancer cells to facilitate angiogenesis of human umbilical vein endothelial cells.39
Conclusion
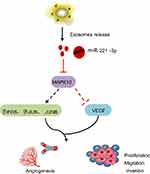
Taken together, our study suggested that miR-221-3p carried by CC cell-secreted exosomes could potentially down-regulate the expression of MAPK10, thereby promoting CC cell invasion, migration and angiogenesis (Figure 6). The present study unravels the effect of exosomal miR-221-3p in CC, thereby providing an insight on the approach of miRNA delivery in CC treatment. However, further studies are still required to illustrate its internal mechanism for this study.
Acknowledgment
This work was supported by the Natural Science Foundation of Shandong Province, China (Grant No. ZR2014HM039) and the Key Research and Development Plan of Shandong Province, China (Grant Nos. 2018GSF118025 and 2019GSF108181).
Author Contributions
Conceptualization: Lu Zhang, Huihui Li; Data curation: Huihui Li, Mingbao Li; Formal analysis: Huihui Li; Investigation: Lu Zhang, Mingbao Li, Shuquan Zhang; Methodology: Lu Zhang, Ming Yuan; Project administration: Shuquan Zhang; Resources: Lu Zhang, Ming Yuan; Software: Huihui Li, Mingbao Li; Validation: Shuquan Zhang; Visualization: Lu Zhang, Ming Yuan; Writing - original draft: Ming Yuan, Mingbao Li, Shuquan Zhang; Writing - review & editing: Lu Zhang, Huihui Li. All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Cohen PA, Jhingran A, Oaknin A, Denny L. Cervical cancer. Lancet. 2019;393(10167):169–182. doi:10.1016/S0140-6736(18)32470-X
2. Vaccarella S, Franceschi S, Zaridze D, et al. Preventable fractions of cervical cancer via effective screening in six Baltic, central, and eastern European countries 2017-40: a population-based study. Lancet Oncol. 2016;17(10):1445–1452. doi:10.1016/S1470-2045(16)30275-3
3. Steenbergen RD, Snijders PJ, Heideman DA, Meijer CJ. Clinical implications of (epi)genetic changes in HPV-induced cervical precancerous lesions. Nat Rev Cancer. 2014;14(6):395–405. doi:10.1038/nrc3728
4. Stevanovic S, Draper LM, Langhan MM, et al. Complete regression of metastatic cervical cancer after treatment with human papillomavirus-targeted tumor-infiltrating T cells. J Clin Oncol. 2015;33(14):1543–1550. doi:10.1200/JCO.2014.58.9093
5. Castiglioni S, Caspani C, Cazzaniga A, Maier JA. Short- and long-term effects of silver nanoparticles on human microvascular endothelial cells. World J Biol Chem. 2014;5(4):457–464. doi:10.4331/wjbc.v5.i4.457
6. Xu ZH, Miao ZW, Jiang QZ, et al. Brain microvascular endothelial cell exosome-mediated S100A16 up-regulation confers small-cell lung cancer cell survival in brain. Faseb J. 2019;33(2):1742–1757. doi:10.1096/fj.201800428R
7. Thind A, Wilson C. Exosomal miRNAs as cancer biomarkers and therapeutic targets. J Extracell Vesicles. 2016;5:31292. doi:10.3402/jev.v5.31292
8. Li X, Corbett AL, Taatizadeh E, et al. Challenges and opportunities in exosome research-Perspectives from biology, engineering, and cancer therapy. APL Bioeng. 2019;3(1):011503. doi:10.1063/1.5087122
9. Bhat A, Sharma A, Bharti AC. Upstream Hedgehog signaling components are exported in exosomes of cervical cancer cell lines. Nanomedicine (Lond). 2018;13(17):2127–2138. doi:10.2217/nnm-2018-0143
10. Shi B, Wang Y, Zhao R, Long X, Deng W, Wang Z. Bone marrow mesenchymal stem cell-derived exosomal miR-21 protects C-kit+ cardiac stem cells from oxidative injury through the PTEN/PI3K/Akt axis. PLoS One. 2018;13(2):e0191616. doi:10.1371/journal.pone.0191616
11. Liu SS, Chan KKL, Chu DKH, et al. Oncogenic microRNA signature for early diagnosis of cervical intraepithelial neoplasia and cancer. Mol Oncol. 2018;12(12):2009–2022. doi:10.1002/1878-0261.12383
12. Zhou CF, Ma J, Huang L, et al. Cervical squamous cell carcinoma-secreted exosomal miR-221-3p promotes lymphangiogenesis and lymphatic metastasis by targeting VASH1. Oncogene. 2019;38(8):1256–1268. doi:10.1038/s41388-018-0511-x
13. Kunde SA, Rademacher N, Tzschach A, et al. Characterisation of de novo MAPK10/JNK3 truncation mutations associated with cognitive disorders in two unrelated patients. Hum Genet. 2013;132(4):461–471. doi:10.1007/s00439-012-1260-5
14. Yoo KH, Park YK, Kim HS, Jung WW, Chang SG. Identification of MAPK10 as a novel epigenetic marker for chromophobe kidney cancer. Pathol Int. 2011;61(1):52–54. doi:10.1111/j.1440-1827.2010.02605.x
15. Tang BB, Liu SY, Zhan YU, et al. microRNA-218 expression and its association with the clinicopathological characteristics of patients with cervical cancer. Exp Ther Med. 2015;10(1):269–274. doi:10.3892/etm.2015.2455
16. Liu J, Sun H, Wang X, et al. Increased exosomal microRNA-21 and microRNA-146a levels in the cervicovaginal lavage specimens of patients with cervical cancer. Int J Mol Sci. 2014;15(1):758–773. doi:10.3390/ijms15010758
17. Zhang B, Wu X, Zhang X, et al. Human umbilical cord mesenchymal stem cell exosomes enhance angiogenesis through the Wnt4/beta-catenin pathway. Stem Cells Transl Med. 2015;4(5):513–522. doi:10.5966/sctm.2014-0267
18. Sharafeldin N, Slattery ML, Liu Q, et al. Multiple gene-environment interactions on the angiogenesis gene-pathway impact rectal cancer risk and survival. Int J Environ Res Public Health. 2017;14:10. doi:10.3390/ijerph14101146
19. Wu Y, Zhou BP. TNF-alpha/NF-kappaB/Snail pathway in cancer cell migration and invasion. Br J Cancer. 2010;102(4):639–644. doi:10.1038/sj.bjc.6605530
20. Rosa MN, Evangelista AF, Leal LF, et al. Establishment, molecular and biological characterization of HCB-514: a novel human cervical cancer cell line. Sci Rep. 2019;9(1):1913. doi:10.1038/s41598-018-38315-7
21. Zhang D, Lee H, Zhu Z, Minhas JK, Jin Y. Enrichment of selective miRNAs in exosomes and delivery of exosomal miRNAs in vitro and in vivo. Am J Physiol Lung Cell Mol Physiol. 2017;312(1):L110–L121. doi:10.1152/ajplung.00423.2016
22. Li F, Xu JW, Wang L, Liu H, Yan Y, Hu SY. MicroRNA-221-3p is up-regulated and serves as a potential biomarker in pancreatic cancer. Artif Cells Nanomed Biotechnol. 2018;46(3):482–487. doi:10.1080/21691401.2017.1315429
23. Zhou X, Wen W, Shan X, et al. A six-microRNA panel in plasma was identified as a potential biomarker for lung adenocarcinoma diagnosis. Oncotarget. 2017;8(4):6513–6525. doi:10.18632/oncotarget.14311
24. Petrozza V, Pastore AL, Palleschi G, et al. Secreted miR-210-3p as non-invasive biomarker in clear cell renal cell carcinoma. Oncotarget. 2017;8(41):69551–69558. doi:10.18632/oncotarget.v8i41
25. Wei WF, Zhou CF, Wu XG, et al. MicroRNA-221-3p, a TWIST2 target, promotes cervical cancer metastasis by directly targeting THBS2. Cell Death Dis. 2017;8(12):3220. doi:10.1038/s41419-017-0077-5
26. Ying J, Li H, Cui Y, Wong AH, Langford C, Tao Q. Epigenetic disruption of two proapoptotic genes MAPK10/JNK3 and PTPN13/FAP-1 in multiple lymphomas and carcinomas through hypermethylation of a common bidirectional promoter. Leukemia. 2006;20(6):1173–1175. doi:10.1038/sj.leu.2404193
27. Li L, Luo Z. Dysregulated miR-27a-3p promotes nasopharyngeal carcinoma cell proliferation and migration by targeting Mapk10. Oncol Rep. 2017;37(5):2679–2687. doi:10.3892/or.2017.5544
28. Zhao L, Zou D, Wei X, et al. MiRNA-221-3p desensitizes pancreatic cancer cells to 5-fluorouracil by targeting RB1. Tumour Biol. 2016;37:16053–16063. doi:10.1007/s13277-016-5445-8
29. Mahata S, Bharti AC, Shukla S, Tyagi A, Husain SA, Das BC. Berberine modulates AP-1 activity to suppress HPV transcription and downstream signaling to induce growth arrest and apoptosis in cervical cancer cells. Mol Cancer. 2011;10:39. doi:10.1186/1476-4598-10-39
30. Shojaei F, Wu X, Malik AK, et al. Tumor refractoriness to anti-VEGF treatment is mediated by CD11b+Gr1+ myeloid cells. Nat Biotechnol. 2007;25(8):911–920. doi:10.1038/nbt1323
31. Hammes LS, Tekmal RR, Naud P, et al. Up-regulation of VEGF, c-fms and COX-2 expression correlates with severity of cervical cancer precursor (CIN) lesions and invasive disease. Gynecol Oncol. 2008;110(3):445–451. doi:10.1016/j.ygyno.2008.04.038
32. de Conti A, Ortega JF, Tryndyak V, et al. MicroRNA deregulation in nonalcoholic steatohepatitis-associated liver carcinogenesis. Oncotarget. 2017;8(51):88517–88528. doi:10.18632/oncotarget.v8i51
33. Peng Q, Deng Z, Pan H, Gu L, Liu O, Tang Z. Mitogen-activated protein kinase signaling pathway in oral cancer. Oncol Lett. 2018;15(2):1379–1388. doi:10.3892/ol.2017.7491
34. Whitmarsh AJ, Davis RJ. Role of mitogen-activated protein kinase kinase 4 in cancer. Oncogene. 2007;26(22):3172–3184. doi:10.1038/sj.onc.1210410
35. Cheng MJ, Cao YG. TMPYP4 exerted antitumor effects in human cervical cancer cells through activation of p38 mitogen-activated protein kinase. Biol Res. 2017;50(1):24. doi:10.1186/s40659-017-0129-4
36. Melo SA, Sugimoto H, O’Connell JT, et al. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell. 2014;26(5):707–721. doi:10.1016/j.ccell.2014.09.005
37. Yan W, Wu X, Zhou W, et al. Cancer-cell-secreted exosomal miR-105 promotes tumour growth through the MYC-dependent metabolic reprogramming of stromal cells. Nat Cell Biol. 2018;20(5):597–609. doi:10.1038/s41556-018-0083-6
38. Sun X, Ma X, Wang J, et al. Glioma stem cells-derived exosomes promote the angiogenic ability of endothelial cells through miR-21/VEGF signal. Oncotarget. 2017;8(22):36137–36148. doi:10.18632/oncotarget.16661
39. Wu XG, Zhou CF, Zhang YM, et al. Cancer-derived exosomal miR-221-3p promotes angiogenesis by targeting THBS2 in cervical squamous cell carcinoma. Angiogenesis. 2019;22:397–410. doi:10.1007/s10456-019-09665-1
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.