Back to Journals » Clinical, Cosmetic and Investigational Dermatology » Volume 15
BIRC7 is Beneficial for Melanoma Progression and Hypoxic Response
Authors Xu H, Liu H, Li Z, Xu Q, Lin N, Li X
Received 15 April 2022
Accepted for publication 14 June 2022
Published 17 June 2022 Volume 2022:15 Pages 1109—1117
DOI https://doi.org/10.2147/CCID.S370969
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Jeffrey Weinberg
Haiting Xu,1 Huazhen Liu,2 Zi Li,3 Qin Xu,3 Nan Lin,1 Xiaoyang Li1
1Department of Hand and Plastic Surgery, The Second Affiliated Hospital and Yuying Children’s Hospital of Wenzhou Medical University, Wenzhou, Zhejiang, 325027, People’s Republic of China; 2Department of Burn, Changhai Hospital Affiliated Naval Medical University, Shanghai, 200433, People’s Republic of China; 3Department of Dermatology, the Second School of Medicine, Wenzhou Medical University, Wenzhou, Zhejiang, 325027, People’s Republic of China
Correspondence: Xiaoyang Li, Department of Hand and Plastic Surgery, The Second Affiliated Hospital and Yuying Children’s Hospital of Wenzhou Medical University, Wenzhou, Zhejiang, 325027, People’s Republic of China, Email [email protected]
Background: Cutaneous melanoma (CM) accounts for about 90% of all melanoma cases. HIF-1α overexpression is associated with poor prognosis in many cancers including CM. Hence, we characterized differentially expressed genes (DEGs) in the response of CM cells to normoxia and hypoxia.
Methods: We first successfully constructed cell hypoxia model and then performed RNA-seq to explore the changes of gene transcription in CM cells during hypoxia. The highest expression of the six genes was detected using qRT-PCR and Western blot assays. The binding sites between BIRC7 promoter and HIF-1α was explored by luciferase assay. Cellular function assays were used to observe the role of BIRC7 in the effect of hypoxia on tumor progression.
Results: Compared with the transcriptome data of the control group, a total of 2601 DEGs were identified in the hypoxic group. There were 1517 genes with significantly higher expression and 1084 genes with lower expression in the hypoxic group. Among them, OSCAR, BIRC7, HBA1, SFN, GOLT1A, and BEX2 were significantly up-regulated in the hypoxic group. BIRC7 expression was most significantly up-regulated and a downstream factor of HIF-1α. We highlighted that knockdown of BIRC7 reversed the positive effects of HIF-1α on A875 and M14 cells.
Conclusion: Our findings demonstrated that BIRC7 was a downstream factor of HIF-1α and reversed the effect of hypoxia on promoting tumor progression of CM cells.
Keywords: apoptosis, BIRC7, HIF-1α, hypoxia, malignant melanomas
Background
Cutaneous melanoma (CM) accounts for about 90% of all melanoma cases (CM, uveal melanoma, and mucosal melanoma).1 In recent years, the global incidence of CM has risen sharply.2 With the development of molecular targeted therapy and immunotherapy, the treatment of patients with unresectable or metastatic melanoma has made great progress, which has changed the current status of non-treatment.2 However, this disease progresses extremely rapidly. At present, the survival time of advanced patients is usually only 6 to 9 months.3
Hypoxia promotes the resistance to various anti-tumor drugs. It is a key factor in the metastasis and treatment response failure of most solid tumors, including CM.2 Due to abnormal blood vessels, oxygen delivery is impaired in the tumor microenvironment. In addition, due to the proliferation requirements of tumor cells, nutrition and oxygen consumption increase.4,5 Hypoxia microenvironment is a sign of most solid tumors. Hypoxia-inducible factor 1 (HIF-1) is a key regulator of tumor cell adaptive response to hypoxic microenvironment and mediates a large number of gene responses. Some preclinical evidence suggests that targeting HIF-1 alone or in combination with other treatments may be beneficial in inhibiting the metastasis of CM.6,7 Therefore, the in-depth exploration of hypoxia regulation pathways in the progress of CM is beneficial to the clinical practice of CM treatment.
BIRC7 is an apoptosis inhibitory protein that is upregulated in many melanoma cell lines and is closely associated with melanoma disease progression, apoptosis resistance, and chemotherapeutic drug resistance.8–10 BIRC7 contains a BIR domain that has been shown to bind and inhibit caspases-3, −7 and −9, playing a key role in inhibiting apoptosis.11 Wang et al also found that knockdown of BIRC7 induced melanoma cell apoptosis.12 Apoptosis-inhibiting proteins including BIRC7 are considered novel targets for anticancer therapy. However, the role of BIRC7 in tumor hypoxia remains unknown.
In this study, CM cells suffering from hypoxia for 12 h were subjected to high-throughput transcriptome sequencing. After analyzing the molecular changes caused by hypoxia, we focused on the role of BIRC7 in CM hypoxia.
Methods
Cell Culture and Treatment
M14 and A875 cells were obtained from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). Cells were cultured in DMEM supplemented with 10% FBS and 1% penicillin-streptomycin with the 5% CO2, 21% O2 and 74% N2 atmosphere at 37 °C. A chamber (MACS V A500 microaerophilic workstation, Don Whitley Scientific, Bingley, UK) was filled with atmosphere of 95% N2 and 5% CO2 to simulate the hypoxia in melanoma cells. Specific siRNA targeting BIRC7 (si-BIRC7) was designed by Genepharm Co. (Shanghai, China), and transfected into cells using Lipofectamine 2000 reagent. si-BIRC7 sequence was as follows: 5′-GGTGAGGTGCTTCTTCTGC-3′.
Western Blot
Total protein from cells was extracted using RIPA lysate, and nuclear-cytoplasmic separation was performed with the Minute (TM) Cytoplasmic and Nuclear Fractionation kit (SC-003, Invent). Equal amounts of protein were run on SDS-PAGE and electroporated onto PVDF membrane. Membrane was sequentially blocked in 5% bovine serum albumin for 1 h, incubated with primary antibodies (anti-HIF-1α, 1:2000, ab1; anti-Lamin B1, 1:5000, ab16048; anti-GAPDH, 1:5000, ab8245; anti-BIRC7, 1:2000, ab97350; Abcam) overnight at 4°C and then incubated with corresponding secondary antibodies (HRP Anti-Rabbit IgG antibody, 1:3000; ab288151; HRP Goat Anti-Mouse IgG H&L, 1:3000; ab205719; Abcam) for 2 h at room temperature.
Immunofluorescence Staining
The cells were fixed with 4% PFA for 15 min. About 0.05% Triton X-100 was used to permeabilize cells. Cells were blocked with 5% BSA, incubated with primary antibodies (anti-HIF-1α, 1:200, ab1, Abcam) overnight at 4 °C and FITC-conjugated secondary antibodies (Goat Anti-Mouse IgG H&L, 1: 200; ab150113; Abcam) at room temperature 1 h, and stained with DAPI for 6 min. Finally, cells were mounted with Aqua Mount.
RNA Sequencing and Data Analysis
Total RNA was isolated from cells entering logarithmic division using Trizol reagent (CWBIO, Beijing, China). An Agilent 2100 Bioanalyzer (Agilent, USA) was used to evaluate RNA quality. cDNA library was constructed using Oligo (dT) magnetic beads (Invitrogen, Carlsbad, CA, USA) and random hexamers, the range of library inserts was detected with the Agilent 2100 bioanalyzer was used to detect, and the library concentration was quantified using the ABI StepOnePlus real-time PCR system. Sequencing was performed using an Illumina HiSeq sequencer. The raw image data file obtained by Illumina HiSeq was converted into readings data through CASAVA base call analysis, and the result was saved in the FASTQ file format. After passing the raw readings through perlscript and discarding low-quality readings to obtain clean readings, HISAT was used to compare the clean readings with the reference genome. The PossionDis algorithm was used for differential gene detection, and genes with |log2(FoldChange)| >1 and q value <0.001 were selected as differentially expressed genes (DEG). Using the FPKM value of the differential gene as the expression level, a hierarchical clustering analysis was performed.
Quantitative Real-Time PCR
Trizol was used to isolate total RNA, and CDNA was synthesized using Revert Aid First Strand cDNA Synthesis Kit (Takara, Beijing, China). RT-qPCR was performed using ABI Prism 7300 sequence detector and SYBR Green reagent (Takara, Beijing, China). The primer sequences are shown in Table 1. β-actin was used as the internal reference.
 |
Table 1 Primers Sequences |
Luciferase Determination
Partial fragment of wild-type BIRC7-3ʹUTR (WT) and mutant (MUT) were amplified and cloned into pGL3 Basic vector (Promega, Madison, WI). All constructs were verified by sequencing. pmirGLO-BIRC7-3ʹUTR wild-type (WT) and mutant (MUT) plasmids were transferred to M14 cells through Lipofectamine 2000 according to the manufacturer’s protocols. Forty-eight hours after transfection, the luciferase activity was measured using the dual-luciferase reporter gene detection system (Promega, Madison, USA) and normalized to Renilla luciferase activity.
CCK-8 Assay
The si-BIRC7 transfected cells were seeded into a 96-well plate at a density of 2000 cells per well. After 48 h of incubation, hypoxia treatment was performed for 12 h. Then, oxygen was restored for 12 h and the CCK-8 experiment was performed. About 10 μL of CCK-8 solution (Beyotime Biotechnology, Shanghai, China) was added to each well, and after incubating for 2 h, the OD value of each well at 450 nm was detected using a microplate reader.
Transwell Assay
About 80 µL of Matrigel (1:6 dilution, Cat # 356234, BD, Franklin Lakes, NJ, USA) was used to coat the upper chamber of the 24-well transwell. After transfection, 1×104 cells were implanted in the upper chamber and incubated in 150 μL serum-free medium. About 500 μL of complete medium was added to the lower chamber. After 12 h of hypoxia, oxygen was restored for 8 h. A cotton swab was used to carefully remove the upper chamber cells, and PBS was used to wash. Cells in the lower chamber were fixed with 4% paraformaldehyde solution for 30 min, stained with 0.1% crystal violet for 10 min, and then the cells were imaged and counted.
Flow Cytometry
Cells from different treatment groups were trypsinized without EDTA and resuspended with 1x binding buffer. The cells were stained with the annexin V/FITC mixture and PI dye, which was ordered from 4A Biotech Company (Cat# FXP018-100, Beijing, People’s Republic of China), according to the manufacturer’s manual. After 15 min of incubation in the dark, the cells were analyzed for apoptosis.
Statistical Analysis
All data were analyzed using SPSS 18.0 software. Student’s t test or one-way ANOVA test was used for the difference analysis of different groups. P < 0.05 was considered statistically significant.
Results
Transcriptome Analysis of M14 Cells Under Hypoxia for 12 h
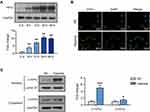
In order to explore the molecular mechanism of hypoxia on melanoma cell behavior, we performed transcriptome sequencing and data analysis on M14 cells suffering hypoxia for 12 h. Figure 1A shows the induced expression of HIF-1α in cells after hypoxia for different periods (0, 6, 12, 24, and 48 h). Combined with the cell status, we chose hypoxia for 12 h as the condition for the follow-up study. The results of immunofluorescence (Figure 1B) and Western blot (Figure 1C) also showed that after 12 h of hypoxia, the expression of HIF-1α was significantly increased and concentrated in the nucleus.
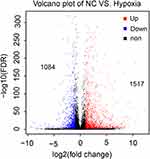
High-throughput RNA-seq on NC and hypoxic (12 h) M14 cells identified 2601 genes that were differentially expressed after hypoxia, of which 1517 genes were significantly up-regulated, and 1084 genes were significantly down-regulated (Figure 2). Compared with the NC group, the top 6 up-regulated genes in the hypoxia group are listed in Table 2. OSCAR, BIRC7, HBA1, SFN, GOLT1A and BEX2 were significantly up-regulated in the hypoxia group (p = 0.000<0.01).
 |
Table 2 Top Six Up-Regulated Genes |
HIF-1α Induces BIRC7 Transcription and Expression
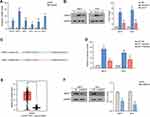
RT-qPCR showed that the mRNA expression levels of OSCAR, BIRC7, HBA1, SFN, GOLT1A, and BEX2 were up-regulated in M14 cells after hypoxia, and BIRC7 was focused (Figure 3A). The BIRC7 protein levels were also increased significantly after hypoxia compared to the NC group (Figure 3B). Furthermore, the addition of HIF-1α inhibitor (Hy+In) reduced the hypoxia-induced BIRC7 protein expression (Figure 3B), and the binding site of HIF-1α on the BIRC7 promoter was predicted with the StarBase software (Figure 3C). After hypoxia, HIF-1α levels increased, HIF-1α bound to the BIRC7 promoter through the putative-binding site and promoted the transcription and expression of BIRC7, so the relative luciferase activity increases significantly (Figure 3D). After the putative-binding site was mutated, the relative luciferase activity was significantly reduced (Figure 3D). In addition, data from GEPIA showed that the expression of BIRC7 in skin cutaneous melanoma (SKCM, n = 461) was significantly higher than that in normal samples (n = 558) (Figure 3E).
Down-Regulation of BIRC7 Expression Inhibits the Proliferation and Invasion of Melanoma Cells Under Normoxia and Hypoxia
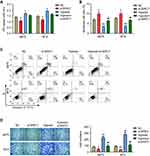
Finally, we used siRNA transfection to down-regulate the expression of BIRC7 in A875 and M14 cells (si-BIRC7) (Figure 3F) and analyzed the effect of low BIRC7 expression on the proliferation and invasion of melanoma cells under normoxia and hypoxia. The results of CCK-8 assay (Figure 4A) showed that si-BIRC7 inhibited the proliferation and viability of A875 and M14 cells under normoxia. Hypoxia promoted the proliferation of A875 and M14 cells, and knockdown of BIRC7 under hypoxia blocked the promoting effect of hypoxia on cell proliferation (Figure 4A). Flow cytometry results suggested that si-BIRC7 increased the percentage of apoptosis in A875 and M14 cells under normoxia (Figure 4B and C). Furthermore, hypoxia inhibited the apoptosis in A875 and M14 cells, whereas knockdown of BIRC7 under hypoxia hindered the inhibitory effect of hypoxia on apoptosis (Figure 4B and C). In addition, compared with the NC group, the number of invaded cells decreased after BIRC7 knockdown, and increased after hypoxia treatment (Figure 4D). However, knockdown of BIRC7 under hypoxia blocked the promotion of cell invasion by hypoxia (Figure 4D).
Discussion
Due to the lack of vasculature, the low level of tissue oxidation is a unique feature of rodent and human epidermis. Both rodent and human epidermis show elevated levels of HIF-1α in the basal epidermal compartment.13 Melanocytes are located on the basement membrane of the basal layer of human epidermis, and exhibit stable HIF-1α expression, which drives melanocytes growth and improves the survival of melanocytes under hypoxic conditions.14 Under physiological conditions, hypoxia triggers an innate cellular stress response to minimize the negative effects of hypoxia on melanocytes. If hypoxia persists, melanocytes will undergo apoptosis. However, due to unique signaling pathways and transcription factors, melanoma cells will survive under constant hypoxic stress.
HIF-1α is a key responder of tumor cells that responds to hypoxia and drives cell migration and invasion. Constitutive expression of HIF-1α was detected in CM tissue sections and in vitro cultured cell lines.15,16 The expression of HIF-1α in CM is higher than that in benign nevi.17 Furthermore, the expression of HIF-1α is correlated with the malignancy degree of CM. HIF1α is hardly expressed in less invasive CM cells but is highly expressed in CM cells that invade the dermis.18 In this study, the expression level of HIF-1α was used as the detection index to detect the hypoxia exposure state of M14 cells cultured in vitro. We found that HIF-1α expression levels were elevated in M14 cells after hypoxia treatment, which can be detected in the nucleus and cytoplasm, mainly concentrated in the nucleus. As a hypoxia response factor and transcription factor, HIF-1α enters the nucleus, activates the transcription of a series of downstream target genes, and initiates the hypoxia stress response.
Subsequently, we performed the transcriptome sequencing and data analysis on M14 cells under hypoxia for 12 h, and identified genes that were differentially expressed after hypoxia, and focused on BIRC7. These results are helpful to analyze the molecular mechanism of hypoxia regulation in CM cells. We found that BIRC7 protein levels were significantly increased after hypoxia. Luciferase experiments and the addition of HIF-1α inhibitors indicated that HIF-1α can act as a transcription factor for BIRC7 and promote its transcription. Consistent with our results, Hsieh et al observed the up-regulation of BIRC7 expression in glioblastoma cells and xenografts under circulating and chronic hypoxic conditions, as well as the binding of HIF-1α to the BIRC7 promoter.19 In addition, targeting BIRC7 is beneficial to the efficacy of radiotherapy and chemotherapy for glioblastoma.19
Basically consistent with previous study,20 we found that BIRC7 was significantly over-expressed in CM tissues, and knockdown of its expression inhibited the proliferation of A875 and M14 cells, and induced apoptosis. However, the present study found that the knockdown of BIRC7 inhibited the invasion of A875 and M14 cells. It has been reported that BIRC7 can bind and deplete C-RAF through the BIR domain, and loss of BIRC7 activates the MAPK cascade and promotes melanoma cell migration.21 The role of BIRC7 in melanoma cell invasion and migration still needs to be further explored. Most importantly, this study found that hypoxia stimulation induced the proliferation and migration of A875 and M14 cells and inhibited apoptosis, whereas BIRC7 knockdown under hypoxia could counteract the effects of hypoxia on cell biological behavior.
Conclusion
In summary, BIRC7 plays a key role in melanoma progression and hypoxic response, and targeting BIRC7 is beneficial to the treatment of melanoma.
Abbreviations
CM, cutaneous melanoma; DEGs, differentially expressed genes; HIF-1, hypoxia-inducible factor 1; BIRC7, baculoviral IAP repeat-containing 7; si-BIRC7, specific siRNA targeting BIRC7; WT, wild-type; MUT, mutant.
Data Sharing Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Funding
This study was funded by Wenzhou Basic Public Welfare Research Project (Y20210177).
Disclosure
The authors declare that they have no competing interests in this work.
References
1. D’Aguanno S, Mallone F, Marenco M, Del Bufalo D, Moramarco A. Hypoxia-dependent drivers of melanoma progression. J Exp Clin Cancer Res. 2021;40(1):159. doi:10.1186/s13046-021-01926-6
2. Jenkins RW, Fisher DE. Treatment of advanced melanoma in 2020 and beyond. J Invest Dermatol. 2021;141(1):23–31. doi:10.1016/j.jid.2020.03.943
3. Sandru A, Voinea S, Panaitescu E, Blidaru A. Survival rates of patients with metastatic malignant melanoma. J Med Life. 2014;7(4):572–576.
4. Manoochehri Khoshinani H, Afshar S, Najafi R. Hypoxia: a double-edged sword in cancer therapy. Cancer Invest. 2016;34(10):536–545. doi:10.1080/07357907.2016.1245317
5. Kumar V, Gabrilovich DI. Hypoxia-inducible factors in regulation of immune responses in tumour microenvironment. Immunology. 2014;143(4):512–519. doi:10.1111/imm.12380
6. Li C, Wang Q, Shen S, Wei X, Li G. HIF-1alpha/VEGF signaling-mediated epithelial-mesenchymal transition and angiogenesis is critically involved in anti-metastasis effect of luteolin in melanoma cells. Phytother Res. 2019;33(3):798–807. doi:10.1002/ptr.6273
7. Park EJ, Lee YM, Oh TI, Kim BM, Lim BO, Lim JH. Vanillin suppresses cell motility by inhibiting STAT3-mediated HIF-1alpha mRNA expression in malignant melanoma cells. Int J Mol Sci. 2017;18(3):3. doi:10.3390/ijms18030532
8. Franklin MC, Kadkhodayan S, Ackerly H, et al. Structure and function analysis of peptide antagonists of melanoma inhibitor of apoptosis (ML-IAP). Biochemistry. 2003;42(27):8223–8231. doi:10.1021/bi034227t
9. Vucic D, Stennicke HR, Pisabarro MT, Salvesen GS, Dixit VM. ML-IAP, a novel inhibitor of apoptosis that is preferentially expressed in human melanomas. Curr Biol. 2000;10(21):1359–1366. doi:10.1016/s0960-9822(00)00781-8
10. Mousavi-Shafaei P, Ziaee AA, Zangemeister-Wittke U. Increased cytotoxicity of cisplatin in SK-MEL 28 melanoma cells upon down-regulation of melanoma inhibitor of apoptosis protein. Iran Biomed J. 2009;13(1):27–34.
11. Kasof GM, Gomes BC. Livin, a novel inhibitor of apoptosis protein family member. J Biol Chem. 2001;276(5):3238–3246. doi:10.1074/jbc.M003670200
12. Wang H, Yang Y, Wang W, et al. Single-chain antibody-delivered Livin siRNA inhibits human malignant melanoma growth in vitro and in vivo. Tumour Biol. 2017;39(5):1010428317701645. doi:10.1177/1010428317701645
13. Boutin AT, Weidemann A, Fu Z, et al. Epidermal sensing of oxygen is essential for systemic hypoxic response. Cell. 2008;133(2):223–234. doi:10.1016/j.cell.2008.02.038
14. Bedogni B, Powell MB. Unique transforming properties of Notch1 in human melanocytes. Pigment Cell Melanoma Res. 2009;22(6):702–703. doi:10.1111/j.1755-148X.2009.00625.x
15. Kuphal S, Winklmeier A, Warnecke C, Bosserhoff AK. Constitutive HIF-1 activity in malignant melanoma. Eur J Cancer. 2010;46(6):1159–1169. doi:10.1016/j.ejca.2010.01.031
16. Mills CN, Joshi SS, Niles RM. Expression and function of hypoxia inducible factor-1 alpha in human melanoma under non-hypoxic conditions. Mol Cancer. 2009;8(1):104. doi:10.1186/1476-4598-8-104
17. Zbytek B, Peacock DL, Seagroves TN, Slominski A. Putative role of HIF transcriptional activity in melanocytes and melanoma biology. Dermatoendocrinol. 2013;5(2):239–251. doi:10.4161/derm.22678
18. Marconi A, Borroni RG, Truzzi F, et al. Hypoxia-Inducible Factor-1alpha and CD271 inversely correlate with melanoma invasiveness. Exp Dermatol. 2015;24(5):396–398. doi:10.1111/exd.12679
19. Hsieh CH, Lin YJ, Wu CP, Lee HT, Shyu WC, Wang CC. Livin contributes to tumor hypoxia-induced resistance to cytotoxic therapies in glioblastoma multiforme. Clin Cancer Res. 2015;21(2):460–470. doi:10.1158/1078-0432.CCR-14-0618
20. Wang H, Tan SS, Wang XY, et al. Silencing livin gene by siRNA leads to apoptosis induction, cell cycle arrest, and proliferation inhibition in malignant melanoma LiBr cells. Acta Pharmacol Sin. 2007;28(12):1968–1974. doi:10.1111/j.1745-7254.2007.00724.x
21. Oberoi-Khanuja TK, Karreman C, Larisch S, Rapp UR, Rajalingam K. Role of melanoma inhibitor of apoptosis (ML-IAP) protein, a member of the baculoviral IAP repeat (BIR) domain family, in the regulation of C-RAF kinase and cell migration. J Biol Chem. 2012;287(34):28445–28455. doi:10.1074/jbc.M112.341297
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.