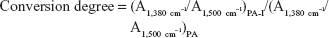Back to Journals » International Journal of Nanomedicine » Volume 10 » Issue 1
Calcium-activated gene transfection from DNA/poly(amic acid-co-imide) complexes
Authors Wu S , Chang L, Peng S, Tsai H
Received 28 October 2014
Accepted for publication 7 December 2014
Published 27 February 2015 Volume 2015:10(1) Pages 1637—1647
DOI https://doi.org/10.2147/IJN.S76502
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Thomas Webster
Szu-Yuan Wu,1–4 Li-Ting Chang,5 Sydeny Peng,6 Hsieh-Chih Tsai5
1Institute of Toxicology, College of Medicine, National Taiwan University, 2Department of Radiation Oncology, Wan Fang Hospital, Taipei Medical University, 3Department of Internal Medicine, School of Medicine, College of Medicine, Taipei Medical University, Taipei, Taiwan; 4Department of Biotechnology, Hungkuang University, Taichung, Taiwan; 5Graduate Institute of Applied Science and Technology, National Taiwan University of Science and Technology, Taipei, Taiwan; 6Department of Chemical Engineering, National Tsing Hua University, Hsinchu, Taiwan
Abstract: In this study, we synthesized a water-soluble poly(amic acid-co-imide) (PA-I) from ethylenediaminetetraacetic dianhydride (EDTA) and 2,2'-(ethylenedioxy)bis(ethylamine) that possesses comparable transfection efficiency to that of polyethylenimine (PEI), when prepared in combination with divalent calcium cations. The polycondensation of monomers afforded poly(amic acid) (PA) precursors, and subsequent thermal imidization resulted in the formation of PA-I. At a polymer/DNA ratio (indicated by the molar ratio of nitrogen in the polymer to phosphate in DNA) of 40, complete retardation of the DNA band was observed by gel electrophoresis, indicating the strong association of DNA with PA-I. A zeta potential of -22 mV was recorded for the PA-I polymer solution, and no apparent cytotoxicity was observed at concentrations up to 500 µg·mL-1. In the presence of divalent Ca2+, the transfection efficiency of PA-I was higher than that of PA, due to the formation of a copolymer/Ca2+/DNA polyplex and the reduction in negative charge due to thermal cyclization. Interestingly, a synergistic effect of Ca2+ and the synthesized copolymer on DNA transfection was observed. The use of Ca2+ or copolymer alone resulted in unsatisfactory delivery, whereas the formation of three-component polyplexes synergistically increased DNA transfection. Our findings demonstrated that a PA-I/Ca2+/DNA polyplex could serve as a promising candidate for gene delivery.
Keywords: gene carrier, poly(amic acid), polyimide
Introduction
Nonviral gene carriers, including polymeric vectors, are considered to be safer alternatives to viral-based gene delivery vehicles. Negatively charged DNA and cationic polymers, such as polyethylenimine (PEI) and poly(lysine), are capable of condensing into a polyplex and efficiently interacting with the negatively charged cell membrane. Upon internalization, the polyplex dissociates, and the DNA is released prior to lysosomal degradation. Unfortunately, their high positive charge density disrupts cell and mitochondrial membranes, leading to cell necrosis and apoptosis; the inherent cytotoxicity of these cationic polymers, both in vitro and in vivo, continues to be an unresolved problem.1,2 Equally of concern is the binding of negatively charged serum proteins to positively charged complexes, initiating aggregation and reducing transfection.3,4 In order to realize the actual clinical potential of these carriers, attempts have been made to modulate the surface charge density by such means as bioconjugation, oxidation, and the addition of anionic materials. In one study, the succinylation of PEI introduced negatively charged carboxylate groups, which lowered the polymer cytotoxicity tenfold while increasing the knockdown percentage of a target luciferase gene.5 In another study, peroxidase oxidization of amine groups lowered the charge density and provided a simple strategy for modification of PEI.6 The incorporation of poly(propylacrylic acid) in commercially available cationic liposomes was shown to increase the delivery of plasmid DNA in vitro and in vivo.7 Schlegal et al identified seven anionic nontoxic polymers, most bearing negatively charged carboxylate groups, including poly(glutamic acid), poly(acrylic acid), and hyaluronic acid, that lowered cellular toxicity and increased gene-silencing efficiencies.8 Furthermore, endosomal escape is a key step in transfection, and when polymer and DNA are too tightly bound, the genetic material cannot be released into the cytosol – one study showed that the addition of anionic liposomes to lipid-based formulations led to membrane destabilization and liberation of cargo DNA.9 Taking together these considerations, it is practical to develop new delivery systems as an alternative to cationic carriers.
The complete substitution of cationic materials with anionic materials is another approach to eliminating strong positive charge.10,11 Divalent cations can be used as mediators to reduce negative charge of DNA, and therefore eliminating the need for high cations charge of polymer.12 Calcium cations, in particular, have been shown to mediate endosomal escape and cytosolic stability, and to enhance DNA uptake by interacting with nuclear pore complexes.13,14 Elsewhere, the use of divalent calcium cations enhanced the transfection efficiency of plasmid DNA-lipid complexes to a variety of cell lines by three- to 20-fold.15 Use of polymers containing carboxylate functional groups is another strategy set forth for driving complex formations through interactions with divalent cations.16
Poly(amic acid) (PA) is a negatively charged copolymer prepared from the polycondensation of diamines and dianhydrides. Imidization of PA, either thermally or chemically, affords poly(imides). Taking advantage of the synthesis procedure, it becomes possible to control the charge density of the final product by varying the degree of imidization, through reaction temperature and time.17,18 Therefore, the modulation of poly(amic acid-co-imide) (PA-I) charge is possible without the addition of a second polymer. PA-I is also thermally and mechanically stable, while exhibiting good biocompatibility.19–21 Strong intermolecular interactions between adjacent donor nitrogen and acceptor carbonyl groups lead to the stacking of PA-Is. Furthermore, the presence of carbonyl groups allows the PA-Is to interact with nearby –NH2 groups, commonly observed in biomaterials, through hydrogen bonding.22,23 However, the biological applications of PA-I have been limited by its insolubility in water. Water solubility is a prerequisite for preparing gene delivery systems, and the modification of water-insoluble polymers, such as chitosan, has initiated extensive related research.24 Recently, the chemical and physical evaluation of a water-soluble PA-I was reported, thereby casting light on the potential of PA-I for gene delivery.25
Herein, we report the synthesis and characterization of a water-soluble PA-I derived from ethylenediaminetetraacetic dianhydride (EDTA) and 2,2′-(ethylenedioxy)bis(ethylamine) (EOEM), through polycondensation and subsequent imidization of its PA precursor. A gel retardation assay was used to verify polyplex formation between the copolymers and DNA. Transfection efficiencies of the synthesized PA-I and PA precursors were evaluated alone and in the presence of divalent calcium cations. These results may serve to elucidate the potential of water-soluble PA-Is as gene delivery vectors.
Methods
Materials
Reagents, such as EDTA dianhydride, EOEM, and dimethyl sulfoxide (DMSO) were purchased from Sigma-Aldrich Corp (St Louis, MO, USA). DMSO was dried over calcium hydride and distilled under vacuum. Plasmid-encoding enhanced green fluorescence protein (GFP) (pEGFP-N1) was donated by Dr Hu at the Department of Chemical Engineering, National Tsing Hua University, Taiwan.
Amplification of plasmid DNA
The pEGFP-N1 plasmid was introduced into Escherichia coli strain DH5α (Invitrogen; Life Technologies, Carlsbad, CA, USA). Briefly, transformed DH5α was grown in 500 mL Luria-Bertani broth containing 250 μg of antibiotics per mL and then incubated at 37°C, with shaking at 200 rpm, for 16 hours. Plasmid DNA was purified from DH5α, using AxyPrep™ Plasmid Maxiprep Kits. The purity of plasmid DNA was certified by an absorbance ratio of 1.8 at OD260/OD280. The plasmid was stored at −20°C before use.
Synthesis of PA and PA-I copolymer
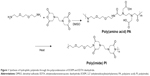
Briefly, EOEM was dissolved in dry DMSO with constant stirring. An equimolar amount of EDTA dianhydride was added, and the mixture was stirred under N2 atmosphere for 24 hours. PA was precipitated in acetone, washed, and filtered three times to remove unreacted monomers. The ring closure reaction was carried out at 110°C for 3 hours to yield the final product, PA-I.
Characterization of copolymers
The molecular structures of the monomers and products were verified by proton nuclear magnetic resonance (1H NMR) spectra, acquired on a Bruker AVANCE III HD 600 MHz spectrometer (Bruker Corporation, Billerica, MA, USA) with DMSO-d6 as solvent. Fourier-transform infrared spectroscopy (FT-IR) spectra were recorded on a Thermo Nicolet Nexus 6700 system (Thermo Fisher Scientific Inc, Waltham, MA, USA), using samples of PA and PA-I prepared on CaF2 disks by the solvent-casting method. Zeta potential measurements of 0.1% polymer solutions (w/v) were carried out with a Horiba SZ-100 nanoparticle analyzer (Horiba Ltd., Kyoto, Japan).
Curve fitting of the contents of PA in the PA-I polymer
All curve fitting was performed and their integrated areas measured by OriginPro 8 software (OriginLab Corporation, Northampton, MA, USA). Overlapping peaks were resolved by using Gaussian function.
Gel retardation assay
Gel electrophoresis was performed with copolymer/DNA ratios of 1:1, 10:1, 30:1, and 40:1. Samples were prepared by mixing nuclease-free polymer solutions with equal volumes of plasmid DNA at various ratios. The samples were allowed to stand for 20 minutes, to allow complex formation, prior to the addition of Tris-acetate-EDTA (TAE) running buffer. Next, the samples were loaded into the wells of a 0.6% agarose gel, and electrophoresis was carried out using 80 V current, in TAE running buffer containing 1 mM EDTA. The DNA bands were visualized with ethidium bromide.
Cytotoxicity
Human embryonic kidney cells (293T) were obtained from the Food Industry Research and Development Institute (Hsinchu, Taiwan) and cultured in Dulbecco’s modified Essential medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin. Cells were seeded in 96-well microtiter plates at a density of 1×104 cells/well, 24 hours prior to incubation with the copolymer solutions, in concentrations ranging from 16 to 1,000 μg·mL−1. Cells were further incubated with 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) reagent for another 4 hours to allow formazan crystal formation. Formazan crystals were dissolved with DMSO (200 μL), and absorbance values were read at 570 nm using a Synergy HT plate reader (Bio Tek Instruments Inc, Winooski, VT, USA). Cell viability was defined by:
|
|
Transfection efficiency
293T cells were seeded in 24-well plates, at a density of 1×105 cells/well, 24 hours prior to testing. The media was withdrawn, and the attached cells were washed with phosphate-buffered saline (PBS). Polyplexes were prepared by mixing appropriate amounts of PA or PA-I with DNA and then adding CaCl2. Two different polymer concentrations, 30 μg/50 μL and 40 μg/50 μL, were prepared with filtered nonserum media and were designated as PA30, PA40, PA-I30, and PA-I40. Plasmid solution, at 1 μg/50 μL, was added dropwise to the copolymer solution and incubated for 2.5 hours. In the Ca2+-containing samples, an equal volume of CaCl2 solution was added and reacted for 1 hour. Then, the complex solution (100 μL) was added to the wells and incubated with 293T cells. After 4 hours transfection, the media was removed from the wells, and serum media (500 μL) was added after washing with PBS. As a positive control, 25 kD PEI (at optimized copolymer-to-DNA ratio of 10) was used, according to the standard protocol. Attached cells were assayed for fluorescence expression using a fluorescence microscope (Leica Microsystems, Wetzlar, Germany).
Calcium-induced decomplexation by gel retardation assay
Polyplexes formed (as described above), at copolymer-to-DNA ratios of 30 and 40, were incubated along with 15 mM of calcium chloride at 37°C for up to 4 hours. As a control, naked plasmid DNA was incubated in 15 mM of calcium chloride for the same time frame. Next, the samples were loaded into the wells of a 0.6% agarose gel, and electrophoresis was carried out using 100 V current in TAE running buffer containing 1 mM EDTA. The DNA bands were visualized with ethidium bromide.
Results and discussion
Synthesis and characterization of PA and PA-I copolymers
The polycondensation of EDTA and EOEM afforded PA polymers, as illustrated in Figure 1. Next, a thermally initiated polycyclodehydration reaction of PA resulted in ring closure and the formation of PA-I copolymers. The characteristics (molecular weight and polydispersity index) of the prepared polymers, which were examined by gel permeation chromotography, are shown in Table 1. The obtained PA and PA-I were characterized by 1H NMR as shown in Figure 2. In Figure 2A, the ethylene protons of EDTA are identified by chemical shifts at 2.71, 3.23, and 3.36 ppm. A sharp peak at 3.5 ppm and the triplet peaks at 3.42 and 3.27 ppm correspond to ethylene protons from the EOEM segment. The amide NH is present further downfield, at 8.0 ppm. In Figure 2B, the imidization of PA is shown to have resulted in cyclic imide groups, which resonate at 3.3 ppm. The resonance at 3.9 ppm represents the ethylene protons in proximity to EDTA, while the ethylene protons of EOEM are present at 3.27, 3.50, and 3.42 ppm. The FT-IR spectra for PA and PA-I are shown in Figure 3. The amide bands at 1,554 and 1,645 cm−1 can be attributed to the amide bonds in PA but were absent in the PA-I spectra, supporting our notion that the preparation conditions led to almost complete cyclization. The C=O groups of the imide rings in PA-I gave rise to bands at 1,669 and 1,776 cm−1, indicating a cyclic five-membered ring and confirming ring closure. The disappearance of the –COOH peak at 1,720 cm−1 further verified the cyclization and formation of the polyimide ring. These results are in agreement with earlier studies.26–28
  | Figure 2 1H NMR spectra of (A) PA and (B) PA-I. |
  | Figure 3 FT-IR spectra of PA and PA-I. |
Zeta potential and DNA binding
The zeta potential is an important consideration in gene delivery because carrier charge is closely related to transfection efficiency and toxicity. The zeta potentials for 0.1% PA and PA-I copolymer solutions were -52.1 and -22.8 mV, respectively. Carboxylate groups on the PA conferred negative charge to the polymer, and the formation of the PA-I ring increased the zeta potential. Unconverted carboxylic acids may have been the contributing factor to the negative zeta potential of PA-I, which is consistent with the FT-IR results. Excessive conversion of PA to PA-I would render the product insoluble due to the high degree of crosslinking between PA-I chains.29 We found that processing conditions of 100°C for 3 hours resulted in acceptable water-soluble copolymers. The conversion degree of PA-I was calculated from relative functional groups of PA and PA-I:30
| (2) |
where A1,380 cm−1 represents the absorption reference imide peak at 1,380 cm−1 due to C-N stretching vibration, and A1,500 cm−1 represents the absorption peak at 1,500 cm−1 due to C-C stretching vibration on the monomer backbone. The relative band area was calculated using curve fitting, in Origin software. The conversion degree of PA-I was 36.9 % and was used for the following study. Complex formation between the copolymers and DNA was examined by agarose gel electrophoresis, as shown in Figure 4. Copolymer to DNA ratios are written as N/P, which indicate the molar ratio of nitrogen (N) in the polymers to phosphate (P) in DNA. As the copolymer/DNA (N/P) ratio increased, the binding strength between the copolymer and DNA increased, thereby limiting the mobility of DNA during electrophoresis. Poor band retardation was observed in the PA samples due to repulsion between the highly negatively charged PA and the like-charged DNA. However, near retardation of DNA was observed at a mixing ratio above 30 as compared with the mobility of free DNA for both PA and PA-I. One possible explanation is that, in addition to electrostatic interactions, hydrophobic interactions and hydrogen bonding drive the formation of stable complexes; therefore, DNA retention is observed despite the negatively charged characters of PA and PA-I. The hydrophobic modification of chitosan increased the retardation of DNA, due to hydrophobic interactions and particle formation.31 Similar success in DNA binding was observed with negatively charged poloxamine nanospheres and was attributed to its amphiphilic nature of DNA.32 In another study, the incorporation of hydrophobic cholesterol into a water-soluble lipoplex enhanced luciferase transfection efficiency by increasing polymer-DNA interaction.33 More directly, hydrophobic groups were conjugated to PEI, and the transfection efficiency positively correlated with the degree of modification, up to a certain level.34 N/P ratios of 30 and 40 displayed complete retardation for both PA and PA-I, and these ratios were selected for subsequent studies.
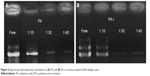  | Figure 4 Agarose gel electrophoresis retardation of (A) PA and (B) PA-I, at various polymer/DNA weight ratios. |
Cytotoxicity of copolymers
Cytotoxicity is a major concern in gene delivery and arises from parameters such as molecular weight and cationic charge density.35 Prior to transfection, it is crucial to verify whether the copolymer alone is toxic to cells. Cytotoxicity studies of the PA and PA-I copolymers, with PEI as a control, were performed on human embryonic kidney (293T) cells, and the effects of various concentrations of copolymers (16–1,000 μg·mL−1) were studied (Figure 5). As anticipated, the PEI copolymer exhibited significant toxicity, with 52.3%±6.6% cell viability observed at a concentration of 16 μg·mL−1. In contrast, greater than 90% cell viability (93.5%±2.5% and 92.5%±3.6% for PA and PA-I, respectively) was observed for the synthesized copolymers at concentrations up to 500 μg·mL−1. This is in agreement with the accepted knowledge that cytotoxicity is most strongly associated with extensive cationic charge density. The zeta potential of PEI was approximately +35 mV. Positively charged carriers are known to aggregate on cell surfaces and disturb cellular functions, thereby leading to significant cell death.36 These results demonstrate that both materials are nontoxic to cells and reveal no significant difference in the cytotoxicities of PA and PA-I.
  | Figure 5 Cytotoxicity of PA, PA-I, and PEI at various concentrations, for 293T cells. |
Transfection efficiency
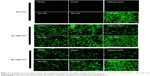
The transfection efficiency of PA and PA-I copolymer-DNA complexes was studied alone and in the presence of divalent cations, with PEI as the positive control, as shown in Figures 6. DNA coupled with Ca2+ exhibited increased transfection compared with naked DNA, in accord with literature reports.13,37,38 Ca2+ was selected because compared with other divalent cations, including Mg2+, a more dramatic increase in transfection efficiency was observed, mainly due to a smaller hydrodynamic radius.39–40 Although PA and PA-I copolymers coupled with DNA alone showed inferior transfection efficiency, the addition of 15 mM Ca2+ resulted in a synergistic effect. In all cases, PA-I exhibited significantly higher transfection efficiency than did PA. The extensive negative charge density present in PA may have prevented complex formation between the like-charge DNA and PA copolymer. In Figure 7, PA-I40 with 15 mM Ca2+ resulted in the highest fluorescence intensity. Since EDTA is a divalent ion-chelating agent, the EDTA derivatives in PA and PA-I may interact strongly with Ca2+, therefore strengthening the copolymer/Ca2+/DNA polyplex. However, the addition of 30 mM Ca2+ led to a decrease in transfection efficiency. This may be attributed to the fact that although calcium was shown to condense copolymer/DNA complexes by interacting with both amines and phosphate, it also acted simultaneously as a competitive inhibitor of amine/phosphate interactions at high concentration.41 Srinivasan et al showed that excessive Ca2+ concentration (beyond 25 mM) led to particle aggregation and decreased transfection efficiency; therefore, the concentration of cations needs to be adequately controlled.39 On the other hand, PA-I significantly outperformed PA in terms of transfection efficiency, in the presence of 15 mM Ca2+. This result, taken with the zeta potential values, conforms to expectations of the effect of moderated negative charge. A higher N/P ratio also corresponded to higher transfection efficiency, due to the formation of more compact polyplexes. This is in agreement with the retardation trend observed in the gel retardation experiment.
DNA-release ability
In order to realize the superior transfection efficiency of PA-I and PA copolymers coupled with DNA in the presence of divalent calcium cations, the release of plasmid DNA from polyplexes was evaluated by agarose gel electrophoresis. Figure 8 shows the migration of DNA, in both PA-I and PA groups, in the presence of divalent calcium cations. These results suggested that DNA can be released from both PA and PA-I polyplexes in Ca2+ solution and that DNA was more efficiently released from PA-I polyplexes. These results were consistent with fluorescence expression assays. These experiments also showed that the cationic part of polyimide promoted the dissociation of polymer/DNA complex (PA-I/DNA) to provide high gene transfection efficiency after incubation with cells. Moreover the particle size and zeta potential of polyplexes were also examined in the presence and absence of calcium ions, as shown in Table 2. The size of polyplexes was approximately 150 nm for DNA/PA and DNA/PA-I. The size changed dramatically when calcium ions was added to the solution. The increase in particle size indicated disruption of polyplexes by calcium ions. In addition, the zeta potential of the polyplexes did not change significantly in the presence of calcium ions.
  | Table 2 The particle size and zeta potential of DNA/PA and DNA/PA-I, in the presence and absence of calcium ion |
Interaction of Ca2+ with PA
The above evidence showed that decomplexation of DNA polyplexes was induced in the presence of calcium ions and therefore resulted in efficient DNA delivery. The interaction of Ca2+ and the copolymer was examined by FT-IR. EDTA is a versatile agent that can form four or six bonds with a metal ion. In the calcium complex, [Ca(EDTA)]2−, EDTA is a tetradentate ligand, and chelation involves two nitrogen atoms and two oxygen atoms. In our study, PA was polymerized from EDTA and EOEM, with EDTA derivatives still present in the polymer chain. Therefore, PA and PA-I could be chelated with calcium ion and release DNA from the complex. The EDTA-chelate-calcium and PA-chelated-calcium structures were shown in Figure 9. In Figure 10, the absorption spectra of Ca2+-free and Ca2+-binding forms of PA was characterized by FT-IR. The amide bonds of PA exhibited bands at 1,569 and 1,652 cm−1. When Ca2+ was added to PA, a new band was found at 1,588 cm−1, which was assigned to the Ca2+-binding COO– group.42 This result proved that calcium ion was coordinated with the COO– group of PA. This coordination between polymer and metal ion is likely the reason why DNA can be released from polymer complexes in the presence of calcium ion.
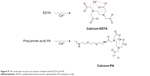  | Figure 9 The molecule structure of calcium chelated with EDTA and PA. |
  | Figure 10 FT-IR spectra of PA and Ca2+-binding PA. |
Conclusion
In summary, we designed a water-soluble PA-I that is nontoxic and exhibits potential to be utilized as a nonviral gene carrier. Through thermal cyclization, the negatively charged PA was converted to the more moderate PA-I, which exhibited low cytotoxicity and high transfection efficiency. The addition of Ca2+ significantly increased transfection, due to the possible formation of a ternary polyplex containing PA-I, Ca2+, and DNA. The devised system exhibited dual control based on the imidization conditions and Ca2+ concentration. In the past, the use of poly(imides) as delivery vehicles for biomolecules has been limited by their poor water solubility. The development of water-soluble PA-I therefore broadens their applications and initiates further explorations of PA-Is as biomaterials.
Acknowledgment
The authors would like to thank the Ministry of Science and Technology of the Republic of China (Taiwan) and National Taiwan University of Science and Technology – Taipei Medical University Joint Research Program, for financially supporting this work (grant numbers NSC 100-2221-E-007-011-030-MY3 and TUM-NTUST-103-08).
Disclosure
The authors report no conflicts of interest in this work.
References
Moghimi SM, Symonds P, Murray JC, Hunter AC, Debska G, Szewczyk A. A two-stage poly(ethylenimine)-mediated cytotoxicity: implications for gene transfer/therapy. Mol Ther. 2005;11(6):990–995. | ||
Grandinetti G, Ingle NP, Reineke TM. Interaction of poly(ethylenimine)-DNA polyplexes with mitochondria: implications for a mechanism of cytotoxicity. Mol Pharm. 2011;8(5):1709–1719. | ||
Zelphati O, Uyechi LS, Barron LG, Szoka FC. Effect of serum components on the physico-chemical properties of cationic lipid/oligonucleotide complexes and on their interactions with cells. Biochim Biophys Acta. 1998;1390(2):119–133. | ||
Zhang Y, Anchordoquy TJ. The role of lipid charge density in the serum stability of cationic lipid/DNA complexes. Biochim Biophys Acta. 2004;1663(1–2):143–157. | ||
Zintchenko A, Philipp A, Dehshahri A, Wagner E. Simple modifications of branched PEI lead to highly efficient siRNA carriers with low toxicity. Bioconjug Chem. 2008;19(7):1448–1455. | ||
Seow WY, Liang K, Kurisawa M, Hauser CA. Oxidation as a facile strategy to reduce the surface charge and toxicity of polyethyleneimine gene carriers. Biomacromolecules. 2013;14(7):2340–2346. | ||
Kyriakides TR, Cheung CY, Murthy N, Bornstein P, Stayton PS, Hoffman AS. pH-sensitive polymers that enhance intracellular drug delivery in vivo. J Control Release. 2002;78(1–3):295–303. | ||
Schlegel A, Largeau C, Bigey P, et al. Anionic polymers for decreased toxicity and enhanced in vivo delivery of siRNA complexed with cationic liposomes. J Control Release. 2011;152(3):393–401. | ||
Harvie P, Wong FM, Bally MB. Characterization of lipid DNA interactions. I. Destabilization of bound lipids and DNA dissociation. Biophys J. 1998;75(2):1040–1051. | ||
Prokop A, Kozlov E, Moore W, Davidson JM. Maximizing the in vivo efficiency of gene transfer by means of nonviral polymeric gene delivery vehicles. J Pharm Sci. 2002;91(1):67–76. | ||
Mozafari MR, Zareie MH, Piskin E, Hasirci V. Formation of supramolecular structures by negatively charged liposomes in the presence of nucleic acids and divalent cations. Drug Deliv. 1998;5(2):135–141. | ||
Khosravi-Darani K, Mozafari MR, Rashidi L, Mohammadi M. Calcium based non-viral gene delivery: an overview of methodology and applications. Acta Med Iran. 2010;48(3):133–141. | ||
Roy I, Mitra S, Maitra A, Mozumdar S. Calcium phosphate nanoparticles as novel non-viral vectors for targeted gene delivery. Int J Pharm. 2003;250(1):25–33. | ||
Patil SD, Rhodes DG, Burgess DJ. Anionic liposomal delivery system for DNA transfection. AAPS J. 2004;6(4):e29. | ||
Lam AM, Cullis PR. Calcium enhances the transfection potency of plasmid DNA-cationic liposome complexes. Biochim Biophys Acta. 2000;1463(2):279–290. | ||
Moradi-Araghi A, Doe PH. Hydrolysis and precipitation of polyacrylamides in hard brines at elevated temperatures. SPE Reservoir Evaluation and Engineering. 1987;2(02):189–198. | ||
Hasegawa M, Arai H, Mita I, Yokota R. Isothermal imidization of an aromatic polyimide precursor studied by fluorescence spectroscopy. Polym J. 1990;22(10):875–882. | ||
Tsai HC, Kuo WJ, Hsiue GH. Highly thermal stable main-chain nonlinear optical polyimide based on two-dimensional carbazole chromophores. Macromol Rapid Commun. 2005;26(12):986–991. | ||
Richardson RR, Miller JA, Reichert WM. Polyimides as biomaterials: preliminary biocompatibility testing. Biomaterials. 1993;14(8):627–635. | ||
Rousche PJ, Pellinen DS, Pivin DP, Williams JC, Vetter RJ, Kipke DR. Flexible polyimide-based intracortical electrode arrays with bioactive capability. IEEE Trans Biomed Eng. 2001;48(3):361–371. | ||
Chung H, Seo J, Kim E, et al. In vivo biocompatibility and stability of polyimide microelectrode array for retinal stimulation. Invest Ophthalmol Vis Sci. 2003;44:5072. Abstract. | ||
Linde HG. Adhesive interface interactions between primary aliphatic amine surface conditioners and polyamic acid/polyimide resins. J Polym Sci A Polym Chem. 1982;20(4):1031–1041. | ||
Zhu Z, Cardin CJ, Gan Y, Colquhoun HM. Sequence-selective assembly of tweezer molecules on linear templates enables frameshift-reading of sequence information. Nat Chem. 2010;2(8):653–660. | ||
Riva R, Ragelle H, des Rieux A, Duhem N, Jérôme C, Préat V. Chitosan and chitosan derivatives in drug delivery and tissue engineering. In: Jayakumar R, Prabaharan M, Muzzarelli RAA, editors. Advances in Polymer Science: Chitosan for Biomaterials II. Dordrecht: Springer-Verlag Berlin Heidelberg; 2011:19–44. | ||
Li B, Liu T, Zhong WH. High modulus aliphatic polyimide from 1, 3-diaminopropane and ethylenediaminetetraacetic dianhydride: Water soluble to self-patterning. Polymer. 2011;52(22):5186–5192. | ||
Lee T, Park SS, Jung Y, et al. Preparation and characterization of polyimide/mesoporous silica hybrid nanocomposites based on water-soluble poly(amic acid) ammonium salt. Eur Polym J. 2009;45(1):19–29. | ||
Yang CP, Chen YC, Hsiao SH, Guo W, Wang HM. Optically transparent and colorless poly(ether imide)s derived from a phenylhydroquinone bis(ether anhydride) and various trifluoromethyl-substituted bis(ether amine)s. J Polym Res. 2010;17:779–788. | ||
Nohara LB, Costa ML, Alves MA, Takahashi MFK, Nohara EL, Rezende MC. Processing of high performance composites based on peek by aqueous suspension prepregging. Mat Res. 2010;13(2):245–252. | ||
Woo Y, Oh SY, Kang YS, Jung B. Synthesis and characterization of sulfonated polyimide membranes for direct methanol fuel cell. J Membr Sci. 2003;220(1–2):31–45. | ||
Nishino T, Kotera M, Inayoshi N, Miki N, Nakamae K. Residual stress and microstructures of aromatic polyimide with different imidization processes. Polymer. 2000;41(18):6913–6918. | ||
Son S, Chae SY, Choi C, et al. Preparation of a hydrophobized chitosan oligosaccharide for application as an efficient gene carrier. Macromol Res. 2004;12(6):573–580. | ||
Pitard B, Bello-Roufaï M, Lambert O, et al. Negatively charged self-assembling DNA/poloxamine nanospheres for in vivo gene transfer. Nucleic Acids Res. 2004;32(20):e159. | ||
Han So, Mahato RI, Kim SW. Water-soluble lipopolymer for gene delivery. Bioconjug Chem. 2001;12(3):337–345. | ||
Teo PY, Yang C, Hedrick JL, et al. Hydrophobic modification of low molecular weight polyethylenimine for improved gene transfection. Biomaterials. 2013;34(32):7971–7979. | ||
Fischer D, Li Y, Ahlemeyer B, Krieglstein J, Kissel T. In vitro cytotoxicity testing of polycations: influence of polymer structure on cell viability and hemolysis. Biomaterials. 2003;24(7):1121–1131. | ||
Kircheis R, Wightman L, Wagner E. Design and gene delivery activity of modified polyethylenimines. Adv Drug Deliv Rev. 2001;53(3):341–358. | ||
Jordan M, Wurm F. Transfection of adherent and suspended cells by calcium phosphate. Methods. 2004;33(2):136–143. | ||
Tang J, Chen JY, Liu J, et al. Calcium phosphate embedded PLGA nanoparticles: a promising gene delivery vector with high gene loading and transfection efficiency. Int J Pharm. 2012;431(1–2):210–221. | ||
Srinivasan C, Burgess DJ. Optimization and characterization of anionic lipoplexes for gene delivery. J Control Release. 2009;136(1):62–70. | ||
Kulkarni VI, Shenoy VS, Dodiya SS, Rajyaguru TH, Murthy RR. Role of calcium in gene delivery. Expert Opin Drug Deliv. 2006;3(2):235–245. | ||
Baoum A, Xie SX, Fakhari A, Berkland C. “Soft” calcium crosslinks enable highly efficient gene transfection using TAT peptide. Pharm Res. 2009;26(12):2619–2629. | ||
Mizuguchi M, Nara M, Kawano K, Nitta K. FT-IR study of the Ca2+-binding to bovine alpha-lactalbumin. Relationships between the type of coordination and characteristics of the bands due to the Asp COO– groups in the Ca2+-binding site. FEBS Lett. 1997;417(1):153–156. |
 © 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.