Back to Journals » OncoTargets and Therapy » Volume 13
CBX6 Promotes HCC Metastasis Via Transcription Factors Snail/Zeb1-Mediated EMT Mechanism
Authors Wang J , He H, Jiang Q, Wang Y, Jia S
Received 8 April 2020
Accepted for publication 22 October 2020
Published 4 December 2020 Volume 2020:13 Pages 12489—12500
DOI https://doi.org/10.2147/OTT.S257363
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr XuYu Yang
Jiamu Wang, Hui He, Qiucheng Jiang, Yu Wang, Shuzhao Jia
Liaocheng Peoples’ Hospital, Liaocheng 252000, People’s Republic of China
Correspondence: Shuzhao Jia
Liaocheng Peoples’ Hospital, Liaocheng 252000, People’s Republic of China
Tel +86 17852420715
Email [email protected]
Purpose: Hepatocellular carcinoma (HCC) is the most common malignant tumor worldwide with high morbidity and mortality rates. We aimed to examine the expression of chromobox 6 (CBX6) in HCC and analyze its correlation with clinicopathological features of HCC patients. Moreover, the role of CBX6 in the HCC cell proliferation, invasion and metastasis and the potential mechanism underlying HCC metastasis were also investigated.
Methods: We used quantitative polymerase chain reaction (qRT-PCR) and Western blot to evaluate the expression levels of CBX6 in HCC cell lines. Furthermore, the expression of CBX6 in HCC and the adjacent non-tumor tissues was assessed by immunohistochemistry (IHC). Cell proliferation was evaluated using MTT assay, cell migration and invasion were measured using wound healing and transwell assays. Finally, we detected the expression of target proteins in HCC cell lines transfected with CBX6 overexpression plasmid or CBX6 shRNA plasmid by Western blot.
Results: We found that the expression of CBX6 was increased in 280 cases of HCC tissues compared that in adjacent non-tumor tissues. HCC patients with high CBX6 expression had a higher tendency to have high growth rate, strong invasion ability, high clinical stage and poor tumor differentiation. Functional study demonstrated that the upregulation of CBX6 promotes proliferation, migration and invasion of HCC cells while silencing CBX6 in HCC cells significantly inhibited cell proliferation, migration and invasion. Furthermore, CBX6 could accelerate the EMT process in HCC cells by upregulating the expression of snail and zeb1.
Conclusion: CBX6 played an important role in the process of tumorigenesis and progression in HCC and enhanced the invasion and metastasis ability of HCC cells through regulating transcription factors snail/zeb1-mediated EMT mechanism, which indicated that the protein could serve as a novel therapeutic target for the treatment of HCC.
Keywords: CBX6, HCC, metastasis, EMT
Introduction
Hepatocellular carcinoma (HCC) is a common malignant tumor of the digestive system, and its morbidity and mortality rank sixth and fourth, respectively, in the world.1 Although great advances have been made in clinical and experimental cancer treatment, the overall prognosis of HCC patients is poor due to a high rate of tumor recurrence and metastasis after surgery.2,3 Previous studies have shown that the progression of HCC might be a multistep process involving a multifactorial etiology and multiple genes alternation, but the specific molecular mechanism underlying HCC metastasis is still unclear. Therefore, a better understanding of the underlying molecular mechanism of HCC metastasis is of great significance for prognosis and targeted therapy of HCC.
Polycomb group proteins (PcG) are important epigenetic regulators that exist in the form of polymeric transcriptional repressor complex PRCs, and it can maintain transcriptional repression.4 The chromobox (CBX) protein is the core component of PRC1 and plays an important role in the transcriptional regulation of polycomb proteins.5 CBX6, a novel identified CBX family member, is a remarkably conserved protein that can read the crucial epigenetic marker, H3K27me3, written by PRC2. CBX6 is an essential regulator of embryonic stem cell identity, its knockdown via shRNA induces spontaneous differentiation of embryonic stem cells.6 Recent studies have reported that CBX6 expression is downregulated in glioblastoma multiforme (GBM) samples, and its overexpression in GBM cells leads to cell growth arrest.7 In addition, CBX6 is significantly downregulated in human breast cancer, it could inhibit cell proliferation and induced the cell cycle G0/G1 arrest in MCF-7 cells, and CXB6 overexpression reduced the breast cancer cell migration and invasion rates.8 Zheng et al represent the first evidence that CBX6 expression is frequently increased in human HCC tissues, high expression of CBX6 promotes cell proliferation and predicts poor prognosis of HCC patients.9 However, there have been no studies concerning the gene functions related to CBX6 in HCC metastasis.
In this study, we elucidated the expression CBX6 in HCC cell lines and a large cohort of clinical samples and further analysed the clinical significance of CBX6 in HCC patients. Moreover, we investigated the potential role of CBX6 in the HCC cell proliferation, migration and invasion, which may provide a novel therapeutic target for the treatment of HCC patients.
Materials and Methods
Cell Culture
The human HCC cell lines HepG2, Hep3B, SMMC-7721, Huh7, MHCC97-L, MHCC97-H, HCCLM3 and human immortalized hepatic cell L02 were purchased from Cell Bank of the Chinese Academy of Sciences (Shanghai, China). All cells were cultured in Dulbecco’s modified Eagle medium (DMEM, Hyclone, Logan, Utah, USA) with 10% fetal bovine serum (FBS, Gibco, Grand Island, NY, USA) at 37°C in a 5% CO2 air atmosphere. MHCC97-H and HCCLM3 cell lines had higher metastasis potential than the rest HCC cell lines.
Plasmid Construction and Cell Transfection
CBX6 overexpression vector plasmid (CBX6-pEX), the empty vector plasmid (vector) and the lentiviral vector with CBX6 shRNA plasmid (LV-CBX6 shRNA) and empty vector (LV-control) were purchased from GeneChem (Shanghai, China). The transfection efficiency was monitored by fluorescence microscopy, overexpression of CBX6 in HCC cells was performed by transfecting 2µg cDNA, the empty vector was used as a negative control. Suppression of CBX6 expression was performed by LV-CBX6 shRNA, the LV-control was used as a negative control. All transfection experiments were performed using Lipofectamine2000 (Invitrogen, Camarillo, CA, USA) according to the manufacturer’s instructions. The stable transfected cells were selected by 1 mg/mL of G418 for 2 weeks.
Tissue Samples
The patients whose tissues were used for this research provided informed consent, in accordance with the Declaration of Helsinki. Two hundred and eighty pairs of paraffin-embedded HCC tissue samples and adjacent non-tumor tissue samples (more than 5 cm from the edge of the tumor) were collected at the Liaocheng People’s Hospital, Shandong, China, between years 2012 and 2014. Among them, 96 cases with metastasis including 57 intra-hepatic metastasis and 39 extra-hepatic metastases. None of the patients had received radiotherapy, chemotherapy or targeted therapy before surgery. Tumor differentiation was assessed according to the World Health Organisation (WHO) classification criteria. All patients were staged according to the 7th edition of Union for International Cancer Control (UICC) Tumor-Node-Metastasis (TNM) staging system. This study was reviewed and approved by the Institutional Medical Ethics Committee of the Liaocheng People’s Hospital.
Quantitative Polymerase Chain Reaction (qRT-PCR)
Total RNA was extracted using TRizol reagent (Invitrogen, Camarillo, CA, USA), and reverse-transcribed complementary DNA was synthesized by PrimeScriptTM kit (TaKaRa, Dalian, China). cDNA was amplified using SYBR green-based qRT-PCR performed on the Bio-Rad CFX96 real-time PCR system. Conditions were set as follows: pre-denaturation at 95°Cfor 30 s, followed by 40 cycles of 95°C for 5 s, 55°C for 30 s and 72°C for 30 s. GAPDH was used as internal controls for mRNA quantification. The relative expression of CBX6 was calculated by the 2−ΔΔCt method.10 The sequences of CBX6 and GAPDH primers were as follows:
CBX6: 5ʹ-AGATGTCACCCTGCTCCAAT-3ʹ (sense);
5ʹ-AGCCACCTTCTCGAAATCCT-3ʹ (antisense);
GAPDH: 5ʹ-GCACCGTCAAGGCTGAGAAC-3ʹ (sense);
5ʹ-TGGTGAAGACGCCAGTGGA-3ʹ (antisense).
Tissue Microarray (TMA)
Two hundred and eighty HCC and adjacent normal tissues were paraffin-embedded, sectioned and stained with hematoxylin and eosin (HE) and representative areas of the paraffin blocks were marked by two experienced pathologists. Each tissue core was punched from the marked areas with a 1.5mm pore diameter, and re-embedded into a recipient paraffin array block.
Immunohistochemistry (IHC)
Paraffin-embedded, formalin-fixed HCC and adjacent non-tumor tissue sections were deparaffinized and rehydrated. The endogenous peroxidase activity was blocked with 3% hydrogen peroxide (H2O2) for 30 mins. The sections were pre-treated using pressure cooker heat mediated antigen retrieval with sodium citrate buffer (pH=6) for 30mins, and incubated overnight at 4°C with the rabbit polyclonal anti-CBX6 antibody (1:100 dilution, Thermo Fisher Scientific, Waltham, MA, USA). After rinsing, the sections were incubated with a secondary antibody (1:4000 dilution, Abcam, Cambridge, UK) for 1hr and stained with 3, 3-diaminobenzidine tetrahydrochloride (DAB). The section was counterstained with haematoxylin, and observed under a microscope. Negative controls were prepared by replacing the primary antibody with PBS, and the immunostaining-positive human tonsil carcinoma tissue was used as positive controls.
The staining result evaluation was made as described previously,11,12 the percent of positive cells was scored as: 0, none; 1, <1/100; 2, 1/100 to 1/10; 3, 1/10 to 1/3; 4, 1/3 to 2/3; and 5, >2/3, and staining intensity was scored as: 0, none; 1, weak; 2, intermediate; and 3, strong. Nucleus immunoreactivity for the CBX6 protein was scored by evaluating the sum of percentage and staining intensity scores, ranging from 0 to 8. Sections were scored by pathologists with no knowledge of the ligand-binding results or patient outcomes.
Western Blot
Total protein was harvested from HCC cells using RIPA lysis buffer, the BCA method was used to determine protein concentration. Forty micrograms of protein was loaded onto 8%-12% SDS-PAGE electrophoreses and transferred to polyvinylidene difluoride (PVDF) membranes. After blocking nonspecific binding by 2% Bovine Serum Albumin, the membranes were incubated overnight at 4 °C with primary antibody against CBX6 (1:1000, E-cadherin, 1:1000, rabbit anti- E-cadherin, Abcam, Cambridge, UK), β-catenin (1:2000, rabbit anti-β-catenin, Abcam, Cambridge, UK), N-cadherin (1:2000, rabbit anti-N-cadherin, Abcam, Cambridge, UK), Vimentin (1:2000, rabbit anti-Vimentin, Abcam, Cambridge, UK), snail (1:1000, rabbit anti-snail, Abcam, Cambridge, UK), zeb1 (1:1000, rabbit anti-zeb1, Abcam, Cambridge, UK), and GAPDH (1:4000, mouse anti-GAPDH, TransGene, Beijing, China). After washing with Tris-buffered saline with Tween20, the membranes were incubated with secondary antibody (1:4000, Abcam, Cambridge, UK) for 1 h at room temperature. The membranes were visualized using enhanced chemiluminescence (ECL) kit.
3-(4,5-Dimethylthiazol-2-Yl)-2,5-Diphenyltetrazolium Bromide (MTT) Assay
After transfection, 2×103 cells per well were seeded into 96-well plates. After 24 hrs, 10ul MTT (Amresco, Solon, OH, USA) was added and the cells were incubated at 37°C for 4 hrs. The supernatants were then removed, and 150μL DMSO was added in the dark. The numbers of cells per well were measured by the absorbance (490 nm) at the indicated time points. The absorbance at 490nm was measured for 1–5 days.
Cell Cycle Assay
Transfected cells were harvested after trypsinized, washed twice with ice-cold PBS. Then, cells were fixed with 75% cold ethanol overnight at 4°C. After washing twice with ice-cold PBS, fixed cells were centrifuged at 1000rpm for 5mins. The cells were resuspended in 500μL PBS containing 12.5μL propidium iodide (PI) (Sangon, Shanghai, China) and 10μL RNase (Beyotime, Shanghai, China), and incubated at 37°C in the dark for 30 mins. Finally, at least 10,000 cells per sample were subjected to flow cytometric analyses.
Wound Healing Assay
Transfected cells were seeded in a 6-well plate at 1×106 cells per well and cultured overnight. The cell layer was wounded using a sterile 200μL tip and washed twice with PBS, and then cultured again with serum-free medium for 48 hrs. The spread of wound closure was observed and photographed at different time points (0 and 48 hrs) under a microscope to assess the migration distance.
Transwell Migration and Invasion Assays
Cell migration and invasion ability were studied using 24-well transwell chambers with an 8-μm pore polycarbonate membrane insert. After transfection, 1×105 cells suspended in 200ul serum-free medium were loaded onto the upper compartment and 600μL medium with 15% FBS was added to the lower compartment. Invasion assay was performed using chamber coated with matrigel (Becton Dickinson, Bedford, MA, USA). The migratory and invasive cells that migrated to the complete medium in the lower compartment were stained with Giemsa (Solarbio, Beijing, China) after incubated for 12hrs for the migration assay and 24hrs for the invasion assay. Five visual fields were randomly chosen from each chamber and counted under a light microscope.
Statistical Analysis
All results were analyzed using SPSS 23.0 software. The data from at least three separate experiments were presented as mean ± standard deviation (SD). Comparisons between groups were analyzed using Student’s t-test. All experiments were repeated at least three times. Value of P<0.05 was considered statistically significant.
Results
The Expression of CBX6 in HCC Cell Lines
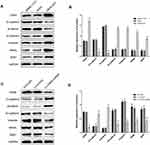
The mRNA and protein expression levels of CBX6 in HCC cell lines (HepG2, Hep3B, SMMC-7721, Huh7, MHCC97-L, MHCC97-H and HCCLM3) and human immortalized hepatic cell L02 were examined by qRT-PCR and Western blot. The results showed that CBX6 mRNA expression was markedly upregulated in the HCC cell lines compared with that in the immortalized hepatic cell line L02. Furthermore, CBX6 mRNA expression was higher in HCC cell lines with higher metastasis potential including MHCC97-H and HCCLM3 than that in the low metastasis potential cell lines (Figure 1A). Consistently, CBX6 showed the same elevated protein expression levels in all HCC cell lines, and it was decreased in the HCC cell lines with lower metastasis potential including HepG2, Hep3B, SMMC-7721, Huh7 and MHCC97-L compared to MHCC97-H and HCCLM3 cell lines (Figure 1B).
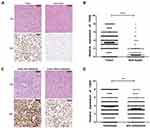
CBX6 Was Highly Expressed in HCC Tissues and Correlated with HCC Metastasis
The expression of CBX6 in 280 pairs of HCC was detected by IHC. The results showed that 253 out of 280 HCC cases (90.36%) were positively expressed, and 55 cases (19.64%) of adjacent normal tissues were positively expressed. The expression level of CBX6 was higher in 239 of 280 (85.36%) cases with HCC than the adjacent non-tumor tissues (Figure 2A). Strong positive staining was mainly observed in the nucleus of tumor cells, and weak staining is observed in the adjacent non-tumor liver cells (mean: 3.42±1.889 vs 0.41±0.957, P<0.001) (Figure 2B). Furthermore, CBX6 was positively expressed in 93 of 96 (96.88%) HCC cases with metastasis and 160 of 184 (86.96%) HCC cases without metastasis (Figure 2C). The expression level of CBX6 was significantly higher (P=0.009) in HCC cases with metastasis (mean: 3.85±1.926) than that in HCC cases without metastasis (mean: 3.22±1.843) (Figure 2D).
Relationship Between CBX6 Expression and Clinicopathological Features of HCC Patients
Clinicopathological analysis of 280 HCC patients showed that CBX6 expression was significantly associated with tumor diameter (P<0.001), vascular invasion (P=0.003), tumor differentiation (P<0.001), TNM stage (P=0.005), and metastasis (P=0.009). There were no statistical correlation between CBX6 expression and the rest of clinicopathological features including age, gender, serum AFP levels, HBsAg, liver cirrhosis, capsule invasion and tumor multiplicity (P>0.05). We also found that CBX6 expression in moderately and poorly differentiated groups was both higher than that in the well-differentiated group, but there was no significant difference in CBX6 expression between moderately differentiated group and poorly differentiated group (Table 1).
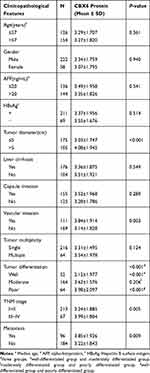 |
Table 1 Correlation Between CBX6 Expression and Clinicopathological Features of HCC Patients |
CBX6 Promotes the Proliferation and G1/S Phase Transition of HCC Cells
Our results showed that higher CBX6 expression was observed in HCCLM3 cells and lower CBX6 expression was exhibited in SMMC7721 cells. To investigate the involvement of CBX6 in the proliferation of HCC cells, we overexpressed CBX6 in SMMC-7721 cells using CBX6-pEX and knocked down CBX6 in HCCLM3 cells using specific shRNA. CBX6-pEX transfection significantly upregulated the expression of CBX6 in SMMC-7721 cells and LV-CBX6 shRNA markedly decreased the expression of CBX6 in HCCLM3 cells (Figure 3A and B). By MTT assay, we found that overexpressing of CBX6 significantly increased the proliferation ability of SMMC-7721 cells, whereas the cell proliferation ability was significantly decreased when knocking down CBX6 in HCCLM3 cells (P<0.05) (Figure 3C and D). To determine the pathway underlying growth promotion by CBX6, we detected the cell cycle distribution of HCCLM3 and SMMC-7721 cells by flow cytometry. Compared with the control cells, overexpression of CBX6 in SMMC-7721 cells resulted in a significant increase in the percentages of cells in the S phase, but a decrease in the percentages of cells in the G0/G1 phase, whereas silencing CBX6 in HCCLM3 cells showed the opposite effects (P<0.05) (Figure 3E and F). These results indicated that CBX6 promoted cell cycle progression of HCC cells, confirming that CBX6 promoted HCC cell proliferation.
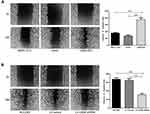
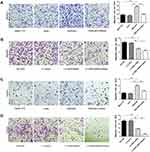
CBX6 Accelerated Cell Migration and Invasion in HCC Cells
Since CBX6 expression was correlated with HCC clinical invasiveness, we further evaluated the invasion and migration ability of HCC cells by wound healing assay and transwell migration and invasion assays. The wound-healing assay showed that overexpression of CBX6 remarkably promoted cell migration at the edge of exposed regions in SMMC-7721 cells compared to control cells, whereas migration distance of HCCLM3 cells transfected with LV-CBX6 shRNA was significantly shorter than control cells at 48hrs after wound time (P<0.05) (Figure 4). Consistently, transwell migration assay also revealed that CBX6 overexpression significantly increased the number of migrated cells, and silencing of CBX6 inhibited the motility behaviours of HCC cells (P<0.05). In addition, proliferation inhibitors could inhibit the motility behaviours of HCC cells (Figure 5A and B). Transwell invasion assay revealed that the invasive cells were significantly increased in SMMC-7721 cells transfected with CBX6-pEX, however proliferation inhibitors could reverse this phenomenon (P<0.05) (Figure 5C). In contrast, the number of invasive cells was decreased when CBX6 expression was knocked down in HCCLM3 cells, and proliferation inhibitors could enhance this effect (P<0.05) (Figure 5D). These data demonstrated that CBX6 could enhance HCC cell migration and invasive ability, but proliferation inhibitors could reverse this result.
CBX6 Induced EMT by Regulating Transcription Factors Snail/Zeb1
As EMT is one of the important mechanisms for cell migration and invasion, we next explore the effect of CBX6 on the progression of EMT. Epithelial markers (E-cadherin and β-catenin), mesenchymal markers (N-cadherin, and Vimentin) and EMT transcription factors (snail and zeb1) were estimated by Western blot in SMMC-7721 and HCCLM3 cells. The results showed that overexpression of CBX6 in SMMC-7721 cells increased the expression of N-cadherin, Vimentin, snail and zeb1, a reduction was noted in E-cadherin and β-catenin levels (Figure 6A and B). Additionally, knockdown of CBX6 in HCCLM3 cells upregulated the expression of E-cadherin and β-catenin, and the increase in the expression of N-cadherin, Vimentin, snail and zeb1 was abrogated upon CBX6 depletion (Figure 6C and D). These results indicated that CBX6 could regulate the expression of transcription factors snail and zeb1-mediated EMT mechanism to induce HCC invasion and metastasis.
Discussion
HCC is one of the most lethal malignant digestive system cancers worldwide with high morbidity and mortality rates.1 HCC has a poor prognosis, with nearly 600,000 deaths occurring each year worldwide.13,14 The high mortality of HCC patients mainly results from high recurrence and metastasis after surgery.15 However, the high metastasis rate of HCC has led to the performance of intense research intended to elucidate the detailed mechanisms underlying HCC development.
The PcG gene was firstly found in Drosophila with a high degree of conservation.16 PcG proteins, a family of master epigenetic regulators, regulate a variety of cellular physiological and pathological processes, such as maintenance of stemness, cell differentiation, regulation of cell cycle, cell senescence, X chromosome inactivation, genomic imprinting, etc.17,18 The deregulation and dysfunction of PcG proteins can mediate multiple biological functions of malignant tumors and lead to blocking or inappropriate activation of developmental pathways, enhancing cellular proliferation, inhibiting apoptosis, and increasing the cancer stem cell population.19–21 CBX protein, as the important member of the PcG proteins, has five family members including CBX2, CBX4, CBX6, CBX7 and CBX8. CBX6 displays significant differences from those of other proteins with respect to their histone peptide binding preferences.22–24 Abnormal expression of CBX6 in HCC has been reported,9 however, its possible roles in the metastasis of HCC have not been studied to date.
CBX6 has been reported to be overexpressed in human HCC tissues and involved in the process of tumorigenesis.9 In accordance with these findings, we demonstrated that CBX6 expression was increased in HCC tissues by IHC. In addition, our results also showed that the expression level of CBX6 was higher in HCC with metastasis than that in HCC without metastasis. Specifically, correlation of CBX6 expression with clinicopathological parameters showed that high CBX6 expression correlated significantly with tumor diameter, vascular invasion, tumor differentiation, TNM stage and metastasis. These results suggested that that CBX6 played an important role in the process of tumorigenesis, and progression of HCC.
To explore the possible biological function of CBX6 in HCC progression, we examined the effect of CBX6 on cell proliferation in SMMC-7721 and HCCLM3 cells using MTT assay. Consistent with the previous study, we found that overexpression of CBX6 effectively promoted tumor growth, whereas silencing of CBX6 led to a significant decrease in cell growth rate. To further determine the pathway underlying growth promotion by CBX6, cell cycle detection was conducted using a flow cytometry. Our results showed that overexpression of CBX6 decreased the proportion of cells in G0/G1 phase and induced S cell cycle arrest in SMMC-7721 cells, and silencing of CBX6 induced G1 cell cycle arrest in HCCLM3 cells. The data revealed that the pro-proliferative effect of CBX6 in HCC cells is associated with G1/S phase transition.
The correlation analysis showed that the expression of CBX6 was closely related to the vascular invasion and metastasis of HCC. Consistent with this, CBX6 expression was significantly higher in HCC cell lines with high metastasis potential than that in low metastatic cell lines. Invasion and metastasis, two of the most important hallmarks of cancer, are the major cause of mortality, particularly for HCC.25 Our studies showed that overexpression of CBX6 markedly increased cell migration and invasion ability in SMMC-7721 cells. We further found the migration and invasion abilities were suppressed in HCCLM3 cells transfected with LV-CBX6 shRNA. Wound healing assay also confirmed that CBX6 could enhance cell migratory ability. In addition, proliferation inhibitors could inhibit the migration and invasion ability of HCC cells. In our study, CBX6, a new metastatic regulator for HCC, was, for the first time, shown to significantly promote HCC invasion and metastasis, and the relationship between cell proliferation and metastasis needs to be further explored.
Invasion and metastasis is a multistep and complex process regulated by a series of genes, involving the adhesion of tumor cells, matrix degradation, tumor cell migration, tumor angiogenesis and so on.26,27 Although numerous genes involved in tumorigenesis and tumor metastasis were identified, the molecular mechanisms underlying these processes are not well understood.28 EMT is one of the most important events during cancer progression, leading to a more invasive, metastatic phenotype in human cancers.29–31 At the molecular level, EMT is related to a variety of factors including epithelia markers and mesenchymal makers. E-cadherin, as a key component of cell-cell adhesion junctions, is an important hallmark of EMT. Snail is the member of the snail family and capable of repressing E-cadherin.32 ZEB1, a key conversion molecule of EMT, is the most relevant repressor for E-cadherin.33–35 Thus, CBX6 overexpression may lead to the upregulation of snail/zeb1, repressing expression of E-cadherin, and thereby triggering EMT. Therefore, we next detected the expression levels of EMT markers in HCC cells by Western blot. Results found that increased expressions of mesenchymal markers (N-cadherin and Vimentin) and decreased epithelial markers (E-cadherin and β-catenin) were observed in SMMC-7721 cells transfected with CBX6-pEX. Consistent results were also observed when CBX6 was silenced in HCCLM3 cells. In addition, the expression of snail and zeb1 was increased in CBX6-overexpressing cells, the reverse results were also observed when CBX6 was silenced by shRNA. These results indicated that the migration and invasion abilities of HCC may be regulated by CBX6 through the EMT mechanism, which was induced by transcription factors snail and zeb1.
Conclusions
CBX6 expression was upregulated in HCC tissues and HCC cell lines, and it was significantly correlated with tumor diameter, vascular invasion, tumor differentiation, TNM stage and metastasis of HCC patients. CBX6 could promote the proliferation and G1/S phase transition in HCC cells. Moreover, CBX6 enhanced the cell migration and invasion and EMT progression induced by snail and zeb1. Our study indicated that CBX6 may function as an oncogene in HCC, and it could serve as a novel therapeutic target for the treatment of HCC patients.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;60:277–300.
2. Claudia A, Weir HK, Helena C, et al. Global surveillance of cancer survival 1995-2009: analysis of individual data for 25,676,887 patients from 279 population-based registries in 67 countries (CONCORD-2). Lancet. 2015;385:977–1010. doi:10.1016/S0140-6736(14)62038-9
3. Serper M, Taddei TH, Mehta R, et al. Association of Provider Specialty and Multidisciplinary Care With Hepatocellular Carcinoma Treatment and Mortality. Gastroenterology. 2017;152(8):S0016508517302652. doi:10.1053/j.gastro.2017.02.040
4. Bajusz I, Sipos L, Pirity MK. Nucleotide substitutions revealing specific functions of Polycomb group genes. Mol Genet Metab. 2015;114:547–556.
5. Bernstein BE, Mikkelsen TS, Xie X, et al. A Bivalent Chromatin Structure Marks Key Developmental Genes in Embryonic Stem Cells. Cell. 2006;125:315–326.
6. Santanach A, Blanco E, Jiang H, et al. The Polycomb group protein CBX6 is an essential regulator of embryonic stem cell identity. Nat Commun. 2016;8:1235.
7. Li G, Warden C, Zou Z, et al. Altered expression of polycomb group genes in glioblastoma multiforme. PLoS One. 2013;8:e80970.
8. Deng H, Guan X, Gong L, et al. CBX6 is negatively regulated by EZH2 and plays a potential tumor suppressor role in breast cancer. Oncotarget. 2019;9:197.
9. Zheng H, Jiang W, Tian T, et al. CBX6 overexpression contributes to tumor progression and is predictive of a poor prognosis in hepatocellular carcinoma. Oncotarget. 2017;8:18872–18884.
10. Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat Protoc. 2008;3(6):1101–1108. doi:10.1038/nprot.2008.73
11. Wang C-Q, Xiang F-G, Li Y-J, et al. Relation between the expression of mitotic centromere–associated kinesin and the progression of squamous cell carcinoma of the tongue. Oral Surgery, Oral Medicine, Oral Pathology and Oral Radiology. 2014;117(3):353–360. doi:10.1016/j.oooo.2013.11.488
12. Kawai H, et al. Estrogen Receptor and are Prognostic Factors in Non-Small Cell Lung Cancer. Clin Cancer Res. 2005;11(14):5084–5089. doi:10.1158/1078-0432.CCR-05-0200
13. Mcglynn KA, Petrick JL, London WT. Global epidemiology of hepatocellular carcinoma: an emphasis on demographic and regional variability. Clin Liver Dis. 2015;19(2):223–238. doi:10.1016/j.cld.2015.01.001
14. Dong YJ, Roberts LR. Hepatocellular carcinoma: A global view. Nat Rev Gastroenterol Hepatol. 2010;7:448–458. doi:10.1038/nrgastro.2010.100
15. Patrick RS. Hepatocellular Carcinoma. N Eng J Med. 2012;363:899.
16. Lewis EB. A Gene Complex Controlling Segmentation in Drosophila. Nature. 1978;276(5688):565–570. doi:10.1038/276565a0
17. Wei W, Jiang-Jiang Q, Sukesh V, et al. Polycomb Group (PcG) Proteins and Human Cancers: multifaceted Functions and Therapeutic Implications. Med Res Rev. 2015;35:1220–1267.
18. Luigi A, Bruno DS, Luciano DC. Polycomb complexes in stem cells and embryonic development. Development. 2013;140:2525–2534. doi:10.1242/dev.091553
19. Tiberi G, Pekowska A, Oudin C. PcG methylation of the HIST1 cluster defines an epigenetic marker of acute myeloid leukemia. Leukemia. 2015;29(5):1202–1206. doi:10.1038/leu.2014.339
20. Zhu X, Yan M, Luo W, et al. Expression and clinical significance of PcG-associated protein RYBP in hepatocellular carcinoma. Oncol Lett. 2017;13(1):141–150. doi:10.3892/ol.2016.5380
21. Wang W, Qin JJ, Voruganti S, et al. Polycomb Group (PcG) Proteins and Human Cancers: multifaceted Functions and Therapeutic Implications. Med Res Rev. 2015;35:1220–1267.
22. Milosevich N, Gignac MC, Mcfarlane J, et al. Selective Inhibition of CBX6: A Methyllysine Reader Protein in the Polycomb Family. ACS Med Chem Lett. 2015;7:139. doi:10.1021/acsmedchemlett.5b00378
23. Lilia K, Hui O, Maria A, et al. Recognition and specificity determinants of the human cbx chromodomains. J Biol Chem. 2011;286:521–529. doi:10.1074/jbc.M110.191411
24. Richly H, Aloia L, Di CL. Roles of the Polycomb group proteins in stem cells and cancer. Cell Death Dis. 2011;2(9):e204. doi:10.1038/cddis.2011.84
25. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi:10.1016/j.cell.2011.02.013
26. Ls P, Dj O. Modeling Cell Migration Mechanics. Adv Exp Med Biol. 2018;1092:159–187.
27. You K, Sun P, Yue Z, et al. NOR1 promotes hepatocellular carcinoma cell proliferation and migration through modulating the Notch signaling pathway. Exp Cell Res. 2017;352(2):375. doi:10.1016/j.yexcr.2017.02.032
28. Li T, Xie J, Shen C, et al. Upregulation of long noncoding RNA ZEB1-AS1 promotes tumor metastasis and predicts poor prognosis in hepatocellular carcinoma. Oncogene. 2016;35(12):1575. doi:10.1038/onc.2015.223
29. Jiaxing Z, Jinhuan W, Jian L, et al. Overexpression of Rab25 contributes to metastasis of bladder cancer through induction of epithelial-mesenchymal transition and activation of Akt/GSK-3β/Snail signaling. Carcinogenesis. 2013;34:2401–2408. doi:10.1093/carcin/bgt187
30. Aiello NM, Brabletz T, Kang Y, et al. Upholding a role for EMT in pancreatic cancer metastasis. Nature. 2017;547(7661):E7. doi:10.1038/nature22963
31. Parvani JG, Gujrati MD, Mack MA, et al. Abstract B45: silencing ß3 integrin by targeted ECO/siRNA nanoparticles inhibits EMT and metastasis of triple-negative breast cancer. Cancer Res. 2017;77:B45–B45.
32. Angela M. N. The snail superfamily of zinc-finger transcription factors. Life Sci Res. 2003;3:155–166.
33. Cano A. Rodrigo The transcription factor Snail controls epithelial–mesenchymal transitions by repressing E-cadherin expression. Nat Cell Biol. 2000;2(2):76–83. doi:10.1038/35000025
34. Wellner UF, Zhu F, Hopt UT, et al. The Role of the Transcription Factor ZEB1 in Epithelial-Mesenchymal Transition and Progression of Pancreatic Carcinoma. 2009.
35. Mazda M, Nishi K, Naito Y, et al. E-cadherin is transcriptionally activated via suppression of ZEB1 transcriptional repressor by small RNA-mediated gene silencing. PLoS One. 2011;6:e28688.
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.