Back to Journals » Cancer Management and Research » Volume 12
CELSR1 Acts as an Oncogene Regulated by miR-199a-5p in Glioma
Authors Wang G, Li Y, Zhang D, Zhao S, Zhang Q, Luo C, Sun X, Zhang B
Received 19 April 2020
Accepted for publication 2 July 2020
Published 23 September 2020 Volume 2020:12 Pages 8857—8865
DOI https://doi.org/10.2147/CMAR.S258835
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Chien-Feng Li
Guang Wang, 1, 2 Yong Li, 3 Dongxia Zhang, 4 Songtao Zhao, 4 Qiong Zhang, 4 Chao Luo, 2 Xiaochuan Sun, 1 Bingqian Zhang 5
1Department of Neurosurgery, First Affiliated Hospital of Chongqing Medical University, Chongqing, People’s Republic of China; 2Department of Neurosurgery, Chongqing Traditional Chinese Medicine Hospital, Chongqing, People’s Republic of China; 3Institute of Pathology and Southwest Cancer Center, Southwest Hospital Affiliated to Third Military Medical University, Chongqing, People’s Republic of China; 4National Key Laboratory of Trauma, Burn and Combined Injury, Southwest Hospital Affiliated to Third Military Medical University, Chongqing, People’s Republic of China; 5Department of Clinical Medicine, Chongqing Engineering Research Center of Pharmaceutical Sciences, Chongqing Medical and Pharmaceutical College, Chongqing, People’s Republic of China
Correspondence: Bingqian Zhang Department of Clinical Medicine
Chongqing Engineering Research Center of Pharmaceutical Sciences, Chongqing Medical and Pharmaceutical College, No. 82 of University-Town Middle Road, Shapingba District, Chongqing 401331, People’s Republic of China
Tel/ Fax +86-23-61969151
Email [email protected]
Xiaochuan Sun
Department of Neurosurgery, First Affiliated Hospital of Chongqing Medical University, No. 1 Youyi Road, Yuanjiagang, Yuzhong District, Chongqing 400016, People’s Republic of China
Tel/ Fax +86-23-89011012
Email [email protected]
Purpose: This study aimed to elucidate the biological function and upstream regulatory mechanism of CELSR1 in glioma.
Materials and Methods: We evaluated the expression of CELSR1 in glioma by TCGA_GEPIA tool, RT-qPCR, and Western blot assays. CCK-8, wound healing, and transwell invasion assays were, respectively, performed to detect the effect of CELSR1 on cell proliferation, migration, and invasion. The upstream regulatory miRNAs of CELSR1 were predicted by TargetScan and validated by luciferase activity reporter assay.
Results: CELSR1 is overexpressed in glioma (P< 0.05). CELSR1 promoted glioma cell proliferation, migration and invasion (P< 0.01). CELSR1 was a direct target of miR-199a-5p. miR199a-5p mimics significantly inhibited CELSR1 mRNA and protein expression (P< 0.01). miR199a-5p mimics reversed the effects of CELSR1 on glioma cell behaviors (P< 0.01).
Conclusion: CELSR1 acts as an oncogene promoting glioma cell proliferation, migration, and invasion, which is regulated by miR199a-5p.
Keywords: CELSR1, miR-199a-5p, glioma, proliferation, migration, invasion
Erratum for this paper has been published
Introduction
Glioma, accounting for about 40% of all brain tumors, is characterized by high morbidity and poor survival.1–4 High-grade gliomas are obviously characterized by high invasiveness, high proliferation rate and poor differentiation.5 Previous research achievements have greatly increased life expectancy and quality of life. However, the molecular mechanisms underlying its pathology remain unclear.
CELSR1 is a member of the flamingo subfamily, which is a part of the cadherin superfamily. CELSR1 is likewise a member of planar cell polarity signaling pathway.6 Mutations of CELSR1 are linked to the severe neural tube defect craniorachischisis.7 Besides, aberrant CELSR1 expression is reported in a variety of tumors (including lung cancer,8 breast cancer,9 hepatocellular carcinoma,10 ductal carcinoma in situ,11 oral squamous cell carcinoma,12 lymphoid leukemia,13,14 and glioma15). However, until now, the biological function of CELSR1 has not been identified in cancers including glioma.
MicroRNAs (miRNAs), a type of non-coding RNA, are single-stranded RNAs of 18~24 nucleotides in length. miRNAs regulate gene expression at a post-transcriptional level by degrading or repressing target mRNAs, resulting in translational repression or mRNA degradation.16,17 Our previous study reported that miR199a-5p suppresses glioma cell proliferation, migration, and invasion by inhibiting MAGT1.18 However, the correlation between CELSR1 and miR-199a-5p has not been reported.
The purpose of our study was to investigate the biological function of CELSR1 and the upstream regulatory mechanism in glioma. Specifically, we evaluated the expression of CELSR1 in glioma by TCGA_GEPIA tool, RT-qPCR, and Western blot assays. Dual-luciferase reporter assay was utilized to determinate the mechanism upstream of CELSR1.
Given that the glioma biomarkers are scarce for clinical, our study on CELSR1 has significant implications in the diagnosis and treatment of glioma.
Methods and Materials
Cell Lines
The human glioma cell lines (U251, U118, U87-MG, and A172) and normal human astrocyte cell line (NHA) were obtained from the Type Cell Culture Collection of the Chinese Academy of Sciences (Shanghai, China). Cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Thermo Fisher Scientific Inc, Rockford, USA) containing 10% fetal bovine serum (FBS), penicillin (100U/mL) and streptomycin (100μg/mL) at 37°C under 5% CO2.
Transfection
GenePharm (Shanghai, China) was responsible for synthesizing miR-199a-5p mimics and inhibitors. CELSR1 overexpression vector was synthesized by Sangon Biotech (Shanghai, China) and cloned into pCDNA3.1 (+) plasmid. Cell transfection was performed by Lipofectamine 2000 reagent (Invitrogen, Carlsbad, USA) according to the manufacturer’s instructions.
Bioinformatics Analysis
The function of CELSR1 in cancer development was explored in Cancer Hallmarks Analytics Tool (CHAT)(http://chat.lionproject.net/).19 GEPIA20 (http://gepia.cancer-pku.cn/index.html) online analysis tool was used to analyze the expression of CELSR1 in glioma. The targets of miR-199a-5p were analyzed by targetscan21 database.
Western Blot
To determine the effect of miR199a-5p on CELSR1, we detected the protein expression levels of CELSR1 according to the following procedures. Lysis Buffer (CST, Danvers, USA) was used to lyse cells on ice for 30 min. Then, the protein samples were centrifuged at 14,000 rpm for 15 min at 4°C. The supernatant protein was collected and normalized, and equal protein samples were electrophoresed in SDS-PAGE gels and transferred onto the polyvinylidene difluoride (PVDF) membranes. The membranes were blocked with 5% fat‐free milk at room temperature for 1 hour and then incubated with primary antibodies (CELSR1: ab225889, GAPDH: ab181602; both from Abcam, Cambridge, USA) at 4°C overnight. Upon being washed three times with washing buffer (Tris buffered saline Tween, TBST), the membranes were incubated with secondary antibody (ab6721, Abcam) diluted in blocking buffer for 60 min at room temperature. The membranes were also washed in TBST. Proteins were then visualized by ECL reagents (KeyGen, Nanjing, China). Image J 1.42 software was used to analyze the relative expression levels.
Quantitative Real-Time PCR
Total RNAs were extracted from cells and specimens using Trizol reagent (Invitrogen, Rockville, USA) following the provided instructions. Complementary DNA (cDNA) was reverse transcribed using the PrimeScript 1st Strand cDNA Synthesis Kit (Takara, Dalian, China). The mRNA expression level was detected using TB Green Fast qPCR Mix (Takara, Dalian, China). The mature miR-199a-5p expression was detected by the premix EX Taq (Takara, Dalian, China). The primers are: miR-199a-5p, 5ʹ-TCAAGAGCAATAACGAAAAATGT-3ʹ (forward), 5ʹ-GCTGTCAACGATACGCTACGT-3ʹ (reverse); U6, 5ʹ-ATTGGAACGATACAGAGAAGATT-3ʹ (forward), 5ʹ-GTCCTTGGTGCCCGAGTG-3ʹ (reverse); CELSR1, 5ʹ-GGCGTTGTTTGAGAACGAACC-3ʹ (forward), 5ʹ-AGAGTCGATTCGGAAGTAGCC-3ʹ (reverse); GAPDH, 5ʹ-GGAGCGAGATCCCTCCAAAAT-3ʹ (forward), 5ʹ-GGCTGTTGTCATACTTCTCATGG-3ʹ (reverse); ROCK1, 5ʹ-AACATGCTGCTGGATAAATCTGG-3ʹ (forward), 5ʹ-TGTATCACATCGTACCATGCCT-3ʹ (reverse); Galectin1, 5ʹ-GGACATCCTCCTGGACTCA-3ʹ (forward), 5ʹ-GTTGAAGCGAGGGTTGAAGT-3ʹ (reverse); MMP2, 5ʹ-CCCACTGCGGTTTTCTCGAAT-3ʹ (forward), 5ʹ-CAAAGGGGTATCCATCGCCAT-3ʹ (reverse); MMP9, 5ʹ-AGACCTGGGCAGATTCCAAAC-3ʹ (forward), 5ʹ-CGGCAAGTCTTCCGAGTAGT-3ʹ (reverse); Snail, 5ʹ-TACAAAAACCCACGCAGACA-3ʹ (forward), 5ʹ-ACCCCACATCCTTCTCACTG-3ʹ (reverse); GAPDH and U6 were used as the standard for normalization. The expression levels were calculated using the 2−ΔΔCT method.
CCK-8 Assay
Cell viability was assessed by Cell Counting Kit-8 (DOJINDO, Japan) assay. U251 and U118 cells (5×103/well) were plated in 96-well plates and incubated 24 h, 48 h, or 72 h. Then, old medium was replaced with 10μL CCK-8 reagent and 100μL fresh medium. The optical density (OD) values were measured at a wavelength of 450 nm after 1 h at 37°C.
Wound Healing Assay
Wound healing assay was performed to evaluate cell migration activity. To make sure the same visual field in the photograph, a mark was left on the back of the 12-well plate. U251 and U118 cells were seeded into plate. When cells approximately 100% covered the plate bottom, a sterilized tip was used to create a wound at the cell surface. Then, we washed cells with phosphate‐buffered saline for three times and add medium. Cells were photographed at a fixed location at 0 h and 24 h.
Transwell Invasion Assay
To evaluate the effect of CELSR1 on glioma cell invasion, a matrix (BD Biosciences, San Jose, USA) precoated transwell chamber was used. In brief, U251 and U118 cells were serum starved for 24 h and then were resuspended in 1% FBS.1×104 cells were seeded on the upper chamber, whereas the lower chamber was supplemented with 20% FBS. After incubation for 48 h, the upper noninvasive cells were gently wiped. The invasion cells were fixed with methanol and stained with 0.5% crystal violet. Cell numbers were averaged under five random fields from triplicate experiments.
Dual-Luciferase Reporter Assay
To determine the binding site between miR-199a-5p and CELSR1, wild-type (wt) or mutated (mut) CELSR1 sequences were cloned into the psiCHECK2 vector (Promega, Madison, USA). Cells were co-transfected with the wt or mut CELSR1 3ʹ-UTR, and the miR-199a-5p mimics/inhibitors/NC. Luciferase activity was measured through Dual-Luciferase Reporter Assay Kit (Promega, Madison, USA) after 48 h.
Statistical Analysis
SPSS 21.0 software (SPSS, Chicago, USA) was used for statistical analysis. Data is presented as mean± standard deviation (SD) of three independent experiments. Differences in measurement data were analyzed by the Student’s t-test and one-way ANOVA. P values <0.05 were considered statistically significant.
Results
CELSR1 is Overexpressed in Glioma
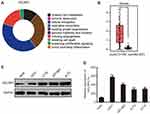
Using Cancer Hallmarks Analytics Tool (CHAT), we found that CELSR1 is associated with genome instability and mutation, evading growth suppressors, inducing angiogenesis, invasion and metastasis, and sustaining proliferative signaling (Figure 1A). To determine the expression levels of CELSR1 in glioma, we analyzed the TCGA datasets by GEPIA tools and detected the protein and mRNA expression levels of CELSR1 in glioma cells. TCGA datasets show that CELSR1 is significantly overexpressed in glioma patients (Figure 1B; P<0.05). Besides, Western blot and RT-qPCR assays, respectively, revealed that the protein and mRNA expression levels of CELSR1 are increased in glioma cells (Figure 1C and D; P<0.001).
CELSR1 Promoted Glioma Cell Proliferation, Migration and Invasion
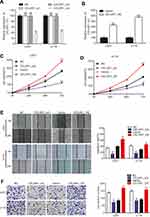
To investigate the biological function of CELSR1 in glioma, we significantly knocked down and overexpressed CELSR1 expression in U251 and U118 cells (Figure 2A and B; P<0.01). CCK-8 assays revealed that CELSR1 significantly increased cell viability, and blockade of CELSR1 significantly inhibited cell viability in U251 and U118 cells (Figure 2C and D; P<0.01). Besides, wound healing assay found that CELSR1 significantly promoted cell migration (Figure 2E; P<0.01). Transwell invasion assay revealed that CELSR1 significantly promoted cell invasion (Figure 2F; P<0.01).
CELSR1 Was a Direct Target of miR-199a-5p
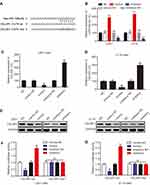
To determinate the upstream regulation mechanism of CELSR1, we predicted the correlated miRNA using targetscan database. We found that CELSR1 probably is a direct target of miR199a-5p (Figure 3A). To further investigate the correlation between miR199a-5p and CELSR1, we transfected miR199a-5p mimics and inhibitors in U251 and U118 cells (Figure 3B; P<0.01) and detected the CELSR1 expression levels in different groups. RT-qPCR assays found that miR199a-5p mimics significantly inhibited CELSR1 mRNA expression (Figure 3C; P<0.01) and miR199a-5p inhibitors significantly promoted CELSR1 mRNA expression (Figure 3D; P<0.01). Western blot assays revealed that miR199a-5p mimics significantly inhibited CELSR1 protein expression and miR199a-5p inhibitors significantly promoted CELSR1 protein expression (Figure 3E; P<0.05). Hence, to further verified the binding sites between miR199a-5p and CELSR1, we detect the relative luciferase activity after co-transfection assays. The dual-luciferase report system revealed that miR-199a-5p mimics significantly inhibit the luciferase activity of CELSR1-wt 3ʹUTR, and miR-199a-5p inhibitors significantly increase the luciferase activity of CELSR1-wt 3ʹUTR among U251 and U118 cells, whereas miR-199a-5p mimics or inhibitors do not regulate the luciferase activity of CELSR1-mut 3ʹUTR among U251 and U118 cells (Figure 3F and G; P<0.05).
CELSR1 Was an Effector for the Role of miR‐199a‐5p
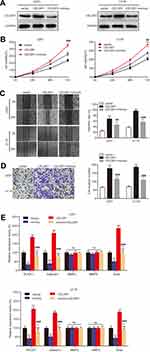
To further illuminate the regulatory process of miR199a-5p/CELSR1 axis on cell biological function, we co-transfected CELSR1 and miR199a-5p mimics (Figure 4A; P<0.05) and detected cell proliferation, migration and invasion in U251 and U118 cells. CCK-8 assays revealed that miR-199a-5p mimics significantly reversed the effect of CELSR1 promoting cell proliferation (Figure 4B; P<0.01). Wound healing assay found that miR-199a-5p mimics significantly reversed the effect of CELSR1 promoting cell migration (Figure 4C; P<0.01). Transwell invasion assay significantly reversed the effect of CELSR1 promoting cell invasion (Figure 4D; P<0.01). Besides, we found that CELSR1 significantly reversed the effect of miR-199a-5p inhibiting the mRNA expression of ROCK1, Galectin1, and Snail.
Discussion
In this study, we found that CELSR1 acts as an oncogene by regulating cell proliferation, cell migration, and cell invasion in glioma, which was negatively regulated by miR199a-5p. The result of Figure 1A indicated that CELSR1 is associated with these cancer hallmarks (such as genome instability and mutation, evading growth suppressors, invasion and metastasis), which provided the research direction. Subsequently, in light of TCGA_GEPIA expression analysis, RT-qPCR and Western blot results, we confirmed that CELSR1 is overexpressed in glioma (Figure 1B–D). These results lead us to detect the role of CELSR1 in tumor cell proliferation, migration and invasion. Our findings revealed that CELSR1 significantly promoted cell proliferation, migration, and invasion (Figure 2C–F). Next, we further explored the upstream regulatory mechanism. We confirmed that miR199a-5p regulates CELSR1 expression at a post-transcriptional level by degrading CELSR1 mRNA (Figure 3A), which indicated that CELSR1 is a direct target of miR-199a-5p.
miR199a-5p regulates cell biological function by multiple target genes. Previous study reported that miR-199a-5p significantly regulated cell proliferation, migration, and invasion.18,22,23 In glioma, Zhang et al reported that miR-199a inhibits tumor growth and attenuates chemoresistance by targeting K-RAS via ATK and ERK signaling.24 Our previous study also found that miR199a-5p suppresses glioma cell proliferation, migration, and invasion by inhibiting MAGT1. Then, we hypothesized that miR199a-5p regulates glioma progression via targeting CELSR1. Our results revealed that miR-199a-5p mimics significantly reversed the effect of CELSR1 promoting cell proliferation, migration, and invasion (Figure 4B–D). Besides, we detected the mRNA expression of several known motility and adhesion regulators including ROCK1, Galectin1, MMP2, MMP9, and Snail.25 Our results found that CELSR1 significantly reversed the effect of miR-199a-5p inhibiting the mRNA expression of ROCK1, Galectin1, and Snail (Figure 4E), which indicated that CELSR1 is an effector for the role of miR‐199a‐5p.
Based on the above results, CELSR1 is an oncogene in glioma, which regulated cell proliferation, migration, and invasion. Besides, we preliminarily demonstrated CELSR1 upstream regulatory mechanism that miR199a-5p inhibits CELSR1 expression at a post-transcriptional level by degrading CELSR1 mRNA. However, there are some limitations to this study. On the one hand, in vivo assays should be performed to verify these results. On the other hand, more experiments should be performed to explore probable mechanisms downstream of CELSR1 regulating glioma progression (such as cell apoptosis, cell cycle). Hence, the next step is to explore the detailed regulatory mechanism of CELSR1 in animal models and patients with glioma.
Above all, our findings validated that CELSR1 promotes cell migration and invasion regulated by miR-199a-5p in glioma, which will provide strong evidence supporting the function of CELSR1 and miR199a-5p in glioma occurrence and development.
Acknowledgments
This work was funded by Medical Scientific Research Joint Funds of Chongqing Science and Technology Bureau and Chongqing Health Commission (General Program) [Grant/Award Number: 2018MSXM053, 2019MSXM042], Science Fund for Creative Research Groups of Chongqing Medical and Pharmaceutical College [Grant/Award Number: ygz2019403], and Open Fund of National Key Laboratory of Trauma, Burn and Combined Injury [Grant/Award Number: SKLKF2019010].
Disclosure
The authors report no conflicts of interest in this work.
References
1. Ostrom QT, Gittleman H, Farah P, et al. CBTRUS statistical report: primary brain and central nervous system tumors diagnosed in the United States in 2006–2010. Neuro Oncol. 2013;15(Suppl 2):ii1–i56. doi:10.1093/neuonc/not151
2. Armstrong TS, Wen PY, Gilbert MR, Schiff D. Management of treatment-associated toxicites of anti-angiogenic therapy in patients with brain tumors. Neuro Oncol. 2012;14(10):1203–1214. doi:10.1093/neuonc/nor223
3. Jansen M, Yip S, Louis DN. Molecular pathology in adult gliomas: diagnostic, prognostic, and predictive markers. Lancet Neurol. 2010;9(7):717–726. doi:10.1016/S1474-4422(10)70105-8
4. Zhang J-Q, Yao Q-H, Kuang Y-Q, et al. Prognostic value of coexistence of abnormal expression of micro-RNA-200b and cyclic adenosine monophosphate-responsive element-binding protein 1 in human astrocytoma. Hum Pathol. 2014;45(10):2154–2161. doi:10.1016/j.humpath.2014.01.025
5. de Groot JF. High-grade gliomas. Continuum. 2015;21(2 Neuro–oncology):332–344. doi:10.1212/01.CON.0000464173.58262.d9
6. Lei Y, Zhu H, Yang W, Ross ME, Shaw GM, Finnell RH. Identification of novel CELSR1 mutations in spina bifida. PLoS One. 2014;9(3):e92207. doi:10.1371/journal.pone.0092207
7. Robinson A, Escuin S, Doudney K, et al. Mutations in the planar cell polarity genes CELSR1 and SCRIB are associated with the severe neural tube defect craniorachischisis. Hum Mutat. 2012;33(2):440–447. doi:10.1002/humu.21662
8. Li Y, Zhang H, Fan L, et al. MiR-629-5p promotes the invasion of lung adenocarcinoma via increasing both tumor cell invasion and endothelial cell permeability. Oncogene. 2020;39(17):3473–3488. doi:10.1038/s41388-020-1228-1
9. Liao S, Desouki MM, Gaile DP, et al. Differential copy number aberrations in novel candidate genes associated with progression from in situ to invasive ductal carcinoma of the breast. Genes Chromosomes Cancer. 2012;51(12):1067–1078. doi:10.1002/gcc.21991
10. Ammerpohl O, Pratschke J, Schafmayer C, et al. Distinct DNA methylation patterns in cirrhotic liver and hepatocellular carcinoma. Int J Cancer. 2012;130(6):1319–1328. doi:10.1002/ijc.26136
11. Geradts J, Groth J, Wu Y, Jin G. Validation of an oligo-gene signature for the prognostic stratification of ductal carcinoma in situ (DCIS). Breast Cancer Res Treat. 2016;157(3):447–459. doi:10.1007/s10549-016-3838-4
12. Marta M, Zhang T, Scognamiglio T, Gudas LJ. Mutations in long-lived epithelial stem cells and their clonal progeny in pre-malignant lesions and in oral squamous cell carcinoma. Carcinogenesis. 2020.
13. Charfi C, Edouard E, Rassart E. Identification of GPM6A and GPM6B as potential new human lymphoid leukemia-associated oncogenes. Cell Oncol. 2014;37(3):179–191.
14. Kaucká M, Plevová K, Pavlová S, et al. The planar cell polarity pathway drives pathogenesis of chronic lymphocytic leukemia by the regulation of B-lymphocyte migration. Cancer Res. 2013;73(5):1491–1501. doi:10.1158/0008-5472.CAN-12-1752
15. Vastrad B, Vastrad C, Godavarthi A, Chandrashekar R. Molecular mechanisms underlying gliomas and glioblastoma pathogenesis revealed by bioinformatics analysis of microarray data. Med Oncol. 2017;34(11). doi:10.1007/s12032-017-1043-x
16. Lv H, Wu X, Ma G, et al. An integrated bioinformatical analysis of miR‑19a target genes in multiple myeloma. Exp Ther Med. 2017. doi:10.3892/etm.2017.5173
17. Rushworth SA, Murray MY, Barrera LN, Heasman S-A, Zaitseva L, MacEwan DJ. Understanding the role of miRNA in regulating NF-κB in blood cancer. Am J Cancer Res. 2012;2(1):65.
18. Wang G, Li Y, Li J, et al. microRNA-199a-5p suppresses glioma progression by inhibiting MAGT1. J Cell Biochem. 2019;120(9):15248–15254. doi:10.1002/jcb.28791
19. Baker S, Ali I, Silins I, et al. Cancer Hallmarks Analytics Tool (CHAT): a text mining approach to organize and evaluate scientific literature on cancer. Bioinformatics. 2017;33(24):3973–3981. doi:10.1093/bioinformatics/btx454
20. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–W102. doi:10.1093/nar/gkx247
21. Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4:e05005.
22. Gao Y, Han D, Sun L, et al. PPARα regulates the proliferation of human glioma cells through miR-214 and E2F2. Biomed Res Int. 2018;2018:1–10.
23. Zhang H, Qin D, Jiang Z, Zhang J. SNHG9/miR-199a-5p/Wnt2 axis regulates cell growth and aerobic glycolysis in glioblastoma. J Neuropathol Exp Neurol. 2019;78(10):939–948. doi:10.1093/jnen/nlz078
24. Li W, Wang L, Ji XB, et al. MiR-199a inhibits tumor growth and attenuates chemoresistance by targeting K-RAS via AKT and ERK signalings. Front Oncol. 2019;9:1071. doi:10.3389/fonc.2019.01071
25. Gerashchenko TS, Novikov NM, Krakhmal NV, et al. Markers of cancer cell invasion: are they good enough? J Clin Med. 2019;8:8. doi:10.3390/jcm8081092
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.