Back to Journals » The Application of Clinical Genetics » Volume 12
Candidate Gene Analysis Of Alopecia Areata In Jordanian Population Of Arab Descent: A Case–Control Study
Authors AL-Eitan LN , Al Momani RO, Al Momani KK, Al Warawrah AM, Aljamal HA , Alghamdi MA , Muhanna AM , Al-Qarqaz FA
Received 9 August 2019
Accepted for publication 31 October 2019
Published 21 November 2019 Volume 2019:12 Pages 221—228
DOI https://doi.org/10.2147/TACG.S226664
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Martin Maurer
Laith N AL-Eitan,1,2 Rawan O Al Momani,1 Khalid K Al Momani,3 Ahmad M Al Warawrah,3 Hanan A Aljamal,1 Mansour A Alghamdi,4 Alsharif M Muhanna,3 Firas A Al-Qarqaz5,6
1Department of Applied Biological Sciences, Jordan University of Science and Technology, Irbid, Jordan; 2Department of Biotechnology and Genetic Engineering, Jordan University of Science and Technology, Irbid, Jordan; 3Department of Dermatology, King Hussein Medical Center (KHMC), Jordan Royal Medical Services (RMS), Amman, Jordan; 4College of Medicine, King Khalid University, Abha, Saudi Arabia; 5Department of Internal Medicine, Jordan University of Science and Technology, Irbid, Jordan; 6Division of Dermatology, Department of Internal Medicine, King Abdullah University Hospital, Irbid, Jordan
Correspondence: Laith N AL-Eitan
Toxicology and Pharmacogenetics, Department of Biotechnology & Genetic Engineering, Faculty of Science and Arts, Jordan University of Science and Technology, PO Box 3030, Irbid 22110, Jordan
Tel +962-2-7201000 ext. 23464
Email [email protected]
Background: Alopecia areata (AA) is a non-cicatricial patchy hair loss on the scalp, face or other parts of the body. AA was found to be responsive to immunosuppressive therapies, a finding that supports an autoimmune basis for the disease. Several genetic studies have shown the significance of immunological factors as key genetic components in AA.
Objective: In this study, we aimed to investigate the genetic association of 7 single-nucleotide polymorphisms (SNPs) within five candidate genes including TAP1, CXCL1, CXCL2, HSPA1B, and TNFα with AA susceptibility in the Jordanian Arab population.
Methods: A case–control genetic association study conducted in 152 patients and 150 healthy individuals was performed using the sequenom MassARRAY system (iPLEX GOLD) to genotype the selected SNPs.
Results: rs1800629 SNP of the TNFα gene was significantly associated with AA in the heterozygous and rare homozygous genotypes (P=0.022 and P=0.0079, respectively) with no linkage of the TAP1, CXCL1, CXCL2 and HSPA1B variants.
Conclusion: This is the first study of its kind among the Jordanian population providing evidence of genetic association of the TNFα with AA susceptibility. Further genetic studies on Arab descent including other variants are required to clarify and strengthen the association of these genes with susceptibility to develop AA.
Keywords: alopecia areata, TAP1, CXCL1, CXCL2, genetic association studies, immunological factors, Jordan
Introduction
Alopecia areata (AA) is a disease characterized by areas of non-scarring hair loss that may take the form of a single round, oval patch or even multiple patches that might become confluent. It could affect both men and women equally at any age.1 Children and young adults are prone more to have the disease, with 30% to 48% of the patients being affected before the age of 20.2
There are several clinical types of alopecia; one favorable division is localized and diffuse types. Localized alopecia can be classified into scarring or non-scarring alopecia.3 Scarring alopecia may be caused by lupus erythematosus, lichen planus, inflammatory tinea capitis, burns or cicatricial basal cell carcinoma. On the other hand, non-scarring alopecia could result from AA, androgenetic alopecia, traction alopecia or non-inflammatory tinea capitis.3
AA is assumed to be a hair-specific autoimmune disease with genetic factors playing a role in disease susceptibility and severity with a concordance rate of 55% that is found in identical twin studies implicating both genetic and environmental factors.4 The etiopathogenesis of AA is not fully understood yet, due to the multifactorial etiology, whereas several genes and environmental factors play a role in triggering its pathology.5
Major histocompatibility complex (MHC) genes have been linked to various autoimmune diseases, including AA.6 A transporter 1 ATP-binding cassette subfamily B gene (TAP1) is located in the MHC class II region forming a heterodimer that plays a key role in the endogenous antigen presentation pathways. Thus, TAP genes represent possible susceptibility for some autoimmune diseases.7 Chemokine (C-X-C motif) ligand 1 (CXCL1) and chemokine (C-X-C motif) ligand 2 (CXCL2) are small cytokines that belong to the CXC chemokine family that plays an important role in modulating T-lymphocyte activities. Therefore, these genes are essential for the pathogenesis of AA as well.8 In addition, HSPA1B protein is known as heat shock 70-kDa protein 1B. It is involved in modulating antigen processing and presentation and contributes to the immune response against pathogens.8,9 In the MHC genes, human leukocyte antigen class III (HLA-III) region contains heat shock proteins such as HSPA1B.11 Furthermore, tumor necrosis factor alpha (TNFα) is a cell signaling protein (cytokine) that involves in systemic inflammation.12 TNF-α is well known as a major player in the pathogenesis of AA.13
This work was focused on non-scarring AA along with alopecia totalis (AT) and alopecia universalis (AU). Our hypothesis pointed to a T-cell-mediated attack on anagen hair follicles after the loss of immune system privilege of the hair follicle which causes the hair to fall in genetically susceptible individuals. Therefore, 7 SNPs within 5 candidate genes were genotyped to reveal the association of AA among the Jordanian population.
Materials And Methods
Participants And Data Source
This study has been established according to the provisions of the Human Ethics Standard and in compliance with the IRB guidelines at Jordan University of Science and Technology (JUST). Approval for patients’ recruitment including blood samples and clinical data collection were also obtained from the Human Ethics Committee at The Jordanian Royal Medical Services (RMS) and King Abdullah II University Hospital (KAUH) with a written informed consent obtained from all participants. History of the disease, clinical diagnosis, medication, treatment follow-up, family history of alopecia and other autoimmune diseases such as thyroid diseases, vitiligo, and psoriasis incidence were obtained.
One hundred and fifty-two patients including 107 males and 45 females were recruited from dermatology clinics of the RMS including Al Hussein Medical City, Prince Rashed bin Al Hasan, Prince Hashem bin Al Hussein, Princes Haya bint Al Hussein and KAUH in addition to 150 unrelated healthy control individuals. The demographic and clinical characteristics of our cohort (302 subjects) are shown in Table 1.
 |
Table 1 Demographics And Clinical Data For Alopecia Areata Patients (n=152) And Healthy Individuals (n=150) |
The average age (±SD) of the patients’ group was 31.144±12.41 with a median of 31 and a range of 13–67 years. Healthy individuals' age (±SD) were 33.9±9.81 with a median of 32 and the range was 17–64 years. Age groups for patients and control were divided into 3 categories, and each category had a range of 18 years. The first category was (13–31), the second category was (32–50), and the third one was (51–69).
DNA Extraction
Genomic DNA was extracted from EDTA blood samples using commercially available Wizard® Genomic DNA Purification Kit (Qiagen, Germany) according to the manufacturer’s protocol.
Candidate Genes And SNPs Selection
In order to detect SNPs that could be associated with AA among the Jordanian population, 5 candidate genes (TAP1, CXCL1, CXCL2, HSPA1B, and TNFα) were selected based on previous association studies or for their position to guarantee the coverage of the entire gene. TAP1 selected SNPs were rs2071480, rs1135216 and rs1057141. For the CXCL1 and CXCL2 chemokines genes, rs3117604 and rs3806792, respectively, rs2763979 for the HSPA1B and rs1800629 for the TNFα. SNPs were selected using available genetic databases such as the National Centre for Biotechnology Information (NCBI) (http://www.ncbi.nlm.nih.gov/SNP/), the US National Library of Medicine (https://ghr.nlm.nih.gov), and The University of California Santa Cruz Genome Browser Database (UCSC) (http://genome.ucsc.edu/).
Genotyping
Extracted DNA samples were sent to be genotyped in collaboration with the Australian Genome Research Facility (AGRF). The Sequenom MassARRAY® system (iPLEX GOLD) (Sequenom, San Diego, CA, USA) was used for genotyping at the AGRF based on the manufacturer’s procedure. Samples were genotyped in duplicate and results were accepted unless the success rate was at least 95%.
Statistical Analysis
Hardy–Weinberg equilibrium (HWE) was calculated to estimate genotyping frequencies including examination for ascertainment bias. The probability of evolutionary event existence that may affect the allele frequency was examined, where p represents the frequency of one allele, while q represents the frequency of the other allele.14 Any significant deviations from HWE may point to a genotyping error. To define the P-values for the allele and genotype association, chi-square test (Pearson χ2 test) was used to test whether there are any significant differences between the expected, seen and calculated genotype frequencies. Genotypic and allelic association was performed to investigate SNP association with AA. Furthermore, haplotype was performed for linkage disequilibrium (LD) analysis. P-value<0.05 was considered statistically significant. The Statistical Package for the Social Sciences (SPSS) version 21.0 and the SNPStat web tool (https://www.snpstats.net/start.htm) were used.
Results
In this study, the age of onset (±SD) was found to be 27.328±12.57 with no significant difference in age and gender between patients and controls (data not shown). Our data showed that about 90.13% of the patients had patchy alopecia areata (PA), 6.57% had AU, and only 3.28% of them had AT. Symptoms associated with AA were revealed in 31.6% of the patients, while 68.4% of them had no symptoms at all. Furthermore, only 7.3% of the patients had nail changes associated with the disease. All polymorphisms met the HWE standards in both case and control groups with minor allele frequencies shown in Table 2. Genotyped SNPs were evaluated (presented in Table 3) by using chi-square test in order to determine if there is a significant linkage between the genotypic and allelic frequencies with AA susceptibility.
 |
Table 2 Genes, SNPs, Their Minor Allele Frequencies And HWE P-Values For Cases And Controls |
 |
Table 3 Genetic Association Of The Selected SNPs With Alopecia Areata Susceptibility |
The genotype frequencies of rs1800629 G/A and A/A within the TNFα gene in chromosome 6 show the highest association with AA susceptibility in the overall estimated effects (P= 0.022 and 0.0079, respectively). However, no significant differences were detected in the allele frequencies between AA cases and controls (P= 0.079 for allele frequency) as shown in Table 4. The heterozygous and rare homozygous (G/A and A/A) genotypes were found to be more frequent among alopecia cases (24% and 3%, respectively) than controls (21% and 0%, respectively). On the other hand, the common homozygous genotype (G/G) showed less frequency among cases (72%) in comparison to the controls (79%) (Table 3). Figure 1 shows a scatter plot of the rs1800629 within the TNFα from sequenom data whereas each point representing the measurement for a single individual. The points were colored according to the genotype calls. Genotypes of the 7 SNPs were highly accurate with an average success rate of 98% and the discrepancy average (±SD) rate across the 7 loci was only 0.02±0.0003 in the whole cohort (302 subjects). Allele frequencies and genotypes of the studied SNPs within TAP1, CXCL1, CXCL2, and HSPA1B genes revealed no significant differences between AA patients and the healthy individuals (Table 2).
 |
Table 4 Genetic Association Of rs1800629 Within The TNFα Gene Between Alopecia Areata And Healthy Individuals Using Different Genetic Models |
Genetic association analysis of the rs1800629 in TNFα gene was investigated using different genetic statistical methods: dominant, additive and recessive genetic models for this associated SNP. The co-dominant form G/A and the rare homozygous A/A were found to be significantly associated depending on the chi-square results. However, no genetic association was found within the common homozygous G/G (P>0.05) (Table 4). For the 3 haplotypes of the TAP1 gene, there was no significant association with AA with P>0.05 (Table 5).
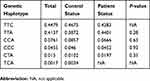 |
Table 5 Genetic Haplotype Frequencies Of TAP1 Gene (rs1057141, rs1135216 And rs2071480) Associated With Alopecia Areata |
Discussion
Alopecia areata is thought to be a tissue-specific autoimmune disease directed against the hair follicle and might be associated with other autoimmune diseases.15 Previous studies showed that MHC genes of chromosome 6 which encodes the human leukocyte antigens (HLAs) are major determining loci for T-cell-mediated diseases including AA.6 A number of organ-specific autoimmune diseases have been identified, including diabetes, autoimmune thyroiditis, psoriasis, inflammatory bowel diseases, rheumatoid arthritis, and systemic lupus to be associated with specific alleles of HLA or MHC.4 However, HLA contribution alone could not be enough for the explanation of the entire genetic basis of AA.16 Several studies found that AA has a polygenic origin where multiple genes intertwine causing a greater susceptibility to develop the disease.17 A recent genome-wide association (GWAS) study revealed the association of HLA in addition to new loci outside the MHC including ACOXL/BCL2L11 and GARP with a nominal association of SH2B3(LNK)/ATXN2.18
So far, IFN-ᵞ, interleukins and TNF-α are cytokines with a major role in the pathogenesis of autoimmune diseases including AA. Previous studies published since the 1980s and early 1990s suggested cytokine and chemokine role in autoimmunity.19–21 A present study indicates a crucial role of the cytokine as an inducer of hair loss with AA.22 TNFα is known to be a risk factor in a variety of inflammatory pathologies.23 Our data show an association of the TNFα with AA that goes in agreement with the study by Gregoriou et al who found that patients with multiple lesions contain extremely high levels of B-cell-activating factor that belongs to the TNF family.13 This indicates an increase in the TNFα levels in patients with AA which has been confirmed in the present study. However, another study performed by Galbraith and Pandey in USA suggested no significant difference in the distribution of TNF-α G/A phenotypes between patient groups.24 Their results provide direct evidence of genetic heterogeneity between the two forms of AA. They also suggested that the TNF-α gene or a closely linked locus within the MHC might play a role in the pathogenesis of the patchy form of the disease.24
In this study, an association between rs1800629 of the TNFα-308G/A and AA was confirmed (P= 0.022) and found to be consistent with the results of Gregoriou et al.13 It is also consistent with another study performed by Cristina et al in Mexico which suggested a credible association between the presence of TNFa-308G/A polymorphism and an increased susceptibility for the development of patchy AA.23 This overproduction of TNFα might lead to a higher risk of facilitating an autoimmune response against the hair follicle. In our data, the variant’s genotype association between AA cases and control was confirmed using different genetic models in order to analyze the genetic association. The association was found in the heterozygous G/A and rare homozygous A/A versus common homozygous G/G. These findings provide evidence of genetic variants in TNFα gene with AA development in the Jordanian population. Upon a recent screening, the EBI-GWAS catalog stated that rs1800629 variant is located in the MHC region and there is an extensive linkage disequilibrium (LD) in the HLA region where the association signal of rs1800629 might be due to LD with neighboring variants associated with AA. EBI-GWAS catalog indicates that rs9275524-C is close to HLA-DQB1 and it is in LD with rs1800629 and associated with AA.25 Therefore, the weak association signal of rs1800629 in this study is likely due to LD and cannot be used to implicate TNFα as the causal gene without considering other correlated variants.
Mast cells and macrophages both conduce in the early stages of recruiting neutrophils into tissues. CXCL1 and CXCL2 are neutrophil chemoattractants produced in response to the neutrophil attack by mast cells and macrophages.26 Chemokines play important roles in the development, homeostasis, and function of the immune system as well as angiogenesis.27 For that reason, this study focused on the role of these genes in the development of AA. A study performed by Kim et al on the Korean population confirmed that genetic sequencing of the rs3117604 and rs3806792 in CXCL1 and CXCL2 gene, respectively, showed a significant correlation between the two SNPs and the development of AA.27 It also confirmed that T/T and C/C haplotypes were significantly associated with an increased risk of AA. In contrast, this study revealed no significant association of the CXCL1 and CXCL2 with AA susceptibility. On the other hand, none of the polymorphisms differed significantly for allele or genotype frequencies among cases and controls.
HSPA1B plays a major role in immunity mainly in MHC class III, and different levels of HSPA1B expression correspond to hair loss and immunologic reactions in anagen (i.e. first stage in hair development) hair follicles, which are common features in AA histopathology.11 In a previous study presented by Seok et al in Korea, rs6457452 was found to be associated with the onset of AA. In their analysis of clinical features obtained for AA, rs6457452 was only weakly related to the age of onset, and rs2763979 was only weakly related to the type of AA lesion.11 In conclusion, they suggested that rs6457452 might be associated with the onset of AA, and the T allele might grant the reduced susceptibility to AA in the Korean population. Contrary to our results that revealed no association of the rs276379 in the Jordanian population.
Investigation of polymorphisms in TAP1 identified an association with several autoimmune diseases and AA is one of them.28 The study of Kim et al was one of the first documented studies in comparing genotypic and allelic frequencies of TAP1 in patients with AA.28 In their study, Korean patients were tested for three selected SNPs (rs2071480, rs1135216, and rs1057141), whereas results suggested an association of a promoter SNP (rs2071480) in the heterozygous G/T co-dominant. In addition, the homozygous T/T genotype showed a significant association with an increased risk of AA. In contrast, none of the three aforementioned SNPs were associated with AA in this study.
Differences in ethnicity groups, type of AA, linkage equilibrium and disease duration could be a probable clarification for such contradictory results reported in the literature. However, most of the studies in Arab populations were restricted by being of a retrospective design since most of them have short follow-up periods. The future direction toward inclusion of a larger cohort and additional genes and loci based on the newly GWAS studies conducted on the association of the AA.
Conclusion
This study further supports the polygenic model of AA. TNFα seems to have an essential role in AA pathogenesis with a statistically significant association. This finding indicates the importance of potential researches for sequencing of the HLA region where TNFα-308G/A could be of linkage equilibrium with other variants as previously performed in other ethnic groups. In the current study, no significant role was identified for genetic variants of chemokines (CXCL1 and CXCL2), heat shock protein (HSPA1B), and ATP-binding cassette (TAP1) to influence the development of AA in the Jordanian population since no genetic association was found with AA susceptibility as well as for the TAP1 haplotypes. Furthermore, no correlation was found between the tested genetic changes and the clinical phenotypes.
Ethics Approval And Informed Consent
All procedures contributing to this work complied with the ethical standards of the relevant national and institutional committees on human experimentation and the Declaration of Helsinki with an IRB no. (48/2017).
Data Availability
The datasets generated and analyzed during the current study are not publicly available. The consent from participants did not cover data sharing but are available from the corresponding author on reasonable request.
Disclosure
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Amin SS, Sachdeva S. Alopecia areata: a review. J Dermatol Dermatol Sur. 2013;17:37–45. doi:10.1016/j.jssdds.2013.05.004
2. Mounsey AL, Reed SW. Diagnosing and treating hair loss. Am Fam Physician. 2009;80:356–362.
3. Weller RB, Hunter HJ, Mann MW. Clinical Dermatology.
4. De Andrade M, Jackow CM, Dahm N, Hordinsky M, Reveille JD, Duvic M. Alopecia areata in families: association with the HLA locus. J Investig Dermatol Symp Proc. 1999;4:220–223. doi:10.1038/sj.jidsp.5640215
5. Da Costa CM, Dupont E, Van der Cruys M, et al. Earlier occurrence of severe alopecia areata in HLA-DRB1* 11-positive patients. Dermatology. 2006;213:2–14.
6. Barahmani N, De Andrade M, Slusser JP, et al. Human leukocyte antigen class II alleles are associated with risk of alopecia areata. J Invest Dermatol. 2008;128:240–243. doi:10.1038/sj.jid.5700973
7. Vinasco J, Fraile A, Nieto A, et al. Analysis of LMP and TAP polymorphisms by polymerase chain reaction-restriction fragment length polymorphism in rheumatoid arthritis. Ann Rheum Dis. 1998;57:33–37. doi:10.1136/ard.57.1.33
8. Zainodini N, Hassanshahi G, Kazemi Arababadi M, Khorramdelazad H, Mirzaei A. Differential expression of CXCL1, CXCL9 CXCL10 and CXCL12 chemokines in alopecia areata. Iran J Immunol. 2013;10:40–46.
9. Milner CM, Campbell RD. Structure and expression of the three MHC-linked HSP70 genes. Immunogenetics. 1990;32:242–251. doi:10.1007/BF00187095
10. Radons J, Multhoff G. Immunostimulatory functions of membrane-bound and exported heat shock protein 70. Exerc Immunol Rev. 2005;11:17–33.
11. Seok H, Jeon HS, Park HJ, et al. Association of HSPA1B SNP rs6457452 with alopecia areata in the Korean population. Immunol Invest. 2014;43:212–223. doi:10.3109/08820139.2013.857351
12. Gahring LC, Carlson NG, Kulmer RA, Rogers SW. Neuronal expression of tumor necrosis factor alpha in the OVOUJI fme brain. Neuroimmunomodulation. 1996;3:289–303. doi:10.1159/000097283
13. Gregoriou S, Papafragkaki D, Kontochristopoulos G, Rallis E, Kalogeromitros D, Rigopoulos D. Cytokines and other mediators in alopecia areata. Mediators Inflamm. 2010;1–5. doi:10.1155/2010/928030
14. Nikoshkov A, Drakenberg K, Wang X, Horvath MC, Keller E, Hurd YL. Opioid neuropeptide genotypes in relation to heroin abuse: dopamine tone contributes to reversed mesolimbic proenkephalin expression. Proc Natl Acad Sci. 2008;105:786–791. doi:10.1073/pnas.0710902105
15. Pforr J, Blaumeiser B, Becker T, et al. Investigation of the p. Ser278Arg polymorphism of the autoimmune regulator (AIRE) gene in alopecia areata. Tissue Antigens. 2006;68:58–61. doi:10.1111/tan.2006.68.issue-1
16. Tazi-Ahnini R, McDonagh AJ, Cox A, et al. Association analysis of IL1A and IL1B variants in alopecia areata. Heredity. 2001;87:215–219. doi:10.1046/j.1365-2540.2001.00916.x
17. Juárez-Rendón KJ, Rivera Sánchez G, Reyes-López MA, et al. Alopecia areata. Current situation and perspectives. Arch Argent Pediatr. 2017;115:e404–e411. doi:10.5546/aap.2017.eng.e404
18. Betz RC, Petukhova L, Ripke S, et al. Genome-wide meta-analysis in alopecia areata resolves HLA associations and reveals two new susceptibility loci. Nat Commun. 2015;22(6):5966. doi:10.1038/ncomms6966
19. Spies T, Morton CC, Nedospasov SA, Fiers W, Pious D, Strominger JL. Genes for the tumor necrosis factors alpha and beta are linked to the human major histocompatibility complex. Proc Natl Acad Sci. 1986;83:8699–8702. doi:10.1073/pnas.83.22.8699
20. Wolpe SD, Sherry B, Juers D, Davatelis G, Yurt RW, Cerami A. Identification and characterization of macrophage inflammatory protein 2. Proc Natl Acad Sci. 1989;1989(86):612–616. doi:10.1073/pnas.86.2.612
21. Haskill S, Peace A, Morris J, et al. Identification of three related human GRO genes encoding cytokine functions. Proc Natl Acad Sci. 1990;87:7732–7736. doi:10.1073/pnas.87.19.7732
22. Shimizu T, Hizawa N, Honda A, et al. Promoter region polymorphism of macrophage migration inhibitory factor is strong risk factor for young onset of extensive alopecia areata. Genes Immun. 2005;6:285–289. doi:10.1038/sj.gene.6364191
23. Cristina CS, Mauricio SS, Armando LR, et al. Tumor necrosis factor alpha promoter‐308G/A polymorphism in Mexican patients with patchy alopecia areata. Int J Dermatol. 2012;51:571–575. doi:10.1111/j.1365-4632.2011.05291.x
24. Galbraith GM, Pandey JP. Tumor necrosis factor alpha (TNF-α) gene polymorphism in alopecia areata. Hum Genet. 1995;96(433–436). doi:10.1007/BF00191802
25. European Bioinformatics Institute (EBI). GWAS catalog. Available from: https://www.ebi.ac.uk/gwas/efotraits/EFO_0004192.
26. De Filippo K, Dudeck A, Hasenberg M, et al. Mast cell and macrophage chemokines CXCL1/CXCL2 control the early stage of neutrophil recruitment during tissue inflammation. Blood. 2013;121:4930–4937. doi:10.1182/blood-2013-02-486217
27. Kim SK, Chung JH, Park HJ, et al. Polymorphisms in the promoter regions of the CXCL1 and CXCL2 genes contribute to increased risk of alopecia areata in the Korean population. Genet Mol Res. 2015;14:9667–9674. doi:10.4238/2015.August.14.29
28. Kim HK, Lee H, Lew BL, et al. Association between TAP1 gene polymorphisms and alopecia areata in a Korean population. Genet Mol Res. 2015;14:18820–18827. doi:10.4238/2015.December.28.31
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.

