Back to Journals » Journal of Inflammation Research » Volume 14
CC Chemokine Receptors in Lung Adenocarcinoma: The Inflammation-Related Prognostic Biomarkers and Immunotherapeutic Targets
Received 23 August 2020
Accepted for publication 26 November 2020
Published 5 February 2021 Volume 2021:14 Pages 267—285
DOI https://doi.org/10.2147/JIR.S278395
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Ning Quan
Fangteng Liu,1,2 Hengyu Wu1
1Department of Breast Surgery, The Third Hospital of Nanchang, Nanchang, Jiangxi, 330009, People’s Republic of China; 2Faculty of Medicine, University of Munich, Munich, 80336, Germany
Correspondence: Hengyu Wu Tel +86-15270280590
Email [email protected]
Background: Lung adenocarcinoma (LUAD) is the most common type of lung cancer with a high incidence and increased mortality. CC chemokine receptors were participating in the modulation of the tumor microenvironment and involved in carcinogenesis and tumor development. However, the potential mechanistic values of CC chemokine receptors as clinical biomarkers and therapeutic targets in LUAD have not been fully clarified.
Methodology: ONCOMINE, UALCAN, GEPIA, Kaplan–Meier Plotter, SurvExpress, MethSurv, SurvivalMeth, cBioPortal, String, GeneMANIA, DAVID, Metascape, TRRUST, LinkedOmics, and Timer were applied in this work.
Results: The transcriptional levels of CCR1/10 in LUAD tissues were significantly reduced while the transcriptional levels of CCR3/6/7/8 were significantly elevated, and the expression of CCR1 was the highest in LUAD among these CC chemokine receptors. A significant correlation was found between the expression of CCR2/4/6/7 and the pathological stage of LUAD patients. There were significant associations between CCR2/3/4/5/6/10 expression levels and OS in LUAD, and LUAD patients with high transcriptional levels of CCR3/4 had inferior first-progression survival. In addition, the prognostic values of CC chemokine receptors signature in LUAD were explored in three independent cohorts, the high-risk group displayed unfavorable OS compared with the low-risk group, and the LUAD cases in the high-risk group also suffered inferior RFS than that in the low-risk group. And for the prognostic value of the DNA methylation of CC chemokine receptors, we found 1 CpG of CCR2, 2 CpGs of CCR3, 1 CpG of CCR4, 3 CpGs of CCR6, 3 CpGs of CCR7, 1 CpG of CCR8, and 3 CpGs of CCR9 were significantly associated with prognosis in LUAD patients. However, the DNA methylation signature analysis showed there was no statistically significant association between the high- and low-risk group. For potential mechanism, the neighbor gene networks, interaction analyses, functional enrichment analyses of CC chemokine receptors in LUAD were performed, the transcription factor targets, kinase targets, and miRNA targets of CC chemokine receptors were also identified in LUAD. We also found significant correlations among CC chemokine receptors expression and the infiltration of immune cells, the tumor infiltration levels among LUAD with different somatic copy number alterations of these chemokine receptors were also assessed. Moreover, the Cox proportional hazard model showed that CCR1/2/10, B_cell, CD4_Tcell were significantly related to the clinical outcome of LUAD patients.
Conclusion: CC chemokine receptors might serve as immunotherapeutic targets and prognostic biomarkers in LUAD.
Keywords: CC chemokine receptors, lung adenocarcinoma, prognostic biomarker, immunotherapeutic target, bioinformatics analysis
Introduction
Lung cancer is the most common cancer worldwide, and it accounts for 18.4% of all deaths from cancer worldwide.1 The 5-year survival rate of lung cancer patients is relatively low, only 19%.2 And lung adenocarcinoma (LUAD) is a major subtype of lung cancer, accounts for approximately 40% of all cases with this disease.3 It is worth noting that up to about 75% of patients were at an advanced stage at the time of diagnosis.4 Although some treatment technologies and drugs have made great progress recently, the long-term survival rate of LUAD patients remains unsatisfactory.5,6 And there is also a big challenge to conduct more accurate risk assessments and optimize treatments for LUAD patients.7,8 Therefore, it is an urgent need to develop and identify more therapeutic targets and prognostic biomarkers for LUAD.
CC chemokine receptors, namely beta chemokine receptors, are a part of the G protein-linked receptor superfamily which is known as seven-(pass)-transmembrane domain receptors.9 CC chemokine receptors are membrane proteins, they could specifically bind to CC chemokine family cytokines. They represent a typical subfamily of chemokine receptors. According to the IUIS/WHO Subcommittee on Chemokine Nomenclature, a total of ten members of CC chemokine receptors, CCR1 to CCR10, are named.10,11 The CC chemokine receptors play multiple important roles in various biological processes, including the regulation of leukocyte chemotaxis, recruitment of immune cells, inflammation and parenchymal remodeling, tumorigenesis, and progression.12–16 Expressed on tumor cells and peripheral blood cells, immune cells, and stromal cells, CC chemokine receptors play prominent roles in determining the composition of tumor stroma, are also closely related to angiogenesis, tumor growth and, metastasis,17–21 thus affecting disease progression, therapeutic effect, and clinical outcomes directly or indirectly. CC chemokine receptors can be activated by binding to the related chemokines and applied as potential therapeutic targets, and prognostic indicators for various kinds of malignant tumors, including lung adenocarcinoma.21–26
Although the expression and functions of certain CC chemokine receptors have been reported in some studies, the general characterizes of all CC chemokine receptors as targets and biomarkers in LUAD are largely unclear. A synthetical analysis of the roles of CC chemokine receptors in LUAD has become urgent at present. Now it is feasible through second-generation sequencing technology and advanced bioinformatics methods. In this study, we performed an in-depth and comprehensive analysis of the potential values of CC chemokine receptors as clinical markers and immunotherapeutic targets in LUAD based on multiple large bioinformatics databases, thus providing clinicians with additional information to help them choose more appropriate drugs and more accurately assess the prognosis of LUAD patients.
Materials and Methods
ONCOMINE
ONCOMINE (www.oncomine.org) is a translational bioinformatics service that provides powerful, genome-wide expression analysis.27 Data were extracted to evaluate the expression of CC chemokine receptors in lung cancer. And the analyses that meet the thresholds for CC chemokine receptors were further reviewed for lung adenocarcinoma (LUAD). In this study, a P 0.05, a fold change of 1.5 were set as the significance thresholds. The data type was selected mRNA. t‑Test was used to analyze the difference in the expression of CC chemokine receptors in LUAD.
UALCAN
UALCAN (http://ualcan.path.uab.edu/analysis.html), a comprehensive and interactive web resource, provides easy access to publicly available cancer OMICS data (TCGA and MET500).28 In our study, CC chemokine receptors level was obtained in the “Expression” links using the “TCGA analysis” module and the “LUAD” dataset.
GEPIA
GEPIA (http://gepia.cancer-pku.cn/index.html) is a developed interactive web server for analyzing the RNA sequencing expression data of 9736 tumors and 8587 normal samples from the TCGA and the GTEx project.29 In this study, we performed the pathological stage analysis and multiple gene comparison analysis of CC chemokine receptors using the “LUAD” dataset. And the correlation between CC chemokine receptors expression and disease-free survival (DFS) was also calculated.
Kaplan–Meier Plotter
Kaplan–Meier Plotter (https://kmplot.com/analysis/) is a useful prognostic biomarker assessment tool that explored the effect of 54k genes on survival in 21 cancer types using the databases from GEO, EGA, and TCGA.30 To analyze the prognostic value of CC chemokine receptors in lung cancer regarding OS (overall survival), FP (first progression), and PPS (post-progression survival), the patient samples were split into two groups by the median expression, with the restricted analysis to subtype histology (adenocarcinoma), the JetSet best probe set was used, and the biased arrays were excluded for array quality control. The hazard ratio with 95% confidence intervals and log-rank P value were calculated.
SurvExpress
SurvExpress (http://bioinformatica.mty.itesm.mx/SurvExpress) is a friendly and versatile free web tool, it could provide a comprehensive analysis of multi-gene biomarkers for gene expression and clinical outcomes in human tumors.31 In this study, two training cohorts (GSE31210, 226 patients; Chitale Lung, 185 patients) and one validation cohort (TCGA, 475 patients) were used to explore the prognostic value of CC chemokine receptors signature in LUAD patients.
MethSurv
MethSurv (https://biit.cs.ut.ee/methsurv/), a web tool for survival analysis based on CpG methylation patterns, was applied to explore the prognostic value of single CpG methylation of the CC chemokine receptors in LUAD patients.32
SurvivalMeth
SurvivalMeth (http://bio-bigdata.hrbmu.edu.cn/survivalmeth/)33 was used to analyze the DNA methylation of CC chemokine receptors signature on LUAD prognosis.
cBioPortal
cBioPortal (www.cbioportal.org) is a comprehensive web resource that could visualize and analyze multidimensional cancer genomics data.34,35 Based on the TCGA database, genetic alterations of CC chemokine receptors were obtained from cBioPortal. Five hundred eighty-six lung adenocarcinoma samples (TCGA, Firehose Legacy) were analyzed. mRNA expression z scores (RNA Seq V2 RSEM) were obtained using a z score threshold of ±2.0. The top 50 most frequently altered neighbor genes associated with CC chemokine receptors were isolated.
String
STRING (https://string-db.org/) is a database of known and predicted protein–protein interactions (PPI).36,37 We conducted a PPI network analysis of CC chemokine receptors to explore their interactions with STRING.
GeneMANIA
GeneMANIA (http://www.genemania.org) is a useful website that provides information for co-expression, co-localization, genetic and protein interactions, pathways, physical interaction, shared protein domains of submitted genes.38
DAVID
DAVID 6.8 (https://david.ncifcrf.gov) is a comprehensive bioinformatics resource for biological functional annotation.39,40 In this study, the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis were performed with the CC chemokine receptors, and closely related neighbor genes were isolated from cBioPortal.
Metascape
Metascape (http://metascape.org) is a reliable public service that provides a biologist-oriented resource for the analysis of systems-level datasets.41 In this study, the “Express Analysis” module was used to further verify the enrichment of CC chemokine receptors and their related most frequently altered neighbor genes.
TRRUST
TRRUST (https://www.grnpedia.org/trrust/) is a manually curated database of human and mouse transcriptional regulatory networks. It contains 8444 and 6552 TF-target regulatory relationships of 800 human TFs and 828 mouse TFs, respectively. TRRUST database was used for the regulatory relationships of CC chemokine receptors genes.42,43
LinkedOmics
LinkedOmics (http://www.linkedomics.org/) is a publicly available portal that includes multi-omics data from all 32 TCGA Cancer types.44 In this study, TCGA_LUAD dataset was used in the “LinkInterpreter” module. Gene Set Enrichment Analysis (GSEA) tool was applied to explore the kinase target and miRNA target enrichment of CC chemokine receptors in the LUAD, with a minimum number of genes (size) of 3 and a simulation of 500. Results were analyzed statistically using the Spearman correlation test. The P-value cutoff was 0.05.
Timer
Timer web server (https://cistrome.shinyapps.io/timer/) is a comprehensive resource for systematical analysis of the infiltration of different immune cells and their clinical impact across diverse cancer types.45,46 “Gene module” and “Survival module” were used in this study to explore the correlation of CC chemokine receptors level and the immune cell infiltration, the clinical outcome in LUAD, respectively. “SCNA module” was used to explore the correlation between somatic copy number alterations and abundance of immune infiltrates.
Results
Expression of CC Chemokine Receptors in the Lung Adenocarcinoma
First, we explored the expression levels of CC chemokine receptors in lung cancer and normal lung tissues using the ONCOMINE database (Figure 1). And the analyses that meet the thresholds for CC chemokine receptors were further reviewed according to the histopathological type (lung adenocarcinoma vs normal). As shown in Supplementary Table 1, the transcriptional levels of CCR1 and CCR2 in LUAD tissues were significantly reduced, while the transcriptional levels of CCR4, CCR7, and CCR8 were significantly increased in LUAD vs normal lung tissue. For CCR6, three datasets suggested that the transcriptional levels of CCR6 in LUAD tissues were significantly elevated, while one dataset showed it was significantly reduced in LUAD tissue.
The expression levels of CC chemokine receptors in LUAD were also assessed with UALCAN. As expected, the transcriptional levels of CCR1 (P = 1.824E-05), and CCR10 (P = 1.528E-08) in LUAD tissues were significantly reduced, while the transcriptional levels of CCR3 (P = 5.402E-03), CCR6 (P = 1.176E-02), CCR7 (P = 4.060E-02) and CCR8 (P < 1E-12) were significantly elevated (Figure 2).
We also compared the relative expression levels of CC chemokine receptors in LUAD and found the relative expression of CCR1 was the highest in LUAD tissues among all CC chemokine receptors (Figure 3).
We then assessed the correlation between the expression of CC chemokine receptors and the pathological stage of LUAD patients, and found a significant correlation between the expression of CCR2 (P = 0.027), CCR4 (P = 0.0336), CCR6 (P = 0.00242), CCR7 (P = 0.00387) and pathological stage (Figure 4), as LUAD progressed, the expression of CCR2, CCR4, CCR6, and CCR 7 decreased. These data suggested that these CC chemokine receptors played significant roles in the development of LUAD.
The Prognostic Value of Individual CC Chemokine Receptor in Patients with LUAD
To evaluate the prognostic value of CC chemokine receptors in LUAD, we assessed the correlation between these chemokine receptors and prognosis using Kaplan–Meier Plotter (Table 1). Overall survival curves are presented in Figure 5. Among them, there were significant associations between these chemokine receptors (CCR2, CCR3, CCR4, CCR5, CCR6, and CCR10) and OS in LUAD. High expression of CCR2, CCR3, CCR4, CCR10 indicated shorter OS, while high expression of CCR5, CCR6 indicated longer OS in LUAD cases.
 |
Table 1 The Relationship Between the Expression Level of CC Chemokine Receptors and Prognosis in Patients with LUAD |
The first-progression survival curves are presented in Figure 6. LUAD patients with high transcriptional levels of CCR3 (P = 0.0026) and CCR4 (P = 3e-04) were significantly associated with inferior first-progression survival. However, no significant difference was found between the CC chemokine receptors and PPS or DFS in LUAD patients (Table 1).
Prognostic Values of CC Chemokine Receptors Signature in Patients with LUAD
The CC chemokine receptors signature was input for prognostic analysis in SurvExpress. For OS, in the training cohort 1 (GSE31210, 226 patients), the mRNA expression of CCR2, CCR6, CCR9 was higher in the low-risk group than that in the high-risk group, while, CCR1, CCR8, and CCR10 were lower expressed in the low-risk group than that in the high-risk group (Figure 7A-B), the high-risk group displayed an unfavorable OS compared with the low-risk group (Figure 7C). Similarly, these results were also displayed in the training cohort 2 (Chitale Lung, 185 patients) (Figure 7D-F), the high-risk group had a worse OS compared with the low-risk group (Figure 7F). And the prognostic value of CC chemokine receptors signature in LUAD was also confirmed in the validation cohort (TCGA, 475 patients) (Figure 7G-I). Besides, for recurrence-free survival (RFS), the LUAD cases in the high-risk group suffered inferior RFS than that in the low-risk group (Supplementary Figure 1).
Prognostic Value of Single CpG of CC Chemokine Receptors Gene
The prognostic value of DNA methylation of CC chemokine receptors in LUAD was analyzed by MethSurv. The heat maps of DNA methylation of the CC chemokine receptor are displayed in Figure 8. Among them, cg01297500 of CCR1, cg11313065 of CCR2, cg11126313 of CCR3, cg11126313 of CCR3, cg21366834 of CCR4, cg15239694 of CCR5, cg19668990 of CCR6, cg11729107 of CCR7, cg11492964 of CCR8, cg14558191 of CCR9, and cg06864083 of CCR10 showed the highest DNA methylation level. And overall, we found that 1 CpG of CCR2, 2 CpGs of CCR3, 1 CpG of CCR4, 3 CpGs of CCR6, 3 CpGs of CCR7, 1 CpG of CCR8, and 3 CpGs of CCR9 were significantly associated with prognosis in LUAD patients (Table 2).
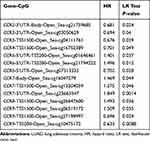 |
Table 2 The Prognostic Value of Single CpG of CC Chemokine Receptors in LUAD by MethSurv (P<0.05) |
Prognostic Value of the DNA Methylation of CC Chemokine Receptor Signature
The gene symbols of ten CC chemokine receptors (CCR1 to CCR10) were input for prognostic analysis in SurvivalMeth. As shown in Figure 9A-C, the significant expression patterns were found in CCR1/2/3/5/7/8/9 between low- and high-risk groups (Figure 9A). And the heatmap showed that the DNA methylation level of CCR3 was the highest, while CCR10 was the lowest (Figure 9B). However, no statistically significant association was found between the high- and low-risk groups (P=0.1664281723) (Figure 9C).
Genetic Alteration, Co-Expression, Protein/Gene Interaction Analyses of CC Chemokine Receptors in Patients with LUAD
A comprehensive analysis of the molecular characteristics of CC chemokine receptors was further performed. First, the genetic alterations of these CC chemokine receptors were analyzed with cBioPortal. As a result, CCR1, CCR2, CCR3, CCR4, CCR5, CCR6, CCR7, CCR8, CCR9, and CCR10 were altered in 3%, 5%, 4%, 4%, 4%, 5%, 2.9%, 5%, 3%, and 0.6% of the LUAD samples, respectively (Figure 10A). Enhanced mRNA expression of CC chemokine receptors, as the most common change, was found in these samples. Then, the potential co-expression of CC chemokine receptors was explored, as displayed in Figure 10B, there was a moderate to strong correlation among the expression of CCR4, CCR5, CCR6, CCR7, and CCR8.
Moreover, PPI network analysis of CC chemokine receptors was conducted with STRING.
As expected, 10 nodes and 36 edges were obtained in the PPI network (Figure 10C). Results of GeneMANIA also revealed that the functions of these CC chemokine receptors were primarily related to cytokine receptor activity, G-protein coupled chemoattractant receptor activity, chemokine-mediated signaling pathway, cytokine receptor activity, and G-protein coupled receptor activity, et al (Figure 10D).
Moreover, the top 50 genes with the highest frequency associated with CC chemokine receptors were got from cBioPortal. The data indicated that CCR1, CCR2, CCR3, CCR4, CCR5, CCR6, CCR7, CCR8, CCR9, CCR10, SEMA3E, CD33, TP53, PCDH15, TTN, ZNF804A, CPS1, KRAS, FLG, RYR2, ZFHX4, GRIN2A, COL15A1, DISP1, IRX1, RRM2, MBD1, ALK, ST6GAL2, ZNF292, TRRAP, MADD, AKAP11, COL6A6, DCDC1, PRDM9, EYS, ADAMTS3, PCNT, TAF1L, ACTN2, PKHD1L1, COL11A1, FMN2, NTRK3, OBSCN, LRP1B, PLXNC1, PPEF2, SKIV2L, EXO1, AGMO, TRPV2, SPHKAP, PDCD11, ZNF479, IQSEC3, PKHD1, KLHL14, and PTPRT were primarily connected with the modulation and function of CC chemokine receptors in LUAD (Figure 10E).
Functional Enrichment Analysis of CC Chemokine Receptors in Patients with LUAD
The functions of CC chemokine receptors and their neighboring genes were analyzed using DAVID 6.8 and Metascape. The significant highly enriched GO items (P<0.05) are shown in Figure 11A-B using DAVID 6.8.
In the biological process (BP) category, the top 10 highly enriched GO items, including chemokine-mediated signaling pathway, chemotaxis, positive regulation of cytosolic calcium ion concentration, dendritic cell chemotaxis, immune response, cellular defense response, cell adhesion, collagen catabolic process, cellular calcium ion homeostasis, and sarcomere organization, were associated with the tumorigenesis and progression of LUAD (Figure 11A). In the cellular component (CC) category, integral component of plasma membrane, cell surface, external side of plasma membrane, Z disc, integral component of membrane, plasma membrane, collagen trimer were the most significantly highly enriched items (Figure 11A). In the molecular function (MF) category, the CC chemokine receptors and their neighboring genes were mainly enriched in chemokine receptor activity and related binding activities (Figure 11A). And as expected in KEGG pathway analysis (Figure 11B), hsa04062 (chemokine signaling pathway), hsa04060 (cytokine-cytokine receptor interaction), hsa05203 (viral carcinogenesis), hsa05223 (non-small cell lung cancer), hsa05230 (central carbon metabolism in cancer) were significantly related to the LUAD formation and progression. Also, the results of the functional enrichment analysis were confirmed using Metascape (Supplementary Figure 2-4).
Transcription Factor Targets, Kinase Targets, and miRNA Targets of CC Chemokine Receptors in Patients with LUAD
The possible transcription factor targets, kinase targets, and miRNA targets of the CC chemokine receptors were explored using the TRRUST and LinkedOmics databases. CCR1, CCR2, CCR3, CCR4, CCR5, CCR6, CCR7, CCR8, CCR9, and CCR10 were included in TRRUST. We found that four transcription factors (NR3C2, KLF2, RELA, and NFKB1) were associated with the regulation of CC chemokine receptors (Table 3). RELA and NFKB1 were the key transcription factors for CCR2, CCR3, CCR5, and CCR7. NR3C2 was the key transcription factor for CCR5 and CCR6. KLF2 was the key transcription factor for CCR5 and CCR7.
 |
Table 3 Key Regulated Factors of CC Chemokine Receptors in LUAD (TRRUST) |
In addition, the top two kinase targets of CC chemokine receptors were identified from LinkedOmics (Supplementary Table 2). Among these, only one kinase target, TNK2, was identified in the CCR5, SYK and LCK were the top two kinase targets in the CCR9, and components of the CCR3 kinase-target network were mainly associated with CDK1 and CHEK1.
The top two miRNA targets of CC chemokine receptors were also identified in LUAD, which is presented in Supplementary Table 3. Among them, no significant miRNA targets were found in CCR6 and CCR8. And there was only one significant miRNA target in the CCR2 (MIR-208), CCR4 (MIR-184), CCR5 (MIR-517), and CCR7 (MIR-525) in LUAD. And two miRNA targets were found in CCR1 (MIR-208 and MIR-371), CCR3 (MIR-202 and MIR-205), CCR9 (MIR-191 and MIR-380-5P), and CCR10 (MIR-323 and MIR-522).
Immune Cell Infiltration of CC Chemokine Receptors in Patients with LUAD
CC chemokine receptors have been involved in cancer-related inflammation and the infiltration of immune cells, thus affecting the clinical outcome of LUAD patients. Therefore, the TIMER database was used to provide a comprehensive analysis of the correlation between CC chemokine receptors and immune cell infiltration. The results are presented in Figure 12. Except for CCR10, nine CC chemokine receptors expression (including CCR1, CCR2, CCR3, CCR4, CCR5, CCR6, CCR7, CCR8, and CCR9) positively correlated with infiltration of four immune cell types (CD8+ T cells, macrophages, neutrophils, and dendritic cells; all P < 0.05). Except for CCR3, the other nine CC chemokine receptors expression positively correlated with the infiltration of B cells, CD8+ T cells, and CD4+ T cells (all P < 0.05).
We also compared the tumor infiltration levels among LUAD with different somatic copy number alterations for the CC chemokine receptors, these results are displayed in Supplementary Figure 5. Moreover, the Cox proportional hazard model was applied for CC chemokine receptors and six tumor-infiltrating immune cells in LUAD. As shown in Table 4, CCR1 (P =0.022), CCR2 (P =0.001), CCR10 (P =0.046), B_cell (P = 0.011), and CD4_Tcell (P = 0.034) were found to be significantly related to the clinical outcome of LUAD patients.
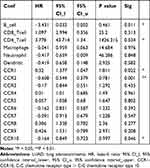 |
Table 4 The Cox Proportional Hazard Model of CC Chemokine Receptors and Six Tumor-Infiltrating Immune Cells in LUAD (TIMER) |
Discussion
As the largest subdivision of the chemokine superfamily, there are ten CC chemokine receptor subtypes, they are all receptors with G protein-coupled, seven-transmembrane region and mainly induce signal transduction via Gi proteins.21 In humans, CCR1, CCR2, CCR3, CCR4, CCR5, CCR8, CCR9, and CCR10 are encoded by a cluster of genes on chromosome 3p21. And CCR6 is encoded by the gene located on the long arm of Chromosome 6 (6q27), and CCR7 is encoded by the gene on chromosome 17q21.2. CCRs are highly differentially expressed on most leukocyte subsets, and mediate different types of immune responses.47 They are closely related to many autoimmune and inflammation-related conditions, such as multiple sclerosis, osteoarthritis, and inflammatory bowel disease.20,48–50 Numerous studies showed that CC chemokine receptors were critical mediators of chronic inflammatory responses, and they were involved in leukocyte recruitment, angiogenesis, tumor growth, proliferation, and metastasis.51–53 CC chemokine receptors were also considered as potential pharmacological targets.21 Some CC chemokine receptor antagonists/inhibitors displayed potent anti-tumor activity in the preclinical models and clinical trials.51,54 For example, CCR1 antagonist BL5923 showed antimetastatic metastatic ability in a mouse model of colon cancer liver metastasis.55 And Mogamulizumab, as a first-in-class CCR4 inhibitor, was considered a promising pharmacotherapy for T-cell lymphomas.56,57 However, the prognostic values and biological functions of CC chemokine receptors in LUAD have not been well characterized.
In this study, we first investigated the expression of CC chemokine receptors in the lung adenocarcinoma and found there were six genes differentially expressed in LUAD compared with the normal tissues (upregulation of CCR3, CCR6, CCR7, and CCR8; downregulation of CCR1 and CCR10). Besides, the relationship between CC chemokine receptors and the pathological stage of LUAD patients was assessed, we found the expression of CCR2, CCR4, CCR6, and CCR 7 decreased as the adenocarcinomas progressed. Furthermore, we investigated the prognostic values of CC chemokine receptors in LUAD cases. The results showed that high expression of CCR2, CCR3, CCR4, and CCR10, while, the low expression of CCR5 and CCR6, was significantly associated with worse OS. And the high expression of CCR3 and CCR4 also indicated inferior FP in LUAD cases. But there was no significant difference between the CC chemokine receptors expression and PPS/DFS in LUAD patients. In addition, the prognostic value of CC chemokine receptors signature was also assessed in three independent cohorts, the LUAD patients in the high-risk group had unfavorable OS and RFS when compared that in the low-risk group. And for the prognostic value of the DNA methylation of CC chemokine receptors, 1 CpG of CCR2, 2 CpGs of CCR3, 1 CpG of CCR4, 3 CpGs of CCR6, 3 CpGs of CCR7, 1 CpG of CCR8, and 3 CpGs of CCR9 were found to be significantly associated with prognosis in LUAD patients. However, the DNA methylation signature analysis showed no statistically significant association between the high- and low-risk groups.
Then, the molecular characteristics of CC chemokine receptors were also explored in LUAD. Since the occurrence and development of LUAD are multi-step and multi-factors, and genetic alteration was also involved in this process, we explored the genetic alterations, and alterations of CCR1, CCR2, CCR3, CCR4, CCR5, CCR6, CCR7, CCR8, CCR9, and CCR10 were observed in 3%, 5%, 4%, 4%, 4%, 5%, 2.9%, 5%, 3%, and 0.6% of the LUAD samples. And we also found a moderate to strong correlation among the expression of CCR4, CCR5, CCR6, CCR7, and CCR8, suggesting that these CC chemokine receptors might play a synergistic role in the tumorigenesis and progression of LUAD.
Then, the function of CC chemokine receptors was explored with GO and KEGG pathway enrichment analyses. As expected, the functions of CCR1 to CCR10 genes are primarily related to the chemokine-mediated signaling pathway, immune response, cytokine-cytokine receptor interaction. We also sought to characterize the transcription factor and kinase targets of CC chemokine receptors, and found that NR3C2, KLF2, RELA, and NFKB1 may be key transcription factors in the regulation of CC chemokine receptors. And the top two kinase and miRNA targets of CC chemokine receptors were also identified. Previous researches have proved that these signaling pathways above played key roles in tumor proliferation, migration, invasion, and metastasis.52,58 And they could also affect treatment effect and clinical outcome by mediating the migration and localization of immune cells, the immune responses, and the balance between immunity and tolerance.59–61 These data suggested that CC chemokine receptors could be used as potential immunotherapy targets.
Moreover, the correlation between CC chemokine receptors and immune cell infiltration of LUAD was also assessed. Eight CC chemokine receptors, including CCR1, CCR2, CCR4, CCR5, CCR6, CCR7, CCR8, and CCR9, were significantly related to all six immune cell types, including B cells, CD8+ T cells, CD4+ T cells, macrophages, neutrophils, and dendritic cells. And there was a positive significant correlation between CCR10 expression and the infiltration of B cells and CD4+ T cells, there was a positive significant correlation between the CCR3 expression and the infiltration of macrophages, neutrophils, and dendritic cells. Furthermore, the tumor infiltration levels among LUAD with different somatic copy number alterations for the CC chemokine receptors were also compared. The Cox proportional hazard model further demonstrated that CCR1, CCR2, CCR10, B_cell, and CD4_Tcell were significantly related to the clinical outcome of LUAD patients. These results indicated that CC chemokine receptors were involved in LUAD progression by affecting immune status.
Recent researches provided possible mechanisms that explained why CC chemokine receptors could act as valuable tumor-markers and immunotherapeutic targets. CC chemokine receptors are involved in tumorigenesis and cancer development.
The ectopic expression of CC chemokine receptors was found in various solid and hematological tumors, including lung cancer, breast cancer, gastric cancer, T- and B-cell lymphomas.62–66 And their altered expression was correlated with clinical outcomes. For example, CCR5 and CCR9 expression were found to be related to lymph node metastasis, clinical stage in head and neck squamous cell carcinomas, and lung adenocarcinoma, respectively.67,68 And CCR7 was related to the development of lymph node metastasis and might be a potential prediction of prognosis in lung cancer.69,70 Furthermore, CC chemokine receptors, activated by binding to cytokines of the CC chemokine family, are closely associated with immune and inflammation response.51,71,72 And the chemokine system plays a key role in the initiation of naive T cells, cell fate determination such as effector and memory cell differentiation, and regulatory T cell functions, and could position cells for host defense and immunity.71 Recent studies in vivo and in vitro also demonstrated important roles for CC chemokine receptors in cancer processes. They exert important roles in apoptosis, proliferation, invasion, migration, and metastasis, and participate in the remodeling of the tumor microenvironment by affecting leukocyte recruitment and activation, angiogenesis, and lymphangiogenesis. For instance, among these for CC chemokine receptors, CCR1 inhibition could reduce tumor growth and metastasis by targeting myeloid cells.55 Activation of the CCL2/CCR2 axis could exert pro-tumoral activities for the increased recruitment of monocytes with pro-tumorigenic and pro-metastatic activities.51 CCR6, combined with its ligand CCL20, could recruit immune cells to the tumor niche, including tumor-associated macrophages (TAMs), T helper 17 (Th17), and regulatory T cells (Tregs).73–76 And CCR10, activated by CCL27 and CCL28, showed an effect on tumor vascularization by recruiting anti-cancer tumor-infiltrating lymphocytes (TILs) and regulatory T cells (Tregs).72
There are some limitations in our study. The biases caused by the confounders might exist since the data for analysis were from multiple online bioinformatics resources. And our results also needed to be validated, researches in vitro and in vivo are required to further explore the exact mechanisms of CC chemokine receptors in the progression of lung adenocarcinoma.
In conclusion, this work provided evidence of the values of CC chemokine receptors as clinical biomarkers and therapeutic targets in LUAD. We hope the results could afford some new inspirations for immunotherapeutic drug development, provide some assistance to the clinicians in the selection of optimal drugs for LUAD patients, and identify the tumor markers that have more accurate prognostic prediction ability in LUAD.
Data Sharing Statement
All data generated or analyzed during this study are included in this article and its supplementary information files.
Acknowledgments
We would like to forward our deepest gratitude to the editors and the anonymous referees for the effort they have invested in reviewing and critiquing our study.
Disclosure
The authors declare that there is no conflict of interest.
References
1. Ferlay J, Colombet M, Soerjomataram I, et al. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int j Cancer. 2019;144(8):1941–1953. doi:10.1002/ijc.31937
2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7–30. doi:10.3322/caac.21590
3. Shi J, Hua X, Zhu B, et al. Somatic genomics and clinical features of lung adenocarcinoma: a retrospective study. PLoS Med. 2016;13(12):e1002162. doi:10.1371/journal.pmed.1002162
4. Liang Y, Wakelee HA. Adjuvant chemotherapy of completely resected early stage non-small cell lung cancer (NSCLC). Transl Lung Cancer Res. 2013;2(5):403–410. doi:10.3978/j.issn.2218-6751.2013.07.01
5. Blandin Knight S, Crosbie PA, Balata H, Chudziak J, Hussell T, Dive C. Progress and prospects of early detection in lung cancer. Open Biol. 2017;7(9). doi:10.1098/rsob.170070
6. Heigener DF, Reck M, Gatzemeier U. [Non-small cell lung cancer - diagnostics and stage-adapted therapy]. Medizinische Klinik (Munich, Germany: 1983). 2007;102(12):
7. Carter BW, Godoy MC, Wu CC, Erasmus JJ, Truong MT. Current controversies in lung cancer staging. J Thorac Imaging. 2016;31(4):201–214. doi:10.1097/rti.0000000000000213
8. Carter BW, Lichtenberger JP
9. Trzaskowski B, Latek D, Yuan S, Ghoshdastider U, Debinski A, Filipek S. Action of molecular switches in GPCRs–theoretical and experimental studies. Curr Med Chem. 2012;19(8):1090–1109. doi:10.2174/092986712799320556
10. Chemokine/chemokine receptor nomenclature. Cytokine. 2003;21(1):48–49. doi:10.1016/s1043-4666(02)00493-3
11. Chemokine/chemokine receptor nomenclature. J Leukoc Biol. 2001;70(3):465–466.
12. Rodríguez-Fernández JL, Criado-García O. The Chemokine Receptor CCR7 uses distinct signaling modules with biased functionality to regulate dendritic cells. Front Immunol. 2020;11:528. doi:10.3389/fimmu.2020.00528
13. Roubeix C, Dominguez E, Raoul W, et al. Mo-derived perivascular macrophage recruitment protects against endothelial cell death in retinal vein occlusion. J Neuroinflammation. 2019;16(1):157. doi:10.1186/s12974-019-1547-8
14. Wang T, Zhou Q, Zeng H, et al. CCR8 blockade primes anti-tumor immunity through intratumoral regulatory T cells destabilization in muscle-invasive bladder cancer. Cancer Immunol Immunother. 2020;69(9):1855–1867. doi:10.1007/s00262-020-02583-y
15. Kadomoto S, Izumi K, Mizokami A. The CCL20-CCR6 axis in cancer progression. Int J Mol Sci. 2020;21. doi:10.3390/ijms21155186.
16. Lokeshwar BL, Kallifatidis G, Hoy JJ. Atypical chemokine receptors in tumor cell growth and metastasis. Adv Cancer Res. 2020;145:1–27. doi:10.1016/bs.acr.2019.12.002
17. Groblewska M, Litman-Zawadzka A, Mroczko B. The role of selected chemokines and their receptors in the development of gliomas. Int J Mol Sci. 2020;21(10):3704. doi:10.3390/ijms21103704
18. White GE, Iqbal AJ, Greaves DR. CC chemokine receptors and chronic inflammation–therapeutic opportunities and pharmacological challenges. Pharmacol Rev. 2013;65(1):47–89. doi:10.1124/pr.111.005074
19. Nishimura M, Kuboi Y, Muramoto K, Kawano T, Imai T. Chemokines as novel therapeutic targets for inflammatory bowel disease. Ann N Y Acad Sci. 2009;1173(1):350–356. doi:10.1111/j.1749-6632.2009.04738.x
20. Trivedi PJ, Adams DH. Chemokines and chemokine receptors as therapeutic targets in inflammatory bowel disease; pitfalls and promise. J Crohn’s & Colitis. 2018;12(suppl_2):S641–s652. doi:10.1093/ecco-jcc/jjx145
21. White G, Iqbal A, Greaves DJPR. CC chemokine receptors and chronic inflammation therapeutic opportunities and pharmacological challenges. 2013;65:47–89.
22. Okayama H, Kohno T, Ishii Y, et al. Identification of genes upregulated in ALK-positive and EGFR/KRAS/ALK-negative lung adenocarcinomas. Cancer Res. 2012;72(1):100–111. doi:10.1158/0008-5472.Can-11-1403
23. Chanrion M, Negre V, Fontaine H, et al. A gene expression signature that can predict the recurrence of tamoxifen-treated primary breast cancer. Clin Cancer Res. 2008;14(6):1744–1752. doi:10.1158/1078-0432.Ccr-07-1833
24. Laurent C, Valet F, Planque N, et al. High PTP4A3 phosphatase expression correlates with metastatic risk in uveal melanoma patients. Cancer Res. 2011;71(3):666–674. doi:10.1158/0008-5472.Can-10-0605
25. Smith JJ, Deane NG, Wu F, et al. Experimentally derived metastasis gene expression profile predicts recurrence and death in patients with colon cancer. Gastroenterology. 2010;138(3):958–968. doi:10.1053/j.gastro.2009.11.005
26. Tomida S, Takeuchi T, Shimada Y, et al. Relapse-related molecular signature in lung adenocarcinomas identifies patients with dismal prognosis. J Clin Oncol. 2009;27(17):2793–2799. doi:10.1200/jco.2008.19.7053
27. Rhodes DR, Yu J, Shanker K, et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia (New York, NY). 2004;6(1):1–6. doi:10.1016/s1476-5586(04)80047-2
28. Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia (New York, NY). 2017;19(8):649–658. doi:10.1016/j.neo.2017.05.002
29. Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res. 2017;45(W1):W98–w102. doi:10.1093/nar/gkx247
30. Nagy Á, Lánczky A, Menyhárt O, Győrffy B. Validation of miRNA prognostic power in hepatocellular carcinoma using expression data of independent datasets. Sci Rep. 2018;8(1):9227. doi:10.1038/s41598-018-27521-y
31. Aguirre-Gamboa R, Gomez-Rueda H, Martínez-Ledesma E, et al. SurvExpress: an online biomarker validation tool and database for cancer gene expression data using survival analysis. PLoS One. 2013;8(9):e74250. doi:10.1371/journal.pone.0074250
32. Modhukur V, Iljasenko T, Metsalu T, Lokk K, Laisk-Podar T, Vilo J. MethSurv: a web tool to perform multivariable survival analysis using DNA methylation data. Epigenomics. 2018;10(3):277–288. doi:10.2217/epi-2017-0118
33. Zhang C, Zhao N, Zhang X, et al. SurvivalMeth: a web server to investigate the effect of DNA methylation-related functional elements on prognosis. Brief Bioinform. 2020. doi:10.1093/bib/bbaa162
34. Cerami E, Gao J, Dogrusoz U, et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2(5):401–404. doi:10.1158/2159-8290.Cd-12-0095
35. Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. doi:10.1126/scisignal.2004088
36. Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–d613. doi:10.1093/nar/gky1131
37. Szklarczyk D, Morris JH, Cook H, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45(D1):D362–d368. doi:10.1093/nar/gkw937
38. Warde-Farley D, Donaldson SL, Comes O, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38(suppl_2):W214–W220. doi:10.1093/nar/gkq537
39. da Huang W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37(1):1–13. doi:10.1093/nar/gkn923
40. da Huang W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4(1):44–57. doi:10.1038/nprot.2008.211
41. Zhou Y, Zhou B, Pache L, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10(1):1523. doi:10.1038/s41467-019-09234-6
42. Han H, Cho JW, Lee S, et al. TRRUST v2: an expanded reference database of human and mouse transcriptional regulatory interactions. Nucleic Acids Res. 2018;46(D1):D380–d386. doi:10.1093/nar/gkx1013
43. Han H, Shim H, Shin D, et al. TRRUST: a reference database of human transcriptional regulatory interactions. Sci Rep. 2015;5:11432. doi:10.1038/srep11432
44. Vasaikar SV, Straub P, Wang J, Zhang B. LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res. 2018;46(D1):D956–d963. doi:10.1093/nar/gkx1090
45. Li B, Severson E, Pignon JC, et al. Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biol. 2016;17(1):174. doi:10.1186/s13059-016-1028-7
46. Li T, Fan J, Wang B, et al. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77(21):e108–e110. doi:10.1158/0008-5472.Can-17-0307
47. Murphy PM. CC Chemokine Receptors. In: Enna SJ, Bylund DB, editors. xPharm: The Comprehensive Pharmacology Reference. Elsevier; 2007:1–2.
48. Fantuzzi L, Tagliamonte M, Gauzzi MC, Lopalco L. Dual CCR5/CCR2 targeting: opportunities for the cure of complex disorders. Cell Mol Life Sci. 2019;76(24):4869–4886. doi:10.1007/s00018-019-03255-6
49. Raghu H, Lepus CM, Wang Q, et al. CCL2/CCR2, but not CCL5/CCR5, mediates monocyte recruitment, inflammation and cartilage destruction in osteoarthritis. Ann Rheum Dis. 2017;76(5):914–922. doi:10.1136/annrheumdis-2016-210426
50. Scheu S, Ali S, Ruland C, Arolt V, Alferink J. The C-C Chemokines CCL17 and CCL22 and their receptor CCR4 in CNS autoimmunity. Int J Mol Sci. 2017;18(11). doi:10.3390/ijms18112306
51. Mollica Poeta V, Massara M, Capucetti A, Bonecchi R. Chemokines and chemokine receptors: new targets for cancer immunotherapy. Mini Rev. 2019;10(379). doi:10.3389/fimmu.2019.00379
52. Rizeq B, Malki MI. The role of CCL21/CCR7 chemokine axis in breast cancer progression. Cancers. 2020;12(4):1036. doi:10.3390/cancers12041036
53. Mollica Poeta V, Massara M, Capucetti A, Bonecchi R. Chemokines and chemokine receptors: new targets for cancer immunotherapy. Front Immunol. 2019;10:379. doi:10.3389/fimmu.2019.00379
54. Vandercappellen J, Van Damme J, Struyf S. The role of CXC chemokines and their receptors in cancer. Cancer Lett. 2008;267(2):226–244. doi:10.1016/j.canlet.2008.04.050
55. Kitamura T, Fujishita T, Loetscher P, et al. Inactivation of chemokine (C-C motif) receptor 1 (CCR1) suppresses colon cancer liver metastasis by blocking accumulation of immature myeloid cells in a mouse model. Proc Natl Acad Sci U S A. 2010;107(29):13063–13068. doi:10.1073/pnas.1002372107
56. Moore DC, Elmes JB, Shibu PA, Larck C, Park SI. Mogamulizumab: an Anti-CC chemokine receptor 4 antibody for T-Cell lymphomas. Ann Pharmacother. 2020;54(4):371–379. doi:10.1177/1060028019884863
57. Makita S, Tobinai K. Mogamulizumab for the treatment of T-cell lymphoma. Expert Opin Biol Ther. 2017;17(9):1145–1153. doi:10.1080/14712598.2017.1347634
58. Bikfalvi A, Billottet C. The CC and CXC chemokines: major regulators of tumor progression and the tumor microenvironment. Am J Physiol Cell Physiol. 2020;318(3):C542–c554. doi:10.1152/ajpcell.00378.2019
59. Förster R, Davalos-Misslitz AC, Rot A. CCR7 and its ligands: balancing immunity and tolerance. Nat Rev Immunol. 2008;8(5):362–371. doi:10.1038/nri2297
60. Kim J, Yang YL, Jang YS. Human β-defensin 2 is involved in CCR2-mediated Nod2 signal transduction, leading to activation of the innate immune response in macrophages. Immunobiology. 2019;224(4):502–510. doi:10.1016/j.imbio.2019.05.004
61. Lee AYS, The KH. CCR6-CCL20 axis in humoral immunity and T-B cell immunobiology. Immunobiology. 2019;224(3):449–454. doi:10.1016/j.imbio.2019.01.005
62. Lee JH, Cho YS, Lee JY, et al. The chemokine receptor CCR4 is expressed and associated with a poor prognosis in patients with gastric cancer. Ann Surg. 2009;249(6):933–941. doi:10.1097/SLA.0b013e3181a77ccc
63. Ishida T, Utsunomiya A, Iida S, et al. Clinical significance of CCR4 expression in adult T-cell leukemia/lymphoma: its close association with skin involvement and unfavorable outcome. Clin Cancer Res. 2003;9(10 Pt 1):3625–3634.
64. Li YL, Shi ZH, Wang X, Gu KS, Zhai ZM. Prognostic significance of monocyte chemoattractant protein-1 and CC chemokine receptor 2 in diffuse large B cell lymphoma. Ann Hematol. 2019;98(2):413–422. doi:10.1007/s00277-018-3522-3
65. Labovsky V, Martinez LM, Davies KM, et al. Prognostic significance of TRAIL-R3 and CCR-2 expression in tumor epithelial cells of patients with early breast cancer. BMC Cancer. 2017;17(1):280. doi:10.1186/s12885-017-3259-8
66. Itakura M, Terashima Y, Shingyoji M, et al. High CC chemokine receptor 7 expression improves postoperative prognosis of lung adenocarcinoma patients. Br J Cancer. 2013;109(5):1100–1108. doi:10.1038/bjc.2013.440
67. González-Arriagada WA, Lozano-Burgos C, Zúñiga-Moreta R, González-Díaz P, Coletta RD. Clinicopathological significance of chemokine receptor (CCR1, CCR3, CCR4, CCR5, CCR7 and CXCR4) expression in head and neck squamous cell carcinomas. J Oral Pathol Med. 2018;47(8):755–763. doi:10.1111/jop.12736
68. Zhong Y, Jiang L, Lin H, et al. Expression of CC chemokine receptor 9 predicts poor prognosis in patients with lung adenocarcinoma. Diagn Pathol. 2015;10:101. doi:10.1186/s13000-015-0341-x
69. Yu J, Tao S, Hu P, et al. CCR7 promote lymph node metastasis via regulating VEGF-C/D-R3 pathway in lung adenocarcinoma. J Cancer. 2017;8(11):2060–2068. doi:10.7150/jca.19069
70. Takanami I. Overexpression of CCR7 mRNA in nonsmall cell lung cancer: correlation with lymph node metastasis. Int j Cancer. 2003;105(2):186–189. doi:10.1002/ijc.11063
71. Griffith JW, Sokol CL, Luster AD. Chemokines and chemokine receptors: positioning cells for host defense and immunity. Annu Rev Immunol. 2014;32:659–702. doi:10.1146/annurev-immunol-032713-120145
72. Korbecki J, Grochans S, Gutowska I, Barczak K, Baranowska-Bosiacka I. CC chemokines in a tumor: a review of pro-cancer and anti-cancer properties of receptors CCR5, CCR6, CCR7, CCR8, CCR9, and CCR10 Ligands. Int J Mol Sci. 2020;21. doi:10.3390/ijms21207619.
73. Chen KJ, Lin SZ, Zhou L, et al. Selective recruitment of regulatory T cell through CCR6-CCL20 in hepatocellular carcinoma fosters tumor progression and predicts poor prognosis. PLoS One. 2011;6(9):e24671. doi:10.1371/journal.pone.0024671
74. Yu Q, Lou XM, He Y. Preferential recruitment of Th17 cells to cervical cancer via CCR6-CCL20 pathway. PLoS One. 2015;10(3):e0120855. doi:10.1371/journal.pone.0120855
75. Zhang CY, Qi Y, Li XN, et al. The role of CCL20/CCR6 axis in recruiting Treg cells to tumor sites of NSCLC patients. Biomedicine & Pharmacotherapy = Biomedecine & Pharmacotherapie. 2015;69:242–248. doi:10.1016/j.biopha.2014.12.008
76. Nandi B, Shapiro M, Samur MK, et al. Stromal CCR6 drives tumor growth in a murine transplantable colon cancer through recruitment of tumor-promoting macrophages. Oncoimmunology. 2016;5(8):e1189052. doi:10.1080/2162402x.2016.1189052
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.