Back to Journals » Pharmacogenomics and Personalized Medicine » Volume 14
BRCA2 3ʹ-UTR Polymorphism rs15869 Alters Susceptibility to Papillary Thyroid Carcinoma via Binding hsa-mir-1178-3p
Authors Guo N, Qu P, Li H, Liu L, Jin H, Liu R, Zhang Z, Zhang X, Li Y, Lu X , Zhao Y
Received 6 January 2021
Accepted for publication 13 April 2021
Published 6 May 2021 Volume 2021:14 Pages 533—544
DOI https://doi.org/10.2147/PGPM.S300783
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Martin H Bluth
Nan Guo,1 Peng Qu,2 Hao Li,2 Liuli Liu,2 Hao Jin,3 Renqi Liu,3 Zhen Zhang,3 Xuan Zhang,2 Yingchun Li,4 Xiaobo Lu,2 Yuejiao Zhao1
1Department of Head and Neck Surgery, Cancer Hospital of China Medical University, Liaoning Cancer Hospital & Institute, Shenyang, People’s Republic of China; 2Department of Toxicology, School of Public Health, China Medical University, Shenyang, People’s Republic of China; 3Jin Zhou Center for Disease Control and Prevention, Jinzhou, People’s Republic of China; 4Department of Central Laboratory, Cancer Hospital of China Medical University, Liaoning Cancer Hospital & Institute, Shenyang, People’s Republic of China
Correspondence: Yuejiao Zhao
Department of Head and Neck Surgery, Cancer Hospital of China Medical University, Liaoning Cancer Hospital & Institute, No. 44 Xiaoheyan Road, Dadong District, Shenyang, 110042, Liaoning Province, People’s Republic of China
Tel +86 24 31916833
Email [email protected]
Xiaobo Lu
Department of Toxicology, School of Public Health, China Medical University, No.77 Puhe Road, Shenyang North New Area, Shenyang, 110122, Liaoning Province, People’s Republic of China
Tel +86 24 31939077
Email [email protected]
Objective: To investigate the associations of polymorphisms in the following DNA double-strand break repair (DSBR) genes with papillary thyroid carcinoma (PTC) risk (including RAD51 rs11852786, RAD51B rs963917, BRCA1 rs12516 and rs8176318, BRCA2 rs15869, XRCC4 rs2035990 and XRCC5 rs2440).
Materials and Methods: A matched case–control study was implemented to examine associations between PTC risk and the above polymorphisms. Subsequently, we evaluated the effects of the potential PTC susceptibility-related variant rs15869 on BRCA2 mRNA secondary structure and BRCA2 expression through bioinformatics analysis and experiment validation. Additionally, luciferase assay was used to identify whether rs15869 polymorphism can substantially affect the binding of hsa-miR-1178-3p to BRCA2 mRNA. Finally, Pearson correlation analysis was performed to determine the correlation between the expression of hsa-miR-1178-3p and BRCA2 mRNA and protein in thyroid tissues harboring rs15869 different genotypes.
Results: BRCA2 rs15869 CC genotype was associated with a higher risk of PTC than its AA genotype. Subsequently, stratified analyses came to the same conclusion in the female or age< 50 population. Furthermore, we confirmed that the A-to-C substitution of rs15869 changed BRCA2 mRNA secondary structure and contributed to a decreased BRCA2 expression. Mechanistically, a significantly decreased luciferase activity verified a greater binding between hsa-miR-1178-3p and rs15869 C allele, but not the A allele, which was evidenced by the significant negative correlation between hsa-miR-1178-3p with BRCA2 mRNA and protein levels in thyroid tissues with AC and CC genotype but not AA genotype at rs15869.
Conclusion: BRCA2 rs15869 is characterized as a potential biomarker associated with PTC risk, highlighting the contribution of the hsa-miR-1178-3p via functional exploration.
Keywords: papillary thyroid carcinoma, DNA double-strand break repair, BRCA2, rs15869 polymorphism, hsa-miR-1178-3p
Introduction
Thyroid cancer, a prevalent malignant tumor of the endocrine system, was seen the highest increase in incidence over the years and has aroused widespread concern. Notably, the diagnostic rate of thyroid cancer in the United States increased by an average of about 3.1% per year;1 thyroid cancer, in 2018, resulted in 41,071 deaths globally up from 24,000 in 1990.2,3 Papillary thyroid carcinoma (PTC) is the most common histological type responsible for up to 70% of thyroid cancer.4 Although patients with PTC respond positively to the surgical resection and thyroid-stimulating hormone suppression with a 5-year survival rate of more than 80%, early onset and lymph node metastasis increase the risk of recurrence or even death of PTC patients.5 Accordingly, patients with thyroid cancer would benefit greatly from further identification of valid early biomarkers, which is of great significance for the prevention and diagnosis of PTC.
Although the exact pathogenesis of PTC is not completely understood, ionizing radiation is known to be the only definitive risk factor for developing thyroid cancer. As a result, a series of DNA damages including base damage, intra- and inter-strand cross-linking, and double-strand breaks (DSBs) are initiated, further inducing mutation and driving tumorigenesis.6 DSBs are particularly hazardous since they can lead to chromosome fragmentation and genome rearrangements. In response to DNA damages induced by ionizing radiation, several complex DNA repair mechanisms are activated, in which some DNA repair genes are essential in the process of combating DNA damages and maintaining genomic stability. Among them, the genes in double-strand break repair (DSBR) are mainly responsible for repairing DSBs, so as to prevent the accumulation of DSBs caused by ionizing radiation and further play a protective role against tumorigenesis.7
Generally, DSBR as a complex DNA repair mechanism is achieved by both pathways including homologous recombination (HR) and non-homologous end joining (NHEJ). It is known that the former requires a homologous sequence to guide repair in mitosis S phase and G2 phase, mainly including RAD51, ATM, RAD51B, BRCA1, BRCA2 and other genes.8 Meanwhile, in NHEJ DNA Ligase IV, a specialized DNA ligase that forms a complex with the cofactor XRCC4, directly joins the two ends, and no template is needed in contrast to HR, mainly including DNA PKcs, XRCC4, XRCC5 genes. NHEJ process can be fixed at any point in time, especially the G0/G1 phase, but it is not considered a very accurate repair mechanism.9
So far, several epidemiology studies have identified some variants of DSBR genes, termed single nucleotide polymorphisms (SNPs), in humans associated with thyroid cancer. As exemplified by ATM, the apical regulators of the response to DSBs, ATM G5557A associated with a decreased risk of PTC (OR=0.69, 95% CI: 0.45–0.86), whereas ATM IVS22-77 T>C increased the risk of sporadic PTC (OR=1.84, 95% CI: 1.10–3.24);10 Fayaz S’s research reminded XRCC3 T241M polymorphism elucidated a significant risk with PTC, which was enhanced in combination with RAD52 2259 C>T and XRCC2 R188H polymorphisms;11 other significant data were found for Ku80 gene (Ex21-238G➝A, and Ex21+466A➝G variants) for papillary tumors (adjusted OR=2.281, 95% CI: 1.063–4.894, P=0.034).12
Although the exact mechanisms of thyroid cancer are not completely understood, the polymorphisms in DNA repair genes, leading to the varied expression of repair proteins, appear to play a crucial role in the genetic instability and progression of cancer.13 Coding variants can affect protein function, while others in the non-coding region may alter gene expression or transcription, respectively. Post-transcriptional regulation, caused by microRNAs binding to the mRNA 3ʹ-untranslated region (UTR), which acts by destabilizing target mRNAs and/or by repressing translation, has provoked great interest.14 The miRNA–mRNA interaction normally requires 6–8 base pairs of perfect complementarity between the miRNA 5ʹterminus (seed sequence) and a cognate miRNA target site in the 3ʹ-UTR.15 Unsurprisingly, polymorphisms located in 3ʹ-UTR, especially in miRNA binding sites, may affect gene expression, and such an alteration in protein activity can further modify the risk of cancer. Given their location in the putative regulatory region of DNA repair genes belonging to DSBR, the polymorphisms may lead to allele-specific changes in repair efficiency of DNA damage, which may be closely related to the hereditary susceptibility of thyroid cancer.16
In the present work, we analyzed the association between SNPs in the 3ʹ-UTR of DSBR genes (RAD51 rs11852786, RAD51B rs963917, BRCA1 rs12516 and rs8176318, BRCA2 rs15869, XRCC4 rs2035990 and XRCC5 rs2440) and the risk of PTC based on a middle-sized case–control study. Since it is necessary to determine which polymorphism out of some identified is a functional variant, in addition to the case–control study, a mechanistic exploration combining bioinformatics prediction and in vitro luciferase assay related to the effects of the candidate SNP on miRNAs binding was further performed. Overall, little is known about the exact biomarker of PTC susceptibility; our current study will contribute to characterize some new susceptibility biomarkers and elucidate the causal relationship between the target SNPs and the risk of thyroid cancer.
Materials and Methods
Study Subjects
In this study, we recruited 206 patients diagnosed with PTC from the Cancer Hospital of China Medical University, Liaoning Cancer Hospital & Institute, from January 2017 to December 2018 as our cases. Meanwhile, 206 healthy volunteers for physical examination were selected as the control. The controls had no history of cancers and were matched to the cases by age (±3 years), gender, and ethnicity and recruited in the same region and period. All activities involving human subjects were done in full compliance with the government policies and the Helsinki Declaration. Informed consent was obtained from each of the participants prior to the study after a detailed explanation about the purpose of our current study. All patients provided written informed consent, and ethical approval was obtained from the Ethics Committees of Liaoning Cancer Hospital & Institute. Each participant donated 2 mL of venous blood, while their demographic data were recorded in questionnaires in detail.
A total of 412 subjects were recruited in the present study, including 206 PTC cases and 206 gender- and age-matched controls (Table S1). The mean age of the cases was 45.5 years old and ranged from 25 to 72 years old, while the controls were characterized by a mean age of 46.6 years old and ranged from 23 to 74 years old. However, there was no significant difference between cases and controls with regard to gender (P=1.000) and age (P=0.267) (Table S1). The smoking and drinking status of the study subjects were also surveyed, while cases and controls did not show any significant difference in smoking and drinking status (Table S1). Additionally, we confirmed that the distribution of genotypes for all SNPs is according to the Hardy–Weinberg equilibrium (Table S2).
Bioinformatics Analysis
Some genes involved in DSBR and related to PTC were predicted by the Genecards database (https://www.genecards.org/). The candidate SNPs located in the 3ʹ-UTR of the above genes were determined by the Website of NCBI (https://www.ncbi.nlm.nih.gov/) and UCSC (https://genome.ucsc.edu/) according to the minor allele frequency (MAF) of the Chinese Han population in Beijing (MAF>0.2). Whether the different genotypes of SNPs can affect the secondary structure of mRNA or not was analyzed by RNAfold (http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi). Furthermore, both PolymiRTs (http://compbio.uthsc.edu/miRSNP/) and miRBase (http://www.mirbase.org) were used to predict the possible miRNAs combined with the significant SNPs; Minimum free energy (MFE) of the binding miRNAs to the specific SNPs was evaluated by RNAhybrid (https://bibiserv.cebitec.uni-bielefeld.de/rnahybrid/). HMDD (http://www.cuilab.cn/hmdd) curated experiment-supported evidence for the candidate miRNAs and disease associations; DAVID (https://DAVID.ncifcrf.gov) was used to perform a gene set from TargetScan (http://www.targetscan.org/vert_72/) prediction enrichment analysis of pathways and biological process targeted by the miRNA.
DNA Extraction and TaqMan® SNP Genotyping Assays
The genomic DNA of the blood sample was routinely extracted by phenol-chloroform extraction; the genomic DNA of thyroid tissues was extracted by DNA purification column (No. 9765, purchased from TaKaRa, Kyoto, Japan). Genotyping for RAD51 (rs11852786: C>G, assay ID C_11305569_20), RAD51B (rs963917: A>G, assay ID C_7571538_10), BRCA1 (rs12516: G>A, assay ID C_29356_10; rs8176318: C>A, assay ID C_318688_10), BRCA2 (rs15869: A>C, assay ID C_807118_10), XRCC4 (rs2035990: T>C, assay ID C_11685999_10) and XRCC5 (rs2440: A>G, assay ID C_3231046_20) were undertaken by the TaqMan SNP genotyping allelic discrimination method on a LightCycler 480 Real-time PCR system (Roche, Foster City, CA, USA). The above TaqMan hydrolysis probes were purchased from ABI Company (ABI, Stagapore, US) and PCR reagents were purchased from Roche Company. All experiments were carried out in strict accordance with the manufacturer’s instructions.
RNA Extraction and QPCR
The total RNA of thyroid tissues was isolated with the GeneJET™ RNA Purification Kit (No. K0731, purchased from TaKaRa, Kyoto, Japan). Subsequently, 1ug total RNA was reverse transcribed into cDNA (RR047a, purchased from TaKaRa, Kyoto, Japan) and QPCR (RR820a, purchased from TaKaRa, Kyoto, Japan) was carried out to amplify the cDNA with specific primers, listed in Table S3.
Western Blotting
Total protein was extracted from the thyroid tissues and its concentration was measured by BCA Protein Assay Kit (Seven, Beijing, China). The protein samples were separated by SDS-PAGE and transferred to polyvinylidene fluoride (PVDF) membranes. After blocking, the membranes were incubated with the specific primary antibody BRCA2 overnight at 4°C (1:500, abclonal, Wuhan, China; and secondary antibody for 1 h at room temperature. Protein expression levels were normalized to α-tublin (1:5000, Proteintech, Wuhan, China). Tanon-2500 automatic digital gel image analysis system was used to determine protein expression, and Image J was used to calculate the band relative intensity.
Dual Luciferase Reporter Assay
Human embryonic kidney 293T (HEK-293T) cells purchased from SIBCB (Cell Bank of the Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences, Beijing, China) were cultured in RPIM-1640 medium (Hyclone, Logan, Utah, USA), supplemented with 10% fetal bovine serum (BioInd, Beit HaEmek, Israel) in a humidified atmosphere of 5% CO2 at 37 °C.
The full-length of BRCA2 3ʹ-UTR, with genetic variation corresponding to A or C (of rs15869), was cloned into the pmirGLO reporter vector (Obio, Shanghai, China). The sequences of hsa-miR-1178-3p mimics involved in the study are as follows: UUGCUCACUGUUCUUCCCUAG (Genepharma, Suzhou, China). HEK293T cells were seeded into 96-well plates with 70% confluence and transfected 24 h later with 100nM hsa-miR-1178-3p mimics or NC was co-transfected with 100 ng wild-type vector (BRCA2-A) or the mutant vector (BRCA2-C) using Lipofectamine 3000 (Firefly: Renilla: Lipofectamine 3000 = 0.1 μg: 0.01 μg: 0.25 μL). At 48 h following transfection, the luciferase activity was measured according to the manufacturer’s protocol. Renilla luciferase activity was normalized to the luminescence of firefly luciferase. Three independent experiments were performed in triplicate.
Statistical Analysis
Data analysis was performed using IBM SPSS 20.0 (IBM Company, Armonk, NY, USA) and visualized using GraphPad Prism 5.0 software (GraphPad Software Inc, San Diego, CA, USA). Chi-square test or Fisher exact probability and logistic regression were used to assess the association of different genotypes of SNPs with the risk of PTC; further, Bonferroni correction was used to perform multiple comparisons. Taking wild type as a reference, OR (95% CI) represents the risk of other genotypes. The luciferase assay data were statistically described using the mean ± standard deviation, and t-test was performed among groups. Two tailed P<0.05 was considered statistically significant. Power analysis and sample size, version 11 (NCSS-PASS, https://www.ncss.com/) software were used to calculate the statistical power of this study. Based on the preliminary association analysis between the candidate SNPs and PTC risk, the power to detect an OR of 2.595 in 412 samples was 99.6% with a two-sided α of 0.05. It indicated the sample size of the present study was deemed sufficient to detect a difference in the group proportions.
Results
Candidate SNPs Selection
Based on the Genecards database, 13 genes in HR and 7 genes in NHEJ highly related to PTC were selected (Table S4). The candidate SNPs located in the 3ʹ-UTR of the above genes were determined by NCBI and UCSC according to MAF (MAF ≥ 0.2 in Chinese Han population) and the sample size of the present study. Accordingly, the following polymorphisms including RAD51 rs11852786, RAD51B rs963917, BRCA1 rs12516, BRCA1 rs8176318, BRCA2 rs15869, XRCC4 rs2035990 and XRCC5 rs2440 were determined in the study (Table S5).
The Effect of Candidate SNPs on the Risk of PTC
First, we evaluated the relationship between those seven polymorphisms located in five selected DSBR genes and the risk of PTC via logistic regression. The results are summarized in Table 1. People carrying rs15869 CC genotype located in the 3ʹ- UTR of BRCA2 showed an increased risk of PTC compared to those carrying homozygous AA genotype (OR=2.595, 95% CI: 1.091–6.171, P=0.031). However, there was no significance after further Bonferroni correction due to setting significance at P=0.017. The other six SNPs (rs11852786, rs963917, rs12516; rs8176318, rs2035990 and rs2440) did not manifest any significant association with PTC risk (Table 1).
 |
Table 1 The Relationship Between Candidate SNPs and PTC Risk |
Further stratified analyses were carried out by gender and age. Notably, rs15869 CC genotype was also demonstrated a significant association with an increased risk of PTC in the female population, in contrast to the reference shown in Table 2 (OR=2.756, 95% CI: 1.024–7.414, P=0.045). Additionally, the age stratified analyses reminded BRCA2 rs15869 was found to show the association with PTC risk in the age<50 population, which means minor CC genotype had a higher risk of PTC than AA genotype (OR=4.400, 95% CI: 1.177–16.444, P=0.028). However, the data were not significant after Bonferroni correction as well. In addition, the correlations between the other SNPs belonging to DSBR genes and the risk of PTC were not observed in the other stratified analyses (Tables 3).
 |
Table 2 The Relationship Between Candidate SNPs and PTC Risk Stratified by Gender Status |
 |
Table 3 The Relationship Between Candidate SNPs and PTC Risk Stratified by Age |
SNP rs15869 A/C Affected BRCA2 mRNA Secondary Structure and BRCA2 Expression
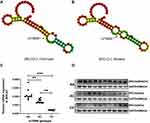
Despite insignificant results after Bonferroni correction, we had decided to perform a more detailed analysis concerning this SNP rs15869. Subsequently, we attempted to evaluate the effect of the locus variation on BRCA2 mRNA secondary structure. As we expected, rs15869 polymorphism located in a 3ʹ-UTR of BRCA2 changed its mRNA secondary structure evaluated by the RNA Fold Webserver program (Figure 1A and B).
In order to clarify whether the changed mRNA secondary structure resulting from rs15869 polymorphism contributes to the regulation of BRCA2 expression, we subsequently examined mRNA levels of the BRCA2 in 30 thyroid tissues of PTC patients harboring rs15869 different genotypes. BRCA2 mRNA expression was significantly higher in thyroid tissues with AA allele than AC and CC allele mRNA (Figure 1C), which was consistent with the Western blotting analysis result (Figure 1D), indicating the A-to-C substitution in rs15869 contributed to the down-regulation of BRCA2.
Bioinformatics Prediction and Functional Mining of the Candidate miRNAs
We speculate that the BRCA2 post-transcriptional expression may be modulated by miRNAs, the crucial regulator of transcription and translation, which can also be affected by the polymorphism rs15869 in target complementary sequence. In order to in-depth mine, some possible miRNAs affected by the polymorphism, both PolymiRTs and RNAhybrid software were used to predict three candidate miRNAs listed in Table 4. The energy changed from −12.8 to −16.8 kcal/mol reminded that the rs15869 polymorphism had the greatest influence on the binding of hsa-miR-1178-3p and BRCA2 (Table 4). Meanwhile, the experiment-supported evidence curated by the HMDD database revealed that hsa-miR-1178-3p acts as an oncomiR in pancreatic cancer cells (Table S6). Therefore, we focused on hsa-miR-1178-3p and further undertook enrichment pathways and biological processes to analyze the commonly predicted targets of hsa-miR-1178-3p. Collectively hsa-miR-1178-3p targets mapped onto multiple tumor-related molecular processes (Figure 2B), mainly including activation of MAPK activity, epidermal growth factor receptor signaling pathway and regulation of transcription. Although hsa-miR-1178-3p was predicted to be primarily involved in non-small cell lung cancer, colorectal cancer, bladder cancer and melanoma (Figure 2A), no evidence, to date, has demonstrated a potential role of hsa-miR-1178-3p in the tumourigenesis of PTC.
 |
Table 4 The Candidate miRNAs Based on PolymiRTs and RNAhybrid Prediction |
rs15869 Created a Binding Site for hsa-miR-1178-3p
The bioinformatics prediction suggested that the A-to-C substitution of BRCA2 rs15869 created the hsa-miR-1178-3p binding site (Figure 3A). The MFE of the binding of miRNA to rs15869 evaluated by RNAhybrid also reminded us that the SNP variant C was the favorable miRNA-mRNA binding pattern. The interaction between hsa-miR-1178-3p with BRCA2 and its potential modulation by rs15869 was further evaluated in vitro using a 3ʹ-UTR dual-luciferase assay. The more significantly decreased luciferase activity up to 21% in the presence of the C allele demonstrated a greater binding between hsa-miR-1178-3p and the C allele than the A allele with a 4% reduction of luciferase activity taking the negative controls as a reference (Figure 3B). Accordingly, it is suggested that rs15869 SNP may be a functional variant.
hsa-miR-1178-3p Targeted BRCA2 Containing rs15869 C Allele in Thyroid Tissues
We next explored the correlation between BRCA2 and hsa-miR-1178-3p in normal thyroid tissues with the different genotype of rs15869 SNP. QPCR assay revealed that hsa-miR-1178-3p expression was negatively related with BRCA2 mRNA levels in thyroid tissues with AC and CC genotype at rs15869, whereas there was no correlation between hsa-miR-1178-3p and BRCA2 expression in the tissues with AA genotype at rs15869 (Figure 4A). We further assessed the effect of hsa-miR-1178-3p on the protein expression of BRCA2 in these tissues harboring varied rs15869 genotypes and came to the same conclusion that hsa-miR-1178-3p could interact with BRCA2 containing rs15869 C allele and decreased BRCA2 expression in normal thyroid tissues (Figure 4B).
Discussion
The incidence of thyroid cancer continues to rise worldwide and the rise in incidence has been due almost entirely to the most common histologic type, PTC.4 In recent years, several research studies have implicated that DNA damage accumulation and genomic instability resulting from deficiencies of key genes related to DNA damage repair help facilitate access to the increased risk of PTC.17 Of note, DSBs, caused by ionizing radiation or other physical factors, contribute to the occurrence of PTC. HR and NHEJ, belonging to DSBR, are involved in the damage repair by competition or cooperation. Not surprisingly, numerous studies have identified polymorphisms located in genes of DSBR associated to the risk of a variety of benign and malignant tumors, for example, XRCC1 R399Q in breast cancer,18 RAD51 135G/C in colorectal cancer,19 XPD Lys751Gln in prostate cancer and so on.20 The present study focuses on the effect of 3ʹ-UTR polymorphisms of DSBR genes on PTC risk based on a medium-sized case–control study.
It is known that BRCA2 specifically participates in the HR pathway for repairing DSBs, together with the BRC motif that mediates binding to the RAD51 recombines for the maintenance of genome stability. Generally, BRCA2 is considered as a tumor suppressor gene due to its positive role in DNA repair. Notably, the BRCA2 gene mutation had been found to lack HR repair function and cause repair defects in DSBs and further loss in cell function, which is more conducive to tumorigenesis.21 Additionally, abnormal expression of BRCA2 is also inextricably associated with malignant tumors, such as breast cancer, prostate cancer and ovarian cancer.22–24 In recent years, several studies demonstrated that a gene abnormal expression may be partially attributed to its polymorphisms, among which coding variants can affect protein function while others in the non-coding region may still affect gene expression via splicing, microRNA binding and mRNA degradation, further modifying the risk of cancer. As exemplified by rs11571836, located in the BRCA2 3ʹ-UTR, it was significantly associated with a lower expression of BRCA2 mRNA and an increased risk of pancreatic cancer (OR=1.30, 95% CI: 1.14–1.47).24 K. Zbuk’s research suggested rs11571836 and rs1799943 in BRCA2 3ʹ-UTR were associated with a lower risk of cardiovascular disease.25 Similarly, our current study found that the people carrying BRCA2 rs15869 CC genotype had a higher risk of PTC compared with AA genotype. Stratification analysis further showed that rs15869 CC genotype carriers had a higher PTC risk in age<50 years old population or in females but not males. The above results were consistent with N.D. Sirisena’s studies: the rs15869 C allele was associated with breast cancer tumor grade (OR=1.600; P=0.041).23 Interestingly, J. Cao’s analysis revealed that heterozygous AC (OR=1.524; 95% CI: 1.141–2.035) of BRCA2 rs15869 could elevate the risk of breast cancer.26 Although the above studies supported the hypothesis that the rs15869 C allele was a hazard to suffer from breast cancer, no study to date has demonstrated a causal relationship with thyroid cancer. Accordingly, our study provided some evidence that rs15869, located in BRCA2 3ʹ-UTR, was weakly associated with PTC (p=0.031, Table 1); however, this association was insignificant after Bonferroni correction. While other candidate SNPs selected in the present study did not show any relationship with the risk of PTC, it further reminded that BRCA2 rs15869 polymorphism out of the many identified may be a functional variant. We subsequently confirmed that the A-to-C substitution of rs15869 changed BRCA2 mRNA secondary structure (Figure 1A and B) and contributed to a decreased BRCA2 expression (Figure 1C and D).
MicroRNAs can regulate mRNA degradation or protein translation by binding to 3ʹ-UTR of target genes and further participate in biological functions including proliferation, apoptosis, DNA repair, and so on. Song found that miR-1245 binding BRCA2 3ʹ-UTR down-regulated the expression of BRCA2 and resulted in more serious DNA damage.27 Additionally, J. Cao demonstrated that miR-627 can down-regulated the expression of BRCA2 in breast cancer cells;26 other studies have uncovered that hsa-miR-1178-3p may serve as an oncomiR and targeted tumor suppressor in pancreatic cancer reported by Z. Cao,28 which was in accordance with H. Liu and J. Bi’s studies in bladder cancer.29,30 In our study, bioinformatics predicted that the seed sequence of hsa-miR-1178-3p could bind to rs15869 SNP in BRCA2 3ʹ-UTR and verified by in vitro dual-luciferase assay (Figure 3). Remarkably, the A-to-C substitution of rs15869 SNP contributed to a greater binding of hsa-miR-1178-3p to BRCA2 3ʹ-UTR and resulted in a decreased expression of BRCA2 through posttranscriptional mechanisms (Figure 4). In addition, Gene set enrichment analysis of pathways and the biological process targeted by hsa-miR-135a-5p, consistent with the above studies, further verified the crucial role of hsa-miR-1178-3p in tumorigenesis and elucidated hierarchical functions in gene regulatory networks to a certain degree (Figure 2).
Collectively, the present study characterized rs15869 as a functional BRCA2 variant associated with the risk of PTC due to the binding of hsa-miR-1178-3p, suggesting a model whereby rs15869 can modify the occurrence of PTC by affecting BRCA2 expression. Although our current study seems to have limited predictive value and applicability in the clinical context due to the lack of in-depth functional studies, it should be kept in mind that rs15869 located in BRCA2 3ʹ-UTR might be associated with PTC risk, as was shown by case-control study, bioinformatics exploration and in vitro assay. Thus, our findings shed light on the potential role of hsa-miR-1178-3p in the development of PTC. Future work should be geared toward a larger sample size study and more precise function exploration in an attempt to find the causal effect of possible biomarkers on the susceptibility of thyroid cancer.
Abbreviations
DSBR, DNA double-strand break repair; DSBs, double-strand breaks; HR, homologous recombination; MAF, minor allele frequency; MFE, minimum free energy; NHEJ, non-homologous end joining; PTC, papillary thyroid carcinoma; SNP, single nucleotide polymorphism; UTR, untranslated region.
Data Sharing Statement
The datasets used and/or analyzed during the current study are available from the corresponding authors on reasonable request.
Ethics Approval and Consent to Participate
Name of the ethics committee: Medical ethics committee of Liaoning Cancer Hospital & Institute.
Acknowledgment
The first three authors Nan Guo, Peng Qu and Hao Li should be considered joint first author.
Author Contributions
Conception: Xiaobo Lu, Yuejiao Zhao, Liuli Li and Nan Guo. Interpretation or analysis of data: Peng Qu, Hao Li, Xuan Zhang, Yingchun Li and Liuli Li. Preparation of the manuscript: Liuli Li, Hao Jin, Renqi Liu, Zhen Zhang and Nan Guo. Revision for important intellectual content: Xiaobo Lu, Yuejiao Zhao and Hao Jin. Supervision: Yuejiao Zhao and Xiaobo Lu. All authors contributed to data analysis, drafting or revising the article, have agreed on the journal to which the article will be submitted, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Funding
This study was supported by grants from the clinical research project of Climbing Fund from the National Cancer Center (No.NCC201806B-009), Liaoning Provincial Key Research and Development Program (No.2017225056) and Liaoning natural fund guidance plan (No.2019-ZD-0595).
Disclosure
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
1. Fitzmaurice C, Akinyemiju TF, Al Lami FH, et al. Global, Regional, and National Cancer Incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 29 Cancer Groups, 1990 to 2016: a systematic analysis for the Global Burden of Disease Study. JAMA Oncol. 2018;4(11):1553–1568. doi:10.1001/jamaoncol.2018.2706
2. Lozano R, Naghavi M, Foreman K, et al. Global and regional mortality from 235 causes of death for 20 age groups in 1990 and 2010: a systematic analysis for the Global Burden of Disease Study 2010. Lancet. 2012;380(9859):2095–2128. doi:10.1016/S0140-6736(12)61728-0
3. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
4. Lloyd RV, Buehler D, Khanafshar E. Papillary thyroid carcinoma variants. Head Neck Pathol. 2011;5(1):51–56. doi:10.1007/s12105-010-0236-9
5. Cabanillas ME, McFadden DG, Durante C. Thyroid cancer. Lancet. 2016;388(10061):2783–2795. doi:10.1016/S0140-6736(16)30172-6
6. Albi E, Cataldi S, Lazzarini A, et al. Radiation and thyroid cancer. Int J Mol Sci. 2017;18:5. doi:10.3390/ijms18050911
7. Broustas CG, Lieberman HB. DNA damage response genes and the development of cancer metastasis. Radiat Res. 2014;181(2):111–130. doi:10.1667/RR13515.1
8. Symington LS, Gautier J. Double-strand break end resection and repair pathway choice. Annu Rev Genet. 2011;45:247–271. doi:10.1146/annurev-genet-110410-132435
9. Burma S, Chen BP, Chen DJ. Role of non-homologous end joining (NHEJ) in maintaining genomic integrity. DNA Repair (Amst). 2006;5(9–10):1042–1048. doi:10.1016/j.dnarep.2006.05.026
10. Akulevich NM, Saenko VA, Rogounovitch TI, et al. Polymorphisms of DNA damage response genes in radiation-related and sporadic papillary thyroid carcinoma. Endocr Relat Cancer. 2009;16(2):491–503. doi:10.1677/ERC-08-0336
11. Fayaz S, Karimmirza M, Tanhaei S, Fathi M, Torbati PM, Fard-Esfahani P. Increased risk of differentiated thyroid carcinoma with combined effects of homologous recombination repair gene polymorphisms in an Iranian population. Asian Pac J Cancer Prev. 2014;14(11):6727–6731. doi:10.7314/APJCP.2013.14.11.6727
12. Gomes BC, Silva SN, Azevedo AP, et al. The role of common variants of non-homologous end-joining repair genes XRCC4, LIG4 and Ku80 in thyroid cancer risk. Oncol Rep. 2010;24(4):1079–1085.
13. Lahtz C, Pfeifer GP. Epigenetic changes of DNA repair genes in cancer. J Mol Cell Biol. 2011;3(1):51–58. doi:10.1093/jmcb/mjq053
14. Chou CK, Liu RT, Kang HY. MicroRNA-146b: a novel biomarker and therapeutic target for human papillary thyroid cancer. Int J Mol Sci. 2017;18(3). doi:10.3390/ijms18030636
15. Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120(1):15–20. doi:10.1016/j.cell.2004.12.035
16. Teo MT, Landi D, Taylor CF, et al. The role of microRNA-binding site polymorphisms in DNA repair genes as risk factors for bladder cancer and breast cancer and their impact on radiotherapy outcomes. Carcinogenesis. 2012;33(3):581–586. doi:10.1093/carcin/bgr300
17. Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi:10.1016/j.cell.2011.02.013
18. Eskandari E, Rezaifar A, Hashemi M. XPG Asp1104His, XRCC2 rs3218536 A/G and RAD51 135G/C gene polymorphisms and colorectal cancer risk: a meta-analysis. Asian Pac J Cancer Prev. 2017;18(7):1805–1813. doi:10.22034/APJCP.2017.18.7.1805
19. Krivokuca AM, Cavic MR, Malisic EJ, et al. Polymorphisms in cancer susceptibility genes XRCC1, RAD51 and TP53 and the risk of breast cancer in Serbian women. Int J Biol Markers. 2016;31(3):e258–e263. doi:10.5301/jbm.5000201
20. Cypriano AS, Alves G, Ornellas AA, et al. Relationship between XPD, RAD51, and APEX1 DNA repair genotypes and prostate cancer risk in the male population of Rio de Janeiro, Brazil. Genet Mol Biol. 2017;40(4):751–758. doi:10.1590/1678-4685-gmb-2017-0039
21. Gudmundsdottir K, Ashworth A. The roles of BRCA1 and BRCA2 and associated proteins in the maintenance of genomic stability. Oncogene. 2006;25(43):5864–5874. doi:10.1038/sj.onc.1209874
22. Lax SF. Hereditary breast and ovarian cancer. Pathologe. 2017;38(3):149–155. doi:10.1007/s00292-017-0298-5
23. Sirisena ND, Adeyemo A, Kuruppu AI, Samaranayake N, Dissanayake VHW. Genetic variants associated with clinicopathological profiles in sporadic breast cancer in Sri Lankan Women. J Breast Cancer. 2018;21(2):165–172. doi:10.4048/jbc.2018.21.2.165
24. Huang L, Wu C, Yu D, et al. Identification of common variants in BRCA2 and MAP2K4 for susceptibility to sporadic pancreatic cancer. Carcinogenesis. 2013;34(5):1001–1005. doi:10.1093/carcin/bgt004
25. Zbuk K, Xie C, Young R, et al. BRCA2 variants and cardiovascular disease in a multi-ethnic study. BMC Med Genet. 2012;13:56. doi:10.1186/1471-2350-13-56
26. Cao J, Luo C, Yan R, et al. rs15869 at miRNA binding site in BRCA2 is associated with breast cancer susceptibility. Med Oncol. 2016;33(12):135. doi:10.1007/s12032-016-0849-2
27. Song L, Dai T, Xie Y, et al. Up-regulation of miR-1245 by c-myc targets BRCA2 and impairs DNA repair. J Mol Cell Biol. 2012;4(2):108–117. doi:10.1093/jmcb/mjr046
28. Cao Z, Xu J, Huang H, et al. MiR-1178 promotes the proliferation, G1/S transition, migration and invasion of pancreatic cancer cells by targeting CHIP. PLoS One. 2015;10(1):e0116934. doi:10.1371/journal.pone.0116934
29. Liu H, Bi J, Dong W, et al. Invasion-related circular RNA circFNDC3B inhibits bladder cancer progression through the miR-1178-3p/G3BP2/SRC/FAK axis. Mol Cancer. 2018;17(1):161. doi:10.1186/s12943-018-0908-8
30. Bi J, Liu H, Dong W, et al. Circular RNA circ-ZKSCAN1 inhibits bladder cancer progression through miR-1178-3p/p21 axis and acts as a prognostic factor of recurrence. Mol Cancer. 2019;18(1):133. doi:10.1186/s12943-019-1060-9
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.