Back to Journals » International Journal of Nanomedicine » Volume 16
CAFs-Derived Exosomal miRNA-130a Confers Cisplatin Resistance of NSCLC Cells Through PUM2-Dependent Packaging
Authors Zhang T, Zhang P, Li HX
Received 13 July 2020
Accepted for publication 29 September 2020
Published 25 January 2021 Volume 2021:16 Pages 561—577
DOI https://doi.org/10.2147/IJN.S271976
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Linlin Sun
Tao Zhang,1 Ping Zhang,2 Hong-Xia Li3
1Department of Tuberculosis, Linyi People’s Hospital, Linyi, Shandong 276034, People’s Republic of China; 2Reproductive Medicine, Linyi People’s Hospital, Linyi, Shandong 276034, People’s Republic of China; 3Endoscopic Room, Linyi Chest Hospital, Linyi, Shandong 276034, People’s Republic of China
Correspondence: Hong-Xia Li
Endoscopic Room, Linyi Chest Hospital, No. 233 Fenghuang Avenue, Hedong District, Linyi, Shandong 276034, People’s Republic of China
Tel +86-13265485365
Email [email protected]
Purpose: Chemoresistance is a significant barrier to the treatment and management of non-small cell lung cancer (NSCLC). Exosomes play an essential role in intercellular communication. Understanding the mechanism underlying the role of tumor stroma, especially cancer-associated fibroblasts (CAFs), during chemoresistance would significantly contribute to the clinical application of chemotherapy agents.
Results: In this study, we demonstrated that NSCLC-derived CAFs were innately resistant to cisplatin treatment and CAFs-conditioned medium significantly promoted the survival rate of NSCLC cells after cisplatin treatment. Additionally, CAFs-derived exosomes were taken up by NSCLC cells. Moreover, exosomal miRNA-130a was transferred from CAFs to recipient NSCLC cells and knockdown of miRNA-130a reversed the effect of CAFs-derived exosomes during chemoresistance of NSCLC cells. Furthermore, pumilio homolog 2 (PUM2), a RNA-binding protein, mediated the packaging of miRNA-130a into exosomes. The overexpression and knockdown of PUM2 promoted and inhibited tumor growth of xenograft mice, respectively.
Conclusion: Taken together, these results suggest that CAFs-derived exosomes confer cisplatin resistance of NSCLC cells through transferring miRNA-130a and that PUM2 is a critical factor for packaging miRNA-130a into exosomes. This study indicates that CAFs-derived exosomal miRNA-130a may be a potential therapeutic target for cisplatin resistance in NSCLC.
Keywords: chemoresistance, non-small cell lung cancer, cancer-associated fibroblasts, exosome, miRNA-130a, pumilio homolog 2
Introduction
Lung cancer has emerged as the leading cause of cancer-associated deaths worldwide, accounting for about 20% of all cancer deaths.1 In 2018, approximately 2.09 million new lung cancer cases took place, ranking first in all cancer types.2,3 In China, lung cancer mortality is higher than in most other countries, and it is estimated that lung cancer mortality may surge nearly 40% between 2015 and 2030.4 As the primary type of lung cancer, non-small cell lung cancer (NSCLC) exhibits multiple histological types, such as bronchoalveolar carcinoma, large cell carcinoma, squamous cell carcinoma, and adenocarcinoma.5 To date, platinum-based chemotherapy is the most effective therapy to treat advanced NSCLC patients,6 with a significantly higher survival and response rate.7,8 Alternatively, cisplatin (cis-Diamminedichloro-platinum(II))-based therapeutic strategy has a relatively low response rate, significant side effects, and acquired or intrinsic chemoresistance.9,10 As such, it is urgent to improve the efficacy of chemotherapy to NSCLC and explore the molecular mechanism underlying cisplatin resistance of NSCLC.
Cancer-associated fibroblasts (CAFs) have been demonstrated to be essential stromal components in the tumor microenvironment, regulating cancer cell growth, migration, invasion, and chemo-response through various mechanisms.11 Chemoresistance, either acquired or intrinsic, is associated with a complicated and multifactorial process, including decreases of intracellular drugs, changes of drug targets, aberrant regulation of cell survival or death, and crosstalks between tumor microenvironment and cancer cells.12,13 Growing evidence recently reveals that CAFs play a critical role in the regulation of chemoresistance in various cancer types. For example, CAFs can secrete cytokines or growth factors to promote tumorigenesis and chemoresistance in breast cancer.14 In addition, CAFs-derived interleukin 6 (IL6) is essential for chemoresistance through increasing C-X-C motif chemokine receptor 7 (CXCR7) expression.15 Moreover, increased secretion of epidermal growth factor sufficiently insensitizes neighboring tumor cells to cetuximab treatment via the mitogen-activated protein kinases (MAPK) pathway.16 Furthermore, CAFs have been reported to promote cisplatin resistance in bladder cancer cells through activating the IGF-1/ERβ/Bcl-2 signaling pathway.17
Given the importance of CAFs in chemoresistance of cancers, the mechanism underlying how CAFs influence the cellular activities, especially the communication between CAFs and cancer cells, has drawn considerable attention from both laboratory and clinical research fields. Over the past decade, it has been demonstrated that exosomes are essential molecule/information transporters in intercellular communication, including the interaction between tumor cells and CAFs.18,19 Exosomes, membrane-derived microvesicles (30–150 nm), are secreted by almost all cell types and are extensively distributed in various body fluids.20 Exosomes can transport bioactive components, such as proteins, non-coding RNAs, DNA fragments, mRNAs, and lipids, from donor cells to neighboring cells, thereby impacting the physiological activities of recipient cells.21,22 Among these functional components, microRNAs (miRNAs), a class of approximately 22-nucleotide non-coding RNAs, have been extensively reported to play a critical role in exosome-mediated cell-to-cell communication.23,24 For example, CAFs-derived exosomal miRNA-106b can promote gemcitabine resistance of pancreatic cancer cells through silencing TP53INP1.25 In addition, exosomal miRNA-522 secreted from CAFs enhances acquired chemoresistance to cisplatin in gastric cancer.26 In addition, exosomal miRNA-21 derived from CAFs contribute to oxaliplatin resistance in colorectal cancer.27
To the best of our knowledge, the role of CAFs-derived exosomal miRNAs during chemoresistance of NSCLC cells has not been thoroughly investigated. Therefore, this study aimed to determine the effect of CAFs-derived exosomes during the chemoresistance of NSCLC cells. In this study, we demonstrated that CAFs-derived exosomes significantly improved the survival rate of NSCLC cells in response to cisplatin treatment. In addition, the experiments suggest that miRNA-130a is a functional miRNA which mediates the role of CAFs-derived exosomes and that pumilio homolog 2 (PUM2) may be a key factor for packaging miRNA-130a into exosomes.
Materials and Methods
Ethics Statement
In this study, written informed consent was offered by the participants before enrollment. The experimental protocols were approved by the Ethics Committee of Linyi chest Hospital and were conducted based on the Declaration of Helsinki. All animal studies were approved by the Institute Animal Care and Use Committee of Linyi chest Hospital and were performed according to the Guide for the Care and Use of Laboratory Animals.28 The mice used in this study were raised in the medicine animal facilities of Linyi chest Hospital.
Samples and Patients
All clinical samples were collected from Linyi chest Hospital. Twenty pairs of tumor and adjacent normal tissues were obtained from twenty patients who were diagnosed with NSCLC based on the evidence of pathological histology from 2016–2018. Table 1 shows the patient information. None of these patients underwent chemotherapy and radiotherapy before the surgery. The medical records and follow-up data of these NSCLC patients were retrospectively reviewed, and pathological and clinical characteristics were also respectively reviewed.
 |
Table 1 Patient Characteristics |
Cell Culture
The NSCLC cell lines, A549 and HCC827, were obtained from the Cell Bank of China Academy of Sciences (Shanghai, China). To establish the cisplatin-resistant cell line, A549-R, A549 cells were treated with a stepwise increasing concentration of cisplatin (Sigma-Aldrich, USA) as previously described.29,30 The CAFs and normal fibroblasts (NFs) were isolated from NSCLC tissues and adjacent normal tissues by primary culture, as previously described, respectively.31,32 Both third-generation purified CAFs and NFs were further identified by detecting CAFs-specific markers, including α-smooth muscle actin (α-SMA; Thermo, USA), fibroblast activation protein (FAP; Thermo, USA), and ferroptosis suppressor protein 1 (FSP1; Abcam, China). The A549 and HCC827 cells were cultured in RPMI 1640 medium (GIBCO-BRL, USA) supplemented with 10% fetal bovine serum (FBS; GIBCO-BRL, USA), 100 U/mL penicillin, and 100 μg/mL streptomycin in humidified air with 5% CO2 at 37 °C. The coculture system was created using transwell membranes with 0.4 μm pores (Corning, USA) in a 6-well plate. The CAFs (1 × 105) were treated with 20 μM GW4869 (Sigma-Aldrich, China) for 24 hours and placed above the membranes. The A549 and HCC827 cells were placed below the permeable membranes. The cells were placed in the coculture system for 72 hours in humidified air with 5% CO2 at 37 °C.
Western Blots
Exosomes or cells were collected at specific times, and total proteins were isolated using SDS lysis buffer (Sigma-Aldrich, China). Total proteins (20 μg) were separated by polyacrylamide gels (15%/10%) and then transferred onto PVDF membranes (Merck Millipore, USA). The blots were blocked with 5% bovine serum albumin (BSA; Sigma-Aldrich, China) at room temperature for 1 hour and incubated with primary antibodies in a microplate shaker overnight at 4 °C. The expression of β-actin was used throughout as a loading control, and all other primary antibodies were used according to the manufacturer’s instructions. Next, the membranes were incubated with corresponding secondary antibodies and the protein bands were detected using the Bio-Rad Chemidoc system (Bio-Rad Laboratories, USA).
Real-Time PCR
Total RNAs were isolated from cells, exosomes, and tissues using the TRIzol reagent (Invitrogen, USA), and first-strand cDNAs were synthesized using the Prime Script RT reagent Kit (Takara, Japan) according to the manufacturer’s instructions. The miRNA cDNAs were synthesized from total RNAs of cells or tissues using the miRNA First-Strand cDNA Synthesis Kit (TIANGEN, China). Real-time PCR for mRNAs was performed using the SYBR Premix Ex Taq Reagent Kit (Takara, Japan) on a 7500 Real-Time PCR Platform (Applied Biosystems, USA). Real-time PCR for miRNA was performed using the miRcute Plus miRNA qPCR Detection Kit (TIANGEN, China). β-actin and U6 were used as internal controls for analyzing relative expressions of RNAs and miRNAs, respectively. The 2−ΔΔCt method was carried out to analyze relative expressions.33 Table 2 summarizes the primer information.
 |
Table 2 Primer Information |
MiRNA Array
The NFs- and CAFs-derived exosomes were used to determine the miRNA expression profile. Exosomal RNAs were isolated as the method mentioned above. The purification and concentration of RNAs were determined by NanoDrop ND-2000 (Thermo Scientific, USA). Exosomal RNA was labeled using FlashTag™ Biotin HSR RNA Labeling Kits (Thermo Scientific, USA) followed by the hybridization of labeled exosomal RNA. After hybridization, the array was scanned and the data was processed. Analysis for miRNA expression profile was performed using the 7G platform Affymetrix GeneChip Command Console Software (version 4.0). The miRNA expression profile was shown in Volcano Plotting. The miRNAs had ≥2-fold change and adjusted P < 0.05 were considered as differentially expressed miRNAs.
Immunofluorescence Assay
Cells (1 × 105) cultured on cover slips were fixed with 4% paraformaldehyde (Thermo Fisher Scientific, China), permeabilized with 0.1% Triton X-100 (Abcam, China), and then blocked in 3% BSA (Sigma-Aldrich, China). Thereafter, processed cells were incubated with primary antibodies at 4 °C overnight and then incubated with corresponding secondary antibodies for 30 mins at room temperature in the dark. Subsequently, samples were treated with 4′,6-diamidino-2-phenylindole (DAPI; Invitrogen, USA) to label the nuclei. Cells were imaged using a SP8 laser-scanning confocal microscope (Leica Microsystems, Germany).
MTT Assay
Cell viability was determined using the MTT assay. Cells were administered a treatment as indicated and then seeded in a 96-well plate (5000 cells per well) in triplicate. Twelve hours later, cells were treated with different concentrations of cisplatin for 72 hours. Next, the viability of cells was determined using the MTT assay kit (Abcam, USA) according to the manufacturer’s instructions. Absorbance was read at 490 nm. Meanwhile, cell proliferation was determined by seeding cells in a 96-well plate (2000 cells per well) in triplicate after pretreatment as indicated. Cell growth was determined daily by recording the absorbance at 490 nm in a plate reader (Molecular Devices, USA).
Conditioned Medium
Cells (1 × 105) were seeded in a culture dish for 24 hours and then the culture medium was replaced with serum-free DMEM (GIBCO-BRL, USA) and incubated for 48 hours. Conditioned medium (CM) with exosomes was prepared by removing exosomes through successively centrifuging at 300×g for 20 mins, 2000×g for 20 mins, and 12,000×g for 70 mins. Cisplatin-treated cells CM was prepared by culturing cells in serum-free DMEM supplemented with 10 μM cisplatin.
Exosome Isolation and Identification
Exosomes were isolated from CM with different pretreatments through successively centrifuging steps.34,35 The morphology and size distribution of exosomes were determined by FEI Tecnai G2 Spirit transmission electron microscopy (Thermo Scientific, USA) and nanoparticle tracking analysis (NTA), respectively, as previously described.36,37 Exosomal protein was determined using the BCA Protein Assay Kit (Abcam, China).
Exosomes Immunofluorescence
Exosomes were labeled with the lipophilic dye, DiO (Biotium, USA), according to the manufacturer’s instructions. The A549 cells (1 × 105) seeded on cover slips were cocultured with labeled exosomes for 24 hours in an 8-well plate. The CAFs with Cy3-labeled miRNA-130a (Biocompare, USA) were cocultured with A549 cells for 48 hours in an 8-well plate. The cytoskeleton of A549 cells was labeled with TRITC Phalloidin or FITC Phalloidin (Sigma-Aldrich, China) according to the manufacturer’s instructions. Cells were then used for the immunofluorescence assay as mentioned above.
Cell Transfection
The cDNA of PUM2 and EIF4B (with 3′ UTR) was synthesized using PrimeSTAR™ HS DNA Polymerase (Takara, Japan) and then were inserted into the PGMLV-6395 vector (Genomeditech, China). The miRNA-130a mimics, miRNA-130a mimics negative control (miRNA-130a mimics-NC), miRNA-130a inhibitor, miRNA-130a inhibitor negative control (miRNA-130a inhibitor-NC) (QIAGEN, China), and siRNA-PUM2 (Genomeditech, China) were transfected to cells using the Lipofectamine™ 3000 Transfection Reagent according to the manufacturer’s instructions (Invitrogen, USA).
MiRNA Pull-down and RNA Immunoprecipitation Chip (RIP) assays
The miRNA Pull-down assay was performed as previously described.38,39 The RNA Immunoprecipitation Chip (RIP) assay was carried out using the Magna RIP RNA-Binding Protein Immunoprecipitation Kit according to the manufacturer’s instructions (Millipore SiGMa, USA).
Xenograft Mouse Model
Male C57BL/6 athymic nude mice (6 weeks old; Jackson Labs, USA) were housed in a pathogen-free animal room at 24 °C with a 12 h dark-light cycle in the medicine animal facilities of Linyi chest Hospital under ad libitum feeding conditions. Mice were subcutaneously injected with a mixture of A549 cells (2 × 105) with PUM2-expressing or PUM2-knockdown CAFs (1 × 105) into the left and right buttocks (n=5). Tumor volume was monitored every three days. Tumor volume was calculated using the formula: V = (L ×W2) × 0.5. After 24 days, mice were sacrificed and the tumor weight was measured.
Statistical Analysis
Data were shown as means ± SEM. The statistical analyses were performed using SPSS 19.0 software (SPSS, USA) and GraphPad Prism (GraphPad Software, USA). The differences between two or more groups were analyzed using one-way ANOVA or Student’s t-test. Tukey’s HSD test was performed for pairwise comparisons when the interaction was significant. P < 0.05 was considered statistically significant.
Results
NSCLC-Derived CAFs are Chemoresistant to Cisplatin
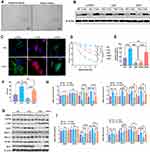
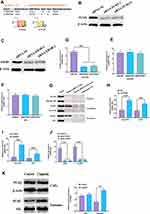
The CAFs from NSCLC tissues and their adjacent tissues were first isolated. The morphology of CAFs and NFs displayed a spindle-like shape and were adherent in culture (Figure 1A). Western blotting immunofluorescence staining assays revealed that CAFs expressed higher levels of the fibroblast markers, α-SMA, FAP, and FSP1, relative to NFs (Figure 1B and C). After treating with the same dosage of cisplatin, CAFs had a higher survival rate than those of NFs, A549, HCC827, and cisplatin-resistant A549 cells (A549-R) (Figure 1D). Also, CAFs retained the highest level of proliferation in response to cisplatin treatment, compared with those of NFs and A549-R, while the most cisplatin-vulnerable cell lines, A549 and HCC827, showed the lowest retained cell proliferation (Figure 1E). Next, the highest cisplatin IC50 was found in CAFs, and A549 cells had the lowest IC50 value (Figure 1F). The mechanism underlying cisplatin resistance is associated with multiple processes, such as reduced drug uptake, increased drug efflux or detoxification, alteration of DNA repair, or antiapoptotic proteins.40,41 Next, Western blots were used to detect the protein expressions of several cisplatin resistance-associated regulators. As shown in Figure 1G and H, the protein expressions of XIAP, ERCC1, ERCC4, GSTK1, and Bcl-2 were higher in CAFs than those of NFs, A549, and A549-R cells, whereas CAFs had the lowest protein level of CTR1 than the other three cell types. Also, the similar mRNA expression profile of these genes was observed in CAFs, NFs, A549, and A549-R cells (Figure 1I). The ERCC1/4 system, nucleotide excision repair factors, displays a negative correlation with cisplatin-based survival response in NSCLC cells.42,43 In this study, ERCC1 and ERCC4 were knockdown by specific siRNAs (Figure 2A, B, D, and E), and the knockdown of ERCC1 or ERCC4 resulted in reduced cisplatin resistance in CAFs (Figure 2C and F). Collectively, these observations suggest that NSCLC-derived CAFs are innately chemoresistant to cisplatin.
CAFs-Derived Exosomes Confer Cisplatin Resistance of NSCLC Cells
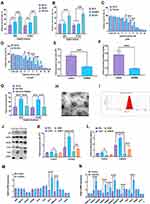
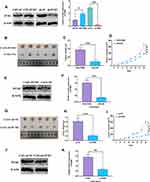
Given the innate cisplatin resistance of CAFs, further investigation of whether CAFs-derived bioactive factors could impact the cellular activities of NSCLC cells was conducted. The present study demonstrated that CAFs-conditioned medium (CM-CAFs) significantly increased the survival rate in A549 and HCC827 cells, compared with CM-NFs and CM-NSCLC cells (CM-CN) (Figure 3A). Also, A549-R-conditioned medium (CM-A549-R) did not exert a similar function to promote the survival rate in NSCLC cells in response to cisplatin treatment (Figure 3B). The effect of CM-CAFs was detected in NSCLC cells in a wide dosage range of cisplatin, namely, 2–8 uM in A549 cells and 1–16 uM in HCC827 cells (Figure 3C and D). Exosomes have emerged as essential message-transporters between CAFs and cancer cells.27,44 Thus, it was speculated that CAFs-derived exosomes might play an essential role in the effect of CM-CAFs on NSCLC cells. First, GW4869, a neutral sphingomyelinase inhibitor for blocking exosome generation45 was applied to CM-CAFs, which showed that GW4869 significantly reduced the number of exosomes as well as exosomal protein in CM-CAFs (Figure 3E and F). Furthermore, the results showed that the addition of GW4869 abolished the effect CM-CAFs on both A549 and HCC827 cells under cisplatin treatment (Figure 3G), which suggests that exosomes participate in CM-CAFs-induced cisplatin resistance in NSCLC cells. Next, the exosomes were isolated from CM-CAFs through the ultracentrifugation assay and nanoparticle tracking analysis. Transmission electron microscopy was applied to determine the morphology and size of the exosomes. As shown in Figure 3H and I, CAFs-derived exosomes displayed typical spherical shape and were 90–130 nm in diameter. Also, Western blots revealed that CAFs-derived exosomes positively expressed exosome markers, including Alix, HSP70, HSP90, CD63, and Rab5, but not the negative marker, GRP94 (Figure 3J). Notably, compared with NFs, A549, and A549-R, CAFs generated the highest number of exosome and exosomal protein in the presence of cisplatin treatment (Figure 3K and L). Given the complexity of the biogenesis of exosomes, this multi-step process includes multivesicular bodies (MVBs) transportation, docking, and fusion with plasma membranes, of which several essential molecule families, such as SNARE and RAB, play an essential role.46,47 In the present study, mRNA expressions of several critical regulators for the generation of exosomes were determined and the results showed that the mRNA expressions of CD63, nSMase2, RAB35, RAB5A, RAB9A, RAB27A, RAB7, and YKT6 were increased in cisplatin-treated CAFs (Figure 3M and N), further indicating that cisplatin may activate an exosome-generation system in CAFs in response to cisplatin treatment.
CAFs-Derived Exosomes are Taken Up by NSCLC Cells
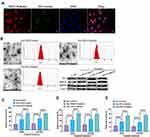
To determine whether CAFs-derived exosomes were taken up by A549 and HCC827 cells, exosomes were labeled with DiO and cocultured with A549 and HCC827 cells for 24 hours. As shown in Figure 4A, DiO-labeled green exosomes were taken up by both A549 and HCC827 cells. By applying the same protocols mentioned above, the characteristics of exosomes derived from CAFs control, cisplatin-treated CAFs and NFs were verified, respectively (Figure 4B). Subsequently, the results revealed that exosomes derived from cisplatin-treated CAFs displayed a more potent effect on the survival rate of A549 and HCC827 cells in the presence of cisplatin, compared with exosomes derived from CAFs without cisplatin treatment (Figure 4C) and A549-R cells (Figure 4D) and NFs with cisplatin treatment (Figure 4E). Together, these data suggest that CAFs or cisplatin-treated CAFs-derived exosomes were internalized by NSCLC cells and promoted the survival rate after cisplatin treatment.
Exosomal miRNA-130a is Transferred from CAFs to NSCLC Cells
Accumulating evidence suggests that exosomes can transfer a group of bioactive molecules such as miRNAs, lncRNAs, and proteins from CAFs to cancer cells, influencing cellular activities of recipient cells.23,27 Next, miRNA array analysis was used to determine the functional molecules, particularly miRNAs that contributed to the effect of CAFs-derived exosomes on NSCLC cells. The results showed that miRNA-130a significantly increased CAFs-derived exosomes, compared with those derived from CAFs without cisplatin treatment (Figure 5A). Meanwhile, real-time PCR revealed that the expression of miRNA-130a was higher in cisplatin-treated CAFs than in CAFs without cisplatin treatment (Figure 5B). Also, the level of miRNA-130a was higher in A549 and HCC827 cells treated with exosomes derived from cisplatin-treated CAFs, relative to those treated with exosomes derived from NSCLC cells own mediums (Figure 5C). Intriguingly, miRNA-130a was increased in NSCLC tissue relative to adjacent normal tissues (Figure 5D). Furthermore, CAFs innately expressed a higher level of miRNA-130a compared with those of NFs, A549, HCC827, and A549-R cells (Figure 5E).
To investigate whether miRNA-130a was transferred from CAFs to NSCLC cells through exosomes, A549 and HCC827 cells were cultured in CM-CAFs or CM-CAFs with the addition of GW4869. The results showed that the expression of miRNA-130a was increased in A549 and HCC827 cells cultured in CM-CAFs, while the increased level of miRNA-130a was reversed by the addition of GW4869 (Figure 5F). Additionally, CAFs transfected with Cy3-labeled miRNA-130a were cocultured with A549 and HCC827 cells for 48 hours and Cy3-labeled green miRNA-130a was found in NSCLC cells (Figure 5G). Also, the expression of miRNA-130a did not change in CM-CAFs with RNaseA, while was dramatically decreased when Triton X-100 was added to CM-CAFs (Figure 5H), suggesting that miRNA-130a was primarily encased within the membrane structures. Moreover, there was no difference in the level of miRNA-130a between CM-CAFs and CAFs-derived exosomes (Figure 5I). Collectively, miRNA-130a was transferred from CAFs to recipient NSCLC cells through exosomes. To determine the function of miRNA-130a in CAFs-derived exosomes, CAFs were transfected with miRNA-130a mimics and inhibitors and the exosomes were isolated and then identified using the same methods mentioned above (Figure 5J). The expression level of miRNA-130a was increased and decreased in exosomes derived from CAFs with overexpression of miRNA-130a or miRNA-130a knockdown, respectively (Figure 5K and L). Functionally, the survival rate was increased in NSCLC cells treated with exosomes derived from CAFs or CAFs with overexpression of miRNA-130a, while exosomes derived from CAFs with miRNA-130a knockdown abolished this effect (Figure 5M and N). These results collectively suggest that CAFs-derived exosomes promote the survival rate of NSCLC cells after cisplatin treatment through exosomal miRNA-130a.
Exosomal miRNA-130a Packaging is Associated with PUM2
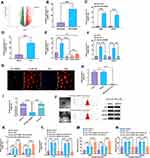
To determine the mechanism underlying the packaging of exosomal miRNA-130a in CAFs, a database of RBP specificities (RBPDB)48 was used to analyze the interaction between miRNA-130a and motifs of RNA-binding proteins (RBPs). The results revealed that PUM2 and eukaryotic translation initiation factor 4B (EIF4B) motifs displayed specific miRNA-130a binding sites (Figure 6A). Next, specific siRNAs were applied to knockdown PUM2 and EIF4B in CAFs (Figure 6B and C) and showed that PUM2 knockdown decreased the level of exosomal miRNA-130a while the level of cellular miRNA-130a did not change (Figure 6D and F). Also, knockdown of EIF4B did not alter the levels of exosomal miRNA-130a (Figure 6E). Furthermore, the mechanical interactions between miRNA-130a and PUM2 in both cytoplasm and exosomes, but not in the nucleus, and these interactions were disrupted when the sequence of miRNA-130a was mutated (Figure 6G). Moreover, the RIP assay was performed in cell and exosome lysates of CAFs which showed that miRNA-130a enriched the PUM2 antibody, relative to the IgG group (Figure 6H). Also, there was more miRNA-130a enrichment in cell and exosome lysates of cisplatin-treated CAFs compared with those of CAFs without cisplatin treatment (Figure 6I). Functionally, the level of exosomal miRNA-130a was decreased in CAFs with PUM2 knockdown (Figure 6J). In addition, cisplatin increased the level of PUM2 protein in exosomes derived from CAFs but did not alter PUM2 protein expression in CAFs (Figure 6K). Collectively, these findings indicate that cisplatin treatment may promote the packaging of miRNA-130a in exosomes of CAFs through the mediation of PUM2.
Effect of CAFs-Derived PUM2 on Tumor Growth in vivo
To determine the effect of CAFs-derived PUM2 on tumor growth, A549 plus CAFs with PUM2 overexpression/knockdown was used in xenograft mice (Figure 7A). The results showed that A549 plus CAFs with PUM2 overexpression was associated with larger tumor weights and faster tumor growth (Figure 7B–D), whereas tumors formed by A549 plus CAFs with PUM2 knockdown displayed smaller tumor weights and slower growth (Figure 7G–I). Also, Western blots revealed that the level of PUM2 was upregulated in tumor tissues treated with A549 plus CAFs with PUM2 overexpression (Figure 7E and F), while was downregulated in those treated with A549 plus CAFs with PUM2 knockdown (Figure 7J and K).
Discussion
Chemoresistance is one of the primary challenges for the treatment and management of cancers. Given its heterogeneous nature, the mechanism related to chemoresistance is complex and is not comprehensively understood, thereby cancer cell resistance to chemotherapeutic agents continues to pose a significant barrier for all oncologists and their patients.49 A previous study revealed that nearly 63% (1409/2227) and 68% (1056/1565) of NSCLC patients’ tumor cultures are extreme or intermediate resistant to cisplatin and carboplatin, respectively.50 Beyond that, these NSCLC samples display a different degree of resistance to other drugs, including doxorubicin, gemcitabine, docetaxel, vinorelbine, etoposide, paclitaxel, and topotecan.50 Thus, all NSCLC patients will ultimately develop chemoresistance to drugs, even with a satisfactory initial response.51 To overcome this barrier, there is an urgent need to develop novel chemotherapy agents as well as to explore the molecular mechanism behind the chemoresistance of NSCLC.
Accumulating evidence has demonstrated that CAFs are the most abundant stromal cells in the tumor microenvironment, playing an essential role in the response of cancer cells to chemotherapy.52,53 In the present study, NSCLC-derived CAFs are innately resistant to cisplatin treatment and that CAFs-CM significantly improved the survival rate and proliferation of NSCLC cell lines, A549 and HCC827, in the presence of cisplatin. Furthermore, after coculturing with CAFs, A549 and HCC827 cells were insensitive to cisplatin. These observations together suggest that CAFs confers with NSCLC cells on the resistance to chemotherapeutic agents. A similar role of CAFs in chemoresistance has also been reported in other cancer types, such as ovarian cancer,54 colorectal cancer,55 and gastric cancer.56 However, due to the complexity and heterogeneity of intercellular communication between CAFs and cancer cells, the molecular mechanism underlying how CAFs regulate chemotherapy resistance in cancers is still not well documented.
Growing evidence has suggested that CAFs can impact cancer progression through various bioactive factors, including growth factors,57 cytokines,31 and chemokines.58 In addition, exosome-mediated intercellular interaction between CAFs and cancer cells has been reported to be involved in the chemoresistance of tumor cells.59,60 In the present study, we found that CAFs-derived exosomes promoted the survival rate of NSCLC cells in response to cisplatin and that exosome secretion inhibitor, GW4869, significantly reversed the effect of CAFs-derived exosomes on chemoresistance of NSCLC cells. Numerous studies reported that CAFs-derived exosomes are essential for the regulation of chemoresistance in various cancers, including bladder cancer,61 ovarian cancer,62 and colorectal cancer.55 Furthermore, cisplatin treatment significantly upregulated the generation of exosomes from CAFs. Therefore, these findings reported may provide a novel CAFs-targeting therapeutic strategy for NSCLC.
To further investigate the mechanism by which CAFs-derived exosomes promoted cisplatin resistance of NSCLC cells, miRNA array analysis was performed to determine the functional miRNAs that contributed to the effect of CAFs-derived exosomes on NSCLC cells. In the present study, CAFs-derived exosomal miRNA-130a played an important role in endowing cisplatin resistance to NSCLC. As a multi-functional factor, miRNA-130a participates in various cellular activities in cancer cells, for example, autophagy,63 proliferation,64 and apoptosis.65 MiRNA-130a has been reported to be an oncogenic factor, whose upregulation in NSCLC tissue is positively correlated to poor prognosis, lymph node metastasis, and stage of tumor node metastasis.66 Meanwhile, miRNA-130a has been reported to play an important role in chemoresistance of various cancer types.67 Particularly, miRNA-130a is upregulated in hepatoma cells in the presence of cisplatin, which is associated with the involvement of RUNX3/Wnt signaling.68 Also, the downregulation of miRNA-130a can attenuate the cisplatin resistance in ovarian cancer cells through MDR1/P-glycoprotein pathways.69 In another study reported by Zhang et al, X-linked inhibitor of apoptosis (XIAP) is identified to be a direct targeting gene of miRNA-130a to confer cisplatin resistance in ovarian cancer.70 These findings collectively demonstrate that the role of miRNA-130a may be involved in a complex regulatory network and displays a cancer type-dependent manner. Given its important role in chemoresistance, miRNA-130a is a potential therapeutic target for developing new drugs for NSCLC treatment.
Given the upregulation of miRNA-130a in CAFs-derived exosomes after cisplatin treatment, the next goal was to investigate how miRNA-130a was packaged into exosomes. It has been demonstrated that extracellular miRNAs are encased in small-size membranous vesicles or packaged with RNA-binding proteins.71 Through bioinformatics analysis, two RBPs, PUM2 and EIF4B, had binding sequences to bind miRNA-130a. Further functional experiments demonstrated that PUM2 knockdown, not EIF4B, in CAFs significantly downregulated the level of miRNA-130a in CAFs-derived exosomes and recipient NSCLC cells after cisplatin treatment. Meanwhile, the upregulation of PUM2 promoted tumor growth of xenograft mice while knockdown of PUM2 exerted the opposite role. These results suggested that PUM2 may play an essential role in packaging miRNA-130a into exosomes, thereby indirectly impacting the abundance and function of miRNA-130a in CAFs, exosomes, and NSCLC cells. Thus, PUM2 may act as a target to inhibit the oncogenic effect of miRNA-130.
Conclusion
In conclusion, the results demonstrate that NSCLC-associated CAFs were innately resistant to cisplatin, and CAFs-derived exosomes conferred chemoresistance of NSCLC cells through transferring miRNA-130a. Also, PUM2 was associated with packaging miRNA-130a into exosomes. Thus, CAFs-derived exosomal miRNA-130a may be a potential therapeutic target for chemoresistance in NSCLC.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Disclosure
The authors declare no conflicts of interest.
References
1. Ferlay J, Ervik M, Lam F, et al. Global Cancer Observatory: Cancer Today. Lyon, France: International Agency for Research on Cancer; 2018.
2. Barta JA, Powell CA, Wisnivesky JP. Global epidemiology of lung cancer. Ann Glob Health. 2019;85(1). doi:10.5334/aogh.2419
3. Cao M, Chen W. Epidemiology of lung cancer in China. Thorac Cancer. 2019;10(1):3–7. doi:10.1111/1759-7714.12916
4. Martín-Sánchez JC, Lunet N, González-Marrón A, et al. Projections in breast and lung cancer mortality among women: a Bayesian analysis of 52 countries worldwide. Cancer Res. 2018;78(15):4436–4442. doi:10.1158/0008-5472.CAN-18-0187
5. Lerouge D, Rivière A, Dansin E, et al. A Phase II study of cisplatin with intravenous and oral vinorelbine as induction chemotherapy followed by concomitant chemoradiotherapy with oral vinorelbine and cisplatin for locally advanced non-small cell lung cancer. BMC Cancer. 2014;14(1):1–9. doi:10.1186/1471-2407-14-231
6. Jakopovic M, Thomas A, Lopez-Chavez A. From platinum compounds to targeted therapies in advanced thoracic malignancies. Anticancer Res. 2014;34(1):477–482.
7. Bunn PA
8. d‘Amato TA, Landreneau RJ, Ricketts W, et al. Chemotherapy resistance and oncogene expression in non–small cell lung cancer. J Thorac Cardiovasc Surg. 2007;133(2):352–363. doi:10.1016/j.jtcvs.2006.10.019
9. Florea A-M, Büsselberg D. Cisplatin as an anti-tumor drug: cellular mechanisms of activity, drug resistance and induced side effects. Cancers (Basel). 2011;3(1):1351–1371. doi:10.3390/cancers3011351
10. D’addario G, Früh M, Reck M, et al. Metastatic non-small-cell lung cancer: ESMO clinical practice guidelines for diagnosis, treatment and follow-up. Ann Oncol. 2010;21(suppl_5):v116–v119. doi:10.1093/annonc/mdq189
11. Xing F, Saidou J, Watabe K. Cancer associated fibroblasts (CAFs) in tumor microenvironment. Front Biosci. 2010;15:166. doi:10.2741/3613
12. Morin PJ. Drug resistance and the microenvironment: nature and nurture. Drug Resist Updat. 2003;6(4):169–172. doi:10.1016/S1368-7646(03)00059-1
13. Shi W-J, Gao J-B. Molecular mechanisms of chemoresistance in gastric cancer. World J Gastrointest Oncol. 2016;8(9):673. doi:10.4251/wjgo.v8.i9.673
14. Kikuchi K, McNamara KM, Miki Y, et al. Effects of cytokines derived from cancer-associated fibroblasts on androgen synthetic enzymes in estrogen receptor-negative breast carcinoma. Breast Cancer Res Treat. 2017;166(3):709–723. doi:10.1007/s10549-017-4464-5
15. Qiao Y, Zhang C, Li A, et al. IL6 derived from cancer-associated fibroblasts promotes chemoresistance via CXCR7 in esophageal squamous cell carcinoma. Oncogene. 2018;37(7):873–883. doi:10.1038/onc.2017.387
16. Garvey CM, Lau R, Sanchez A, et al. Anti-EGFR therapy induces EGF secretion by cancer-associated fibroblasts to confer colorectal cancer chemoresistance. Cancers (Basel). 2020;12(6):1393. doi:10.3390/cancers12061393
17. Long X, Xiong W, Zeng X, et al. Cancer-associated fibroblasts promote cisplatin resistance in bladder cancer cells by increasing IGF-1/ERβ/Bcl-2 signalling. Cell Death Dis. 2019;10(5):1–16. doi:10.1038/s41419-019-1581-6
18. Yang X, Li Y, Zou L, Zhu Z. Role of exosomes in crosstalk between cancer-associated fibroblasts and cancer cells. Front Oncol. 2019;9:356. doi:10.3389/fonc.2019.00356
19. Liu L, Zhang Z, Zhou L, et al. Cancer associated fibroblasts-derived exosomes contribute to radioresistance through promoting colorectal cancer stem cells phenotype. Exp Cell Res. 2020;391(2):111956. doi:10.1016/j.yexcr.2020.111956
20. Pascual M, Ibáñez F, Guerri C. Exosomes as mediators of neuron-glia communication in neuroinflammation. Neural Regen Res. 2020;15(5):796. doi:10.4103/1673-5374.268893
21. Zhang J, Li S, Li L, et al. Exosome and exosomal microRNA: trafficking, sorting, and function. Genomics Proteomics Bioinformatics. 2015;13(1):17–24. doi:10.1016/j.gpb.2015.02.001
22. Schorey JS, Bhatnagar S. Exosome function: from tumor immunology to pathogen biology. Traffic. 2008;9(6):871–881. doi:10.1111/j.1600-0854.2008.00734.x
23. Sun Z, Shi K, Yang S, et al. Effect of exosomal miRNA on cancer biology and clinical applications. Mol Cancer. 2018;17(1):147. doi:10.1186/s12943-018-0897-7
24. Melo SA, Sugimoto H, O’Connell JT, et al. Cancer exosomes perform cell-independent microRNA biogenesis and promote tumorigenesis. Cancer Cell. 2014;26(5):707–721. doi:10.1016/j.ccell.2014.09.005
25. Fang Y, Zhou W, Rong Y, et al. Exosomal miRNA-106b from cancer-associated fibroblast promotes gemcitabine resistance in pancreatic cancer. Exp Cell Res. 2019;383(1):111543. doi:10.1016/j.yexcr.2019.111543
26. Zhang H, Deng T, Liu R, et al. CAF secreted miR-522 suppresses ferroptosis and promotes acquired chemo-resistance in gastric cancer. Mol Cancer. 2020;19(1):1–17. doi:10.1186/s12943-020-01168-8
27. Yang F, Ning Z, Ma L, et al. Exosomal miRNAs and miRNA dysregulation in cancer-associated fibroblasts. Mol Cancer. 2017;16(1):1–10. doi:10.1186/s12943-017-0718-4
28. Albus U. Guide for the Care and Use of Laboratory Animals.
29. Plasencia C, Martinez-Balibrea E, Martinez-Cardus A, Quinn D, Abad A, Neamati N. Expression analysis of genes involved in oxaliplatin response and development of oxaliplatin-resistant HT29 colon cancer cells. Int J Oncol. 2006;29(1):225–235.
30. De Caceres II, Cortes-Sempere M, Moratilla C, et al. IGFBP-3 hypermethylation-derived deficiency mediates cisplatin resistance in non-small-cell lung cancer. Oncogene. 2010;29(11):1681–1690. doi:10.1038/onc.2009.454
31. Qin X, Yan M, Wang X, et al. Cancer-associated fibroblast-derived IL-6 promotes head and neck cancer progression via the osteopontin-NF-kappa B signaling pathway. Theranostics. 2018;8(4):921. doi:10.7150/thno.22182
32. Ji X, Ji J, Shan F, Zhang Y, Chen Y, Lu X. Cancer-associated fibroblasts from NSCLC promote the radioresistance in lung cancer cell lines. Int J Clin Exp Med. 2015;8(5):7002.
33. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–408. doi:10.1006/meth.2001.1262
34. Hu Y, Yan C, Mu L, et al. Fibroblast-derived exosomes contribute to chemoresistance through priming cancer stem cells in colorectal cancer. PLoS One. 2015;10(5):e0125625. doi:10.1371/journal.pone.0125625
35. Chairoungdua A, Smith DL, Pochard P, Hull M, Caplan MJ. Exosome release of β-catenin: a novel mechanism that antagonizes Wnt signaling. J Cell Biol. 2010;190(6):1079–1091. doi:10.1083/jcb.201002049
36. Wei Y, Lai X, Yu S, et al. Exosomal miR-221/222 enhances tamoxifen resistance in recipient ER-positive breast cancer cells. Breast Cancer Res Treat. 2014;147(2):423–431. doi:10.1007/s10549-014-3037-0
37. Qu J-L, Qu X-J, Zhao M-F, et al. Gastric cancer exosomes promote tumour cell proliferation through PI3K/Akt and MAPK/ERK activation. Dig Liver Dis. 2009;41(12):875–880. doi:10.1016/j.dld.2009.04.006
38. Santangelo L, Giurato G, Cicchini C, et al. The RNA-binding protein SYNCRIP is a component of the hepatocyte exosomal machinery controlling microRNA sorting. Cell Rep. 2016;17(3):799–808. doi:10.1016/j.celrep.2016.09.031
39. Tan SM, Lieberman J. Capture and identification of miRNA targets by biotin pulldown and RNA-seq. In: Post-Transcriptional Gene Regulation. New York: Humana Press; 2016:211–228.
40. Ferreira JA, Peixoto A, Neves M, et al. Mechanisms of cisplatin resistance and targeting of cancer stem cells: adding glycosylation to the equation. Drug Resist Updat. 2016;24:34–54. doi:10.1016/j.drup.2015.11.003
41. Galluzzi L, Senovilla L, Vitale I, et al. Molecular mechanisms of cisplatin resistance. Oncogene. 2012;31(15):1869–1883. doi:10.1038/onc.2011.384
42. Fujii T, Toyooka S, Ichimura K, et al. ERCC1 protein expression predicts the response of cisplatin-based neoadjuvant chemotherapy in non-small-cell lung cancer. Lung Cancer. 2008;59(3):377–384. doi:10.1016/j.lungcan.2007.08.025
43. Ning J, Jiao Y, Xie X, et al. miR‑138‑5p modulates the expression of excision repair cross‑complementing proteins ERCC1 and ERCC4, and regulates the sensitivity of gastric cancer cells to cisplatin. Oncol Rep. 2019;41(2):1131–1139.
44. Donnarumma E, Fiore D, Nappa M, et al. Cancer-associated fibroblasts release exosomal microRNAs that dictate an aggressive phenotype in breast cancer. Oncotarget. 2017;8(12):19592. doi:10.18632/oncotarget.14752
45. Essandoh K, Yang L, Wang X, et al. Blockade of exosome generation with GW4869 dampens the sepsis-induced inflammation and cardiac dysfunction. Biochim Biophys Acta Mol Basis Dis. 2015;1852(11):2362–2371. doi:10.1016/j.bbadis.2015.08.010
46. Kowal J, Tkach M, Théry C. Biogenesis and secretion of exosomes. Curr Opin Cell Biol. 2014;29:116–125. doi:10.1016/j.ceb.2014.05.004
47. van Niel G, D’Angelo G, Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol. 2018;19(4):213.
48. Cook KB, Kazan H, Zuberi K, Morris Q, Hughes TR. RBPDB: a database of RNA-binding specificities. Nucleic Acids Res. 2010;39(suppl_1):D301–D308. doi:10.1093/nar/gkq1069
49. Sarkaria JN, Kitange GJ, James CD, et al. Mechanisms of chemoresistance to alkylating agents in malignant glioma. Clin Cancer Res. 2008;14(10):2900–2908. doi:10.1158/1078-0432.CCR-07-1719
50. d’Amato TA, Landreneau RJ, McKenna RJ, Santos RS, Parker RJ. Prevalence of in vitro extreme chemotherapy resistance in resected nonsmall-cell lung cancer. Ann Thorac Surg. 2006;81(2):440–447. doi:10.1016/j.athoracsur.2005.08.037
51. Chang A. Chemotherapy, chemoresistance and the changing treatment landscape for NSCLC. Lung Cancer. 2011;71(1):3–10. doi:10.1016/j.lungcan.2010.08.022
52. Pietras K, Östman A. Hallmarks of cancer: interactions with the tumor stroma. Exp Cell Res. 2010;316(8):1324–1331. doi:10.1016/j.yexcr.2010.02.045
53. Mahale J, Smagurauskaite G, Brown K, Thomas A, Howells LM. The role of stromal fibroblasts in lung carcinogenesis: a target for chemoprevention? Int J Cancer. 2016;138(1):30–44. doi:10.1002/ijc.29447
54. Leung CS, Yeung T-L, Yip K-P, et al. Cancer-associated fibroblasts regulate endothelial adhesion protein LPP to promote ovarian cancer chemoresistance. J Clin Invest. 2018;128(2):589–606. doi:10.1172/JCI95200
55. Ren J, Ding L, Zhang D, et al. Carcinoma-associated fibroblasts promote the stemness and chemoresistance of colorectal cancer by transferring exosomal lncRNA H19. Theranostics. 2018;8(14):3932. doi:10.7150/thno.25541
56. Ma J, Song X, Xu X, Mou Y. Cancer-associated fibroblasts promote the chemo-resistance in gastric cancer through secreting IL-11 targeting JAK/STAT3/Bcl2 pathway. Cancer Res Treat. 2019;51(1):194. doi:10.4143/crt.2018.031
57. Straussman R, Morikawa T, Shee K, et al. Tumour micro-environment elicits innate resistance to RAF inhibitors through HGF secretion. Nature. 2012;487(7408):500–504. doi:10.1038/nature11183
58. Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med. 2013;19(11):1423–1437. doi:10.1038/nm.3394
59. Richards KE, Zeleniak AE, Fishel ML, Wu J, Littlepage LE, Hill R. Cancer-associated fibroblast exosomes regulate survival and proliferation of pancreatic cancer cells. Oncogene. 2017;36(13):1770–1778. doi:10.1038/onc.2016.353
60. Deng X, Ruan H, Zhang X, et al. Long noncoding RNA CCAL transferred from fibroblasts by exosomes promotes chemoresistance of colorectal cancer cells. Int J Cancer. 2020;146(6):1700–1716. doi:10.1002/ijc.32608
61. Zhuang J, Shen L, Yan J, Guo H. MP58-08 cancer-associated fibroblasts secreted exosomal miR-146a promotes bladder cancer chemoresistance. J Urol. 2018;199(4S):e775–e776. doi:10.1016/j.juro.2018.02.1840
62. Yeung CLA, Co -N-N, Tsuruga T, et al. Exosomal transfer of stroma-derived miR21 confers paclitaxel resistance in ovarian cancer cells through targeting APAF1. Nat Commun. 2016;7(1):1–14.
63. Kovaleva V, Mora R, Park YJ, et al. miRNA-130a targets ATG2B and DICER1 to inhibit autophagy and trigger killing of chronic lymphocytic leukemia cells. Cancer Res. 2012;72(7):1763–1772. doi:10.1158/0008-5472.CAN-11-3671
64. Lee SH, Jung YD, Choi YS, Lee YM. Targeting of RUNX3 by miR-130a and miR-495 cooperatively increases cell proliferation and tumor angiogenesis in gastric cancer cells. Oncotarget. 2015;6(32):33269. doi:10.18632/oncotarget.5037
65. Lu K, Shen H, Zhu S, Bi S, Wu S. Effects of miRNA-130a on the proliferation and apoptosis of glioma cell lines. Oncol Lett. 2018;16(2):2478–2482.
66. Wang X-C, Du L-Q, Zhang H, et al. Expression of miRNA-130a in nonsmall cell lung cancer. Am J Med Sci. 2010;340(5):385–388. doi:10.1097/MAJ.0b013e3181e892a0
67. Jiang L-H, Sun D-W, Li J, Ji Z-L. The role of miR-130a in cancer. Breast Cancer. 2017;24(4):521–527. doi:10.1007/s12282-017-0776-x
68. Xu N, Shen C, Luo Y, et al. Upregulated miR-130a increases drug resistance by regulating RUNX3 and Wnt signaling in cisplatin-treated HCC cell. Biochem Biophys Res Commun. 2012;425(2):468–472. doi:10.1016/j.bbrc.2012.07.127
69. Yang L, Li N, Wang H, Jia X, Wang X, Luo J. Altered microRNA expression in cisplatin-resistant ovarian cancer cells and upregulation of miR-130a associated with MDR1/P-glycoprotein-mediated drug resistance. Oncol Rep. 2012;28(2):592–600. doi:10.3892/or.2012.1823
70. Zhang X, Huang L, Zhao Y, Tan W. Downregulation of miR-130a contributes to cisplatin resistance in ovarian cancer cells by targeting X-linked inhibitor of apoptosis (XIAP) directly. Acta Biochim Biophys Sin. 2013;45(12):995–1001. doi:10.1093/abbs/gmt113
71. Liang H, Zhang J, Zen K, Zhang C-Y, Chen X. Nuclear microRNAs and their unconventional role in regulating non-coding RNAs. Protein Cell. 2013;4(5):325–330. doi:10.1007/s13238-013-3001-5
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.