Back to Archived Journals » Current Biomarker Findings » Volume 4
Biomarkers in the Ewing sarcoma family of tumors
Authors Machado I, López-Guerrero JA, Llombart-Bosch A
Received 4 March 2014
Accepted for publication 12 April 2014
Published 18 July 2014 Volume 2014:4 Pages 81—92
DOI https://doi.org/10.2147/CBF.S40803
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Isidro Machado,1 José Antonio López-Guerrero,2 Antonio Llombart-Bosch3
1Pathology Department, Instituto Valenciano de Oncología, Valencia, Spain; 2Molecular Biology Department, Instituto Valenciano de Oncología, Valencia, Spain; 3Pathology Department, University of Valencia, Valencia, Spain
Abstract: This review provides a concise analysis of the well-characterized phenotypic and biological biomarkers having diagnostic and prognostic significance in the Ewing sarcoma family of tumors. Of particular interest, and potentially high clinical relevance, are studies of cell cycle biomarkers, growth signaling pathway expression, biomarkers related with angiogenesis, epithelial mesenchymal transition, and tumor microenvironment. In addition, we discuss findings of specific interest from recent biomarker studies, including copy number alterations, minimal residual disease and predictive biomarkers, and biological targets such as CD133, insulin-like growth factor 1, and poly(adenosine diphosphate-ribose) polymerase 1. In conclusion, several diagnostic and prognostic biomarkers for the Ewing sarcoma family of tumors have been published in recent years; nevertheless, these all need to be validated in a series of prospective studies. The findings that only a small proportion of patients with relapsed Ewing sarcoma family of tumors respond to targeted therapies serve as an example of the critical need for predictive biomarkers in this disease.
Keywords: Ewing sarcoma, biomarkers, immunohistochemistry, molecular biology
Introduction
Ewing sarcoma (ES) is a solid tumor of bone and soft tissue that primarily affects adolescents and young adults.1 These tumors can arise in any bone of the body, although the most common sites of disease include the pelvis, ribs, and long bones of the extremities. Approximately 25% of patients have metastatic disease at diagnosis, most often in the lungs, bone, and bone marrow. The treatment of ES relies on a multidisciplinary approach, coupling highly intensive chemotherapy with surgery and/or radiotherapy to control the primary site of disease and metastatic lesions.2 Patients with advanced disease have less than 30% disease-free survival, whereas those with disease localized to a single site generally have more than 70% disease-free survival at 5 years after diagnosis.3 In addition, no improvements in outcomes have been experienced for patients with metastatic disease in more than 30 years.1,4 Apart from metastasis, other clinicopathological features have been considered as markers of high-risk disease. Although increasing tumor size, decreased tumor necrosis after neoadjuvant chemotherapy, central tumor site, and increasing patient age have all been implicated as independent prognostic factors of poor outcome, none of these are as significant as the presence of metastatic disease at diagnosis.5 Nowadays, we have no significant evidence about which patients with localized disease are at risk for recurrence or which of those with metastatic disease may be curable with conventional therapy. In addition, with the incorporation of new directed therapies within the clinical context, it will also be important to introduce them rationally through the selection of optimal cohorts of patients who will be most likely to respond.
The aim of this review is to provide a detailed description of those biomarkers most recognized as having a clinical prognostic and predictive effect in ES.
What do we expect from biomarkers?
A National Health Institutes working group defines a biomarker as a characteristic that can be objectively measured and evaluated as an indicator of normal biologic or pathogenic processes or pharmacologic responses to a therapeutic intervention.6 To consider a biomarker as useful in the clinical context, it should provide a clear risk/benefit ratio to facilitate clinical decisions, be available in an efficient and cost-effective manner, and should be assessed on available technological platforms from easily obtainable samples.7
Biomarkers can be divided into three types: diagnostic, prognostic, and predictive. Most of the biomarkers studied in ES are prognostic and provide information about the disease outcome after standard therapy. 8
Diagnostic and prognostic biomarkers in ES
Immunohistochemical panel
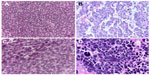
ESFT (Ewing sarcoma family of tumors) constitutes a group of small round cell sarcomas with specific molecular alterations showing a spectrum of neuroectodermal differentiation.9 The histopathology reveals a diffuse proliferation of small round cells fluctuating between undifferentiated patterns (conventional ES) and neoplasms that show neuroectodermal differentiation with Homer-Wright pseudorosettes (so-called primitive neuroectodermal tumors [PNET]; Figure 1A and B).10 Geographic necrosis is frequent, apoptosis/mitosis varies from case to case, and commonly the tumor cells display abundant Periodic acid–Schiff-positive glycogen.10 Three larger categories appear according to their predominant morphological criteria: conventional/classical/typical ES, PNET with neuroectodermal features, and atypical ES comprising subtypes distinct from the other two types. Atypical ES includes large cell variant, tumor with spindle cells, hemangioendothelial ES, adamantinoma-like ES, and sclerotic ES (Figure 1B and C). At present, some issues remain as to whether or not unusual patterns (atypical ES) may be seen in combination with the classical pattern and whether the morphological phenotype of recurrent tumors can be modified by treatment regimens.11
The atypical variant of ESFT seems to be associated with less-favorable prognosis when compared with the conventional and PNET variant;10 however, these results should be confirmed in prospective multicenter studies. Tumor size greater than 8 cm, the presence of metastasis, and poor histological response to neoadjuvant therapy are potential predictors and are significantly related to a negative outcome.12
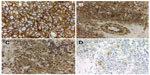
Immunohistochemistry constitutes a useful tool in the diagnosis and identification of ESFT, especially in the differential diagnosis with other small round cell tumors (SRCT) of bone and soft tissue.13 CD99, friend leukemia integration 1 (Fli-1), and human natural killer 1 (HNK-1) represent the more traditional antibodies, and recently, caveolin 1 (CAV-1) has been described as an additional marker that can be useful in the diagnosis of ESFT (Figure 2A–D).10
CD99 immunoexpression is usually membranous and/or cytoplasmic and is, as a rule, moderately (++) or intensely (+++) positive in ESFTs.10,11 However, the specificity of CD99 is not exclusive for ESFT because it can be expressed in other SRCTs, including small cell osteosarcoma, rhabdomyosarcomas, poorly differentiated synovial sarcoma, desmoplastic small round cell tumors, undifferentiated carcinomas, neuroendocrine carcinomas, and lymphomas.14–16 CD99 immunoexpression in SRCT (non-ESFT) is generally cytoplasmic, weak, and focal, although there are cases in which the expression is membranous and intense.10,14–18
At first, it was thought that the immunoreactivity with FLI1 and HNK-1 would be of great support in the diagnosis of ESFT; however, several authors agree that this marker lacks specificity, and its sensitivity is quite variable.9,10 In contrast, CAV-1 is overexpressed in a high proportion of ESFTs.10 When the four antibodies described earlier (CD99, Fli-1, HNK-1, and CAV-1) are combined, they cover the diagnosis of more than 99% of ESFTs.10 Almost all ESFTs with an absence of CD99 expression reveal CAV-1 immunoreactivity; therefore, the use of these antibodies offers additional support in the diagnosis of these neoplasms, regardless of the histological subtype or clinical stage (Table 1; Figure 2A–D).10
  | Table 1 Potential use of morphological, immunohistochemical and molecular biomarkers in the diagnosis of ESFT |
Other immunohistochemical markers considered of use in the diagnosis of EFSTs are NKX2.2 and ERG. NKX2.2 has revealed a sensitivity and specificity of 93% and 89%, respectively, suggesting this marker could be used in the differential diagnosis of SRCT.19 In contrast, ERG is expressed not only in ESTF bearing the ERG rearrangement20 but also in a wide variety of tumors, including angiosarcomas, prostate carcinoma, myeloid sarcomas, and epithelioid sarcomas.21–23 For this reason, specificity for ERG staining in sarcomas should be considered with caution, and the type of immunoexpression is undoubtedly influenced by antibody selection.
Morphological and immunohistochemical neural and neuroendocrine differentiation in ESFT has been widely described, but at present it does not provide additional data for the diagnosis, differential diagnosis, or prognosis of this family of tumors.10,11,13,24–27 Neural differentiation can be demonstrated in ESFTs by use of biomarkers such as neuron-specific enolase, S-100, neurofilaments, CD56, chromogranin A, and synaptophysin.10,11,13,24–27
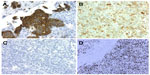
Epithelial differentiation in ESFT has been described in several studies and is particularly observed in the atypical variant characterized by extensive epithelial differentiation (adamantinoma-like ES).9–11,17,18,28–30 Epithelial differentiation has also been described in conventional and variants of PNET.17,18,28 Cytokeratin (AE/AE3), epithelial membrane antigen (EMA), and carcinoembryonic antigen (CEA) are the most widely used antibodies for detecting epithelial differentiation in ESFTs. In addition, many adhesion molecules such as desmoplakin, desmoglein, claudin, occludin ZO-9, and E-cadherin have been also explored.30 Our group reported (AE1/AE3) immunoexpression in 20% of a series of 415 ESFTs (Figure 3A and B), mainly in the atypical variant.17,18 Similarly, Elbashier et al,29 in a series of 43 cases, demonstrated cytokeratin expression in 39.5% of tumors, finding no significant association between cytokeratin immunoreactivity and patient age, sex, and skeletal and extraskeletal primary location, as well as primary, metastatic, or recurrent tumors or chemotherapy treatment. Both studies support the evidence for epithelial differentiation in ESFTs and emphasize that the expression of cytokeratin does not exclude ESFT in the differential diagnosis of SRCT.9–11,17,18,28–30 Other epithelial biomarkers such as epithelial membrane antigen and carcinoembryonic antigen immunoexpression are not usually described in ESFT (Figure 3B). However, in the same recently published study, we also found that up to 6.6% and 20.8% of ESFT can show epithelial membrane antigen or carcinoembryonic antigen expression, respectively, a fact that complicates the differential diagnosis with other tumors (sarcomas, lymphomas, and carcinomas).17,18
Unlike epithelial biomarkers, muscular differentiation markers such as desmin and myogenin are consistently negative in ESFT (Figure 3C). In fact, muscular differentiation in SRCT virtually excludes ESFT;14,15,31 nevertheless, biphenotypic tumors (neural/neuroectodermal and muscular differentiation) have been reported in the literature.31–33 Molecular analysis has provided important diagnosis clues in such neoplasms (Table 1).
Genetics
From the genetic point of view, ESs are specifically characterized by the presence of recurrent chromosomal translocations. In more than 85% of cases, the translocation occurs between chromosomes 11 and 22, resulting in the creation of a pathognomonic chimeric fusion gene, EWSR1/FLI1, that encodes the EWS/FLI1 protein.34 Alternative chromosomal translocations between members of the EWSR1 and ETS gene families such as t(21;22) and EWSR1/ERG have been identified in the majority of cases that lack the classic EWSR1/FLI1 fusion.4 The most common fusion type joins exon 7 of EWSR1 with exon 6 of FLI1, also known as the type 1 fusion. However, numerous, less-common breakpoints between both genes have been identified. Furthermore, about 10% of cases involve alternate ETS family genes such as the 3′ translocation partner.35
Associations between fusion type and prognosis were observed in the late 1990s, such that tumors harboring a type 1 fusion had a significantly better outcome compared with those with other fusion types.36 However, these results have not been further validated. Prospective sets of tumors from equivalently treated patients enrolled in the Children’s Oncology Group and Euro-Ewing studies were evaluated for fusion status and outcome and failed to reproduce the original findings.37 Hence, current evidence strongly suggests that among the more than 90% of ES tumors that harbor EWSR1 rearrangements, the fusion type is no longer a reliable prognostic marker and should not be used to stratify therapy or instruct treatment decisions.8
Ewing-like sarcomas
Those tumors make up a subgroup of sarcomas with a histopathology similar to ES, but with the CIC/DUX4 or BCOR/CCNB3 gene fusion instead of EWSR1 or FUS rearrangement.38,39 Sarcomas with CIC/DUX4 gene fusion are usually extraskeletal lesions that affect young adults with an aggressive clinical evolution and metastases (lung and lymph nodes). Morphologically, CIC/DUX4 sarcoma resembles an atypical ES with variable expression of CD99 and cytokeratin.38 Sarcomas with the BCOR/CCNB3 gene fusion also reveal a morphology very similar to atypical ES and frequently arise in bone. Strong nuclear immunoexpression of CCNB3 characterizes these tumors; however, molecular studies are needed to confirm the final diagnosis 39
Cell cycle biomarkers
The cell cycle pathway and its multiple protein components are frequently altered in cancer. In ESFT, genetic alterations affecting the regulators of the G1-phase cell cycle transition have been described, including deletion of both CDKN2A (INK4/ARF locus) and RB1. Kovar et al were the first to describe CDKN2A, finding it in 30% of primary tumors and 52% of ES cell lines.40 After this work, several retrospective studies have demonstrated an association between CDKN2A alterations and clinical outcome in ESFT patients.41–44 Patients in these studies were found to have a worse disease-specific survival in univariate and multivariate analysis. A meta-analysis examining the prognostic significance of CDKN2A alterations in ESFT based on six separated studies (N=188 patients) concluded that the pooled risk ratio for worse outcome with CDKN2A alterations was 2.17 (95% confidence interval, 1.55–3.03; P<0.0001).45 Hence, the evidence to support CDKN2A loss as a negative prognostic marker is strong and worthy of prospective validation.
The prognostic significance of TP53 point mutations in ESFTs has also been evaluated in retrospective series by immunohistochemistry and/or sequencing. TP53 overexpression is detected in approximately 10% of ESFT and has been associated with advanced disease at diagnosis, poorer treatment response, and a worse overall survival.46,47 Huang et al,48 in a series of 60 patients, reported TP53 mutations in 13% of cases, as well as CDKN2A homozygous deletions in another 13% of cases.48 These authors demonstrated that TP53 mutations and/or CDKN2A alterations were significantly associated with poor response to chemotherapy, and in a multivariate analysis, TP53 and/or CDKN2A alterations as a single, combined variable were identified as the most significant prognostic factors. Finally, our group, using immunohistochemistry and fluorescent in situ hybridization analyzed a series of cell cycle regulation markers (p53, p21, p27, and Ki-67) in a series of 324 genetically well-characterized ESs.49 In this study, we found that expression of p53 (P=0.025), p21 (P=0.015), and p27 (P=0.013) was higher in disseminated than in localized disease. In addition, Ki67 was expressed in 34% of cases (Figure 3D) and constituted an independent prognostic factor for progression-free and overall survival, independent of the type of treatment. In fact, the study demonstrated that Ki67 expression represents a valuable indicator of poor prognosis in localized ESFT.
As a whole, compelling data from several retrospective studies implicate alterations of TP53, CDKN2A, and Ki67 as negative prognostic biomarkers in ESFT. We are still waiting for the analysis of prospectively collected samples from clinical studies such as the AEWS0031 from the Children’s Oncology Group.8 If studies such as this confirm prior observations, then analysis of these cell cycle proteins will become a strong candidate for inclusion as a prognostic biomarker that can inform treatment decisions in future clinical trials.
Growth signaling pathway expression in ESFT
Mora et al50 described the relationship between clinical outcome and the immunohistochemical expression of proteins involved in active growth signaling pathways such as c-KIT (tyrosine–protein kinase KIT), AKT (protein kinase B), p-mTOR (phosphorylated mTOR), IGF-1R (insulin-like growth factor receptor 1), IGFBP-3 (IGF-binding protein 3), MAPK (MAP-Kinase), p-AKT, p27KIP1, and p70S6 kinase. These authors analyzed 45 ESFTs, finding that of all the variables analyzed, only presence of metastasis, p-mTOR, and p27KIP1 immunostaining were independent prognostic factors of outcome, with p-mTOR and p27KIP1 protein overexpression being associated with better survival.
Ahmed et al51 also studied the expression of therapeutic targets in 72 ESFTs. They performed immunohistochemical (IHC) staining of intracellular molecules mTOR, vascular endothelial growth factor (VEGF), Akt, nuclear factor κB, and BRAF and correlated these results with survival data. The tumors revealed heterogeneous protein expression with poor BRAF immunoreactivity. No prognostic significance was observed, except for VEGF, where decreased VEGF expression was significantly associated with better prognosis.
Expression of KIT protein is an uncommon event in ESFT. An immunohistochemical and mutational study of 71 ESFTs revealed positive expression by IHC in 38% of cases; however, activating mutations were found in only 2.6% of the tumors.52,53 This study confirms that c-kit mutations are not necessarily concordant with CD117 IHC expression in ESFT, as in other neoplasms.52,53 As far as we know, c-kit status has neither diagnostic nor prognostic significance in ESFT.
NOTCH pathway alterations have been described in ESFTs and are associated with abnormalities in tumor cell growth.54,55 Kovar et al54,55 studied the Notch pathway by gene expression profiling and immunohistochemistry in ESFT and found that although Notch receptors were highly expressed, Notch did not appear to be active. High expression of the Notch effector HES1 transcription factor, usually used as a surrogate marker for active Notch, was also restricted to the outside of the nucleus in the majority of ESFTs. This indicates that HES1 is uncoupled from Notch in ESFT and supports a role for Notch in ESFT tumor suppression.
ERBB4 immunohistochemical expression has been studied by Mendoza-Naranjo et al56 in a cohort of 19 paired biopsies of primary and metastatic ESFTs from the same patients, as well as 94 (78 primary and 16 metastatic) nonpaired ESFTs. The immunohistochemical study revealed significantly higher ERBB4 expression in metastatic lesions (78.9%) compared with primary tumors (42.1%), and the Kaplan–Meier survival analysis demonstrated a direct correlation between high ERBB4 expression and poor outcome. These findings confirm a link between high ERBB4 expression and metastatic disease in ES patient samples. In addition, the same group had previously reported that activation of the ERBB4 tyrosine kinase suppresses anoikis, or detachment-induced cell death, and induces chemoresistance in ESFT cell lines in vitro.57
Considering the probability of aberrant expression of ganglioside antigen GD2 in undifferentiated/primitive neuroectodermal tumors related with stem cells, Kailayangiri et al58 explored the GD2 expression in ESFT cell lines and primary tumors, using immunofluorescence. Surface GD2 was detected in ten of ten ESFT cell lines and three of three primary cell cultures. Furthermore, diagnostic biopsies from 12 of 14 patients also revealed GD2 expression. In the same study, they found that GD2-specific T cells had further activity against ES xenografts. In conclusion, GD2 surface expression is common in ESFT and provides a suitable target antigen for immunotherapeutic strategies to prevent metastasis and relapse.
Biomarkers related with angiogenesis
Angiogenesis is essential for sustained tumor growth and provides the systemic network that stimulates metastasis. Without the formation of supporting vasculature, tumor cells would be unable to obtain the nutrients and oxygen necessary for proliferation and would not be able to mediate metastatic spread. A delicately controlled balance between pro- and antiangiogenic factors typically regulates this process; environmental stressors or genetic changes such as hypoxia, acidosis, oncogene activation, and loss of tumor suppressor genes lead to dysfunction of this balance and result in angiogenesis.59
To consider the potential utility of antiangiogenic and vascular targeting therapies, it would be helpful to understand the clinical effect of angiogenesis biomarkers in patients with EFST. Unfortunately, these data are relatively scarce and difficult to interpret. At least three groups have investigated tumor microvascular density as a prognostic factor in EFST. One group demonstrated a significant association between elevated microvascular density and poor event-free survival and overall survival (OS) in 29 patients with ES. In contrast, two other groups reported that tumor microvascular density does not correlate with metastatic status, disease-free survival, or OS.60
VEGF is the best characterized proangiogenic factor and is considered the most important factor involved in the development of the vasculature. There are a number of different VEGF molecules (VEGFA–VEGFE) that bind to VEGF receptors (VEGFR1–VEGF3).
Patients with ESFTs have increased circulating VEGF levels compared with controls.61 Interestingly, EWS-ETS fusion oncoproteins drive the expression of VEGF and may contribute to the increased VEGF levels observed in patients. In addition, a study of angiogenesis using immunohistochemistry62 demonstrated that VEGF expression is correlated with poorer overall survival; however, confirmation with a larger series is needed.
Other biomarkers of angiogenesis have not been studied systematically in patients with EFST. These markers include soluble VEGFR, thrombospondin, placental growth factor, and circulating endothelial cells. Larger studies that specifically include patients with ESFT will be required to further assess the utility of angiogenic biomarkers for prognostic and therapeutic guidance.60
Biomarkers related to epithelial mesenchymal transition
An immunohistochemical study performed in a large series of ESFTs revealed that desmoplakin expression was correlated with improved PFS. In addition, expression of ZO-1 (zona occludens 1) or Snail was correlated with improved OS, and cytokeratin 8/18 immunoreactivity was associated with a poorer prognosis.18
ZEB2 is a transcription factor that represses epithelial gene expression in ES cells, and this, in turn, represses the epithelial phenotype and facilitates metastasis. ZEB2 reduction in ES cells results in a decreased metastatic potential, using a mouse metastasis model.63
Ezrin, a member of the ezrin/radixin/moesin protein family, plays a pivotal role in tumor invasion and metastasis. The prognostic significance of this protein has been studied in a large series of ESFTs (415 cases),64 in which IHC ezrin expression was found to represent a favorable prognostic factor in these tumors.
Tumor protein D52 (TPD52) is overexpressed in cancers of diverse cellular origins, and increased TPD52 expression is associated with increased proliferation and invasive capacity in different cell types.65 Although ESFT can reveal TPD52 immunoexpression, no prognostic significance has been described.66
Biomarkers related to the tumor microenvironment
The Leiden (Holland) working group of ESFT has demonstrated that the CXCR4/CXCL12 axis is frequently altered in ESFTs and affects tumor progression and patient survival by promoting cell growth.67–69 They demonstrated an expression level-dependent negative effect of CXCR4 protein expression on ESFT overall survival and provide evidence for a crucial role of the CXCR4–CXCL12 axis in promoting ESFT cell growth.67 The same group indicated that increased tumor infiltration of CD8+ T-cells is correlated with improved OS.68 In addition, Fujiwara et al,70 using IHC analysis of macrophage infiltration reported that high macrophage infiltration was correlated with poorer OS.
Immunohistochemical analysis of papillomavirus binding factor (PBF), previously defined as an osteosarcoma-associated antigen, was performed by Yabe et al71 in a series of 20 ESFTs. Ninety percent of the tumors were positive for PBF, and in the prognostic analysis, PBF overexpression was significantly associated with decreased disease-free and OS. Their finding demonstrates that PBF overexpression is a poor prognostic factor in ESFT. This result should be validated in large and prospective series.
CCN3 is a family of proteins that stimulate mitosis, adhesion, apoptosis, extracellular matrix production, growth arrest, and migration of multiple cell types.72 Perbal et al73 examined the expression of CCN3 protein by IHC in a large series of ESFTs, and their results showed that expression of the full-length CCN3 in ESFTs is associated with a worse prognosis.
Lectin galactoside-binding soluble binding protein (LGALS3BP) is a matrixcellular protein with a role in tumor progression and metastasis. Zambelli et al74 explored the possible correlation between LGALS3BP and event-free survival and overall survival in ESFT. In their study, IHC expression of this protein was correlated with improved EFS and OS. The association between LGALS3BP and prognosis was confirmed at the protein level, when expression of the molecule was determined in tumor tissues, but not in serum, indicating a role for the protein in the local tumor microenvironment.
Xg is a member of the CD99 family, expressed in ESFT cell lines and ESF primary tumors. Meynet et al75 concluded that Xg immunoexpression in ESFT defines a subgroup of patients with worse prognosis compared with those with Xg-negative localized tumors, indicating a clinical relevance of Xg expression in ESFT.
Connexins are building unit proteins of gap junctions that are prognostic markers in carcinomas.76 Bui et al77 studied Connexin 43 immunoexpression in a series of 36 ESFTs and found cytoplasmic reactivity in 78% of tumors. In addition, high expression was correlated with poorer OS.
The expression in ESFT of proteins, such as lysine demethylase specific, has recently been demonstrated, although studies are ongoing with the goal of defining whether or not there is a prognostic implication of these proteins in the ESFT.78
The six-transmembrane epithelial antigen of the prostate 1 (STEAP1) is a membrane-bound mesenchymal stem cell marker highly expressed in ESFT.79 Grunewal et al80,81 investigated the possible association between STEAP1 immunoexpression and outcome in a series of 114 primary ESFTs. STEAP1 was positive in 62.3% of tumors, and high membranous STEAP1 immunoreactivity was correlated with better overall survival; thus, high membranous STEAP1 expression predicts improved outcome.
Copy number alterations
Genomic instability with subsequent copy number alterations (CNAs) has been well-characterized in ESFTs.82 The most commonly reported CNAs in ESFTs are trisomy of chromosome 8, trisomy of chromosome 12, and gain of chromosome 1q. Using a variety of platforms, several recurring regions of gains and losses with clinical relevance have been described.8 However, most of these results are derived from retrospective studies that use different approaches to identify CNAs. A prospective analysis of CNAs and clinical outcome has not yet been undertaken.
Minimal residual disease
Assessment of minimal residual disease (MRD) has been considered a critical part of therapeutic decision-making in many leukemic processes. Attempts to validate methodologies and prognostic correlations for MRD detection in ESFTs have primarily used reverse-transcriptase polymerase chain reaction and flow cytometry.8
Reverse-transcriptase polymerase chain reaction assays for MRD are designed to identify specific ESFT-related fusion transcripts both in blood and/or bone marrow as evidence of occult micrometastatic disease or persistent disease after systemic therapy. The largest published study examined EWSR1-FLI1 and EWSR1-ERG transcript levels in a total of 172 patients from a study of the French Society of Pediatric Oncology.83 Circulating transcripts were identified in 20% of patients at diagnosis and were more frequently observed in patients with large tumor burdens. In localized disease, the presence of fusion gene transcripts in blood or bone marrow correlated with significantly poorer outcomes.
To rigorously address whether the detection of circulating tumor transcript is of prognostic significance, the multicenter European Ewing Tumor Working Initiative of National Groups Ewing Tumour Studies 1999 trial prospectively collected bone marrow samples for more than 10 years. As the first large prospective trial examining MRD by reverse-transcriptase polymerase chain reaction in ESFT patients, the findings of this study will be critical for evaluating the feasibility and usefulness of this modality as a biomarker for ES.
Flow cytometry has also been used for some studies to define the presence of MRD in ESFT.84,85 These studies use the cell-surface glycoprotein CD99 to identify tumor cells. These studies confirm the feasibility of the approach and provide preliminary support for the potential prognostic significance of MRD.
Predictive biomarkers in ES
CD133 has been described as a putative marker of mesenchymal stem cell in ESFTs, and its immunoexpression has been associated with chemoresistance in at least some cases of primary ESFT.86 Jiang et al86 have explored whether high levels of CD133 are associated with drug resistance in ESFT; their study revealed heterogeneity in PROM1/CD133 expression in tumors and cell lines and confirmed that high levels of PROM1 expression were associated in some cases with chemoresistant disease. Nevertheless, they suggested that further studies are required to elucidate the contribution of PROM1/CD133 expressing cells to therapeutic resistance in a large, prospective cohort of primary ESFT.
A number of biological targets and potentially promising novel agents have been identified for ESFT,87 with insulin growth factor receptor 1 (IGF1R) and poly(adenosine diphosphate-ribose) polymerase 1 (PARP1) being the better-identified targets in the context of ESFT. IGF1R is highly expressed by ES cells, and many studies have demonstrated the importance of the IGF1R pathway in ES tumor models.88–90 Clinical applications of IGF1R-directed antibodies resulted in dramatic responses in a few patients with refractory disease.91 However, in subsequent trials in unselected ES populations, response rates have been only about 10%, albeit in heavily pretreated patients.92,93 Thus, whether IGF1R targeted therapy has failed to provide significant response rates because of a lack of intended biologic activity against the tumor remains unknown.
The ability to predict whether a patient is likely to respond to a novel agent greatly increases the chance of success of a targeted therapy and encourages personalized medicine more generally. Such a biomarker does not exist for IGF1R-directed therapy in ESFT, although some studies have suggested that differential expression and activation of the insulin receptor and nuclear localization of phosphorylated IGF1R may be useful predictors of treatment response.94–96
Another potential treatment to emerge from recent preclinical research is PARP1 inhibition. PARP1 is a key enzyme involved in single-strand repair DNA.97 In late 1990, it was demonstrated that PARP1 is overexpressed in ESFT, that its expression is regulated by ETS transcription factors, and that in vitro and in vivo models of ESFT are highly sensitive to PARP1 inhibitors alone and in combination with temozolomide.98,99 Because PARP1 inhibition has already been evaluated in numerous different adult-onset tumor types, a variety of potential biomarkers of DNA repair currently exist (PARP1 expression or γ-H2AX) and could be incorporated for evaluation in future pediatric trials.
Conclusion
Several diagnostic and prognostic biomarkers for ESFT have been published in recent years; Table 2 summarizes the biomarkers with references. Of particular relevance are studies of cell cycle proteins, epithelial components, and MRD; nevertheless, they all need to be validated in a series of prospective studies.
The findings that only a small proportion of patients with relapsed ESFT respond to targeted therapies serve as an example of the critical need for predictive biomarkers in this disease.
Acknowledgment
The authors thank David Harrison for assistance with English editing.
Disclosure
The authors report no conflicts of interest in this work.
References
Karski EE, Matthay KK, Neuhaus JM, Goldsby RE, Dubois SG. Characteristics and outcomes of patients with Ewing sarcoma over 40 years of age at diagnosis. Cancer Epidemiol. 2013;37(1):29–33. | |
Borinstein SC, Beeler N, Block JJ, et al; COG Ewing Sarcoma Biology Committee. A decade in banking ewing sarcoma: A Report from the Children’s Oncology Group. Front Oncol. 2013;3:57. | |
Womer RB, West DC, Krailo MD, et al. Randomized controlled trial of interval-compressed chemotherapy for the treatment of localized Ewing sarcoma: a report from the Children’s Oncology Group. J Clin Oncol. 2012;30(33):4148–4154. | |
Balamuth NJ, Womer RB. Ewing’s sarcoma. Lancet Oncol. 2010; 11(2):184–192. | |
Lee J, Hoang BH, Ziogas A, Zell JA. Analysis of prognostic factors in Ewing sarcoma using a population-based cancer registry. Cancer. 2010;116(8):1964–1973. | |
De Gruttola VG, Clax P, DeMets DL, et al. Considerations in the evaluation of surrogate endpoints in clinical trials. Summary of a National Institutes of Health workshop. Control Clin Trials. 2001;22(5):485–502. | |
Hodgson DR, Whittaker RD, Herath A, Amakye D, Clack G. Biomarkers in oncology drug development. Mol Oncol. 2009;3(1):24–32. | |
Shukla N, Schiffman J, Reed D, et al; COG Ewing Sarcoma Biology Committee. Biomarkers in Ewing sarcoma: the promise and challenge of personalized medicine. A report from the Children’s Oncology Group. Front Oncol. 2013;3:141. | |
Folpe AL, Goldblum JR, Rubin BP, et al. Morphologic and immunophenotypic diversity in Ewing family tumors: a study of 66 genetically confirmed cases. Am J Surg Pathol. 2005;29(8):1025–1033. | |
Llombart-Bosch A, Machado I, Navarro S, et al. Histological heterogeneity of Ewing’s sarcoma/PNET: an immunohistochemical analysis of 415 genetically confirmed cases with clinical support. Virchows Arch. 2009;455(5):397–411. | |
Pinto A, Dickman P, Parham D. Pathobiologic markers of the Ewing sarcoma family of tumors: state of the art and prediction of behaviour. Sarcoma. 2011;2011:856190. | |
van Maldegem AM, Hogendoorn PC, Hassan AB. The clinical use of biomarkers as prognostic factors in Ewing sarcoma. Clin Sarcoma Res. 2012;2(1):7. | |
Llombart-Bosch A, Contesso G, Peydro-Olaya A. Histology, immunohistochemistry, and electron microscopy of small round cell tumors of bone. Semin Diagn Pathol. 1996;13(3):153–170. | |
Machado I, Traves V, Cruz J, Llombart B, Navarro S, Llombart-Bosch A. Superficial small round-cell tumors with special reference to the Ewing’s sarcoma family of tumors and the spectrum of differential diagnosis. Semin Diagn Pathol. 2013;30(1):85–94. | |
Machado I, Noguera R, Mateos EA, et al. The many faces of atypical Ewing’s sarcoma. A true entity mimicking sarcomas, carcinomas and lymphomas. Virchows Arch. 2011;458(3):281–290. | |
Machado I, Alberghini M, Giner F, et al. Histopathological characterization of small cell osteosarcoma with immunohistochemistry and molecular genetic support. A study of 10 cases. Histopathology. 2010;57(1):162–167. | |
Machado I, Navarro S, López-Guerrero JA, Alberghini M, Picci P, Llombart-Bosch A. Epithelial marker expression does not rule out a diagnosis of Ewing’s sarcoma family of tumours. Virchows Arch. 2011;459(4):409–414. | |
Machado I, López-Guerrero JA, Navarro S, et al. Epithelial cell adhesion molecules and epithelial mesenchymal transition (EMT) markers in Ewing’s sarcoma family of tumors (ESFTs). Do they offer any prognostic significance? Virchows Arch. 2012;461(3):333–337. | |
Yoshida A, Sekine S, Tsuta K, Fukayama M, Furuta K, Tsuda H. NKX2.2 is a useful immunohistochemical marker for Ewing sarcoma. Am J Surg Pathol. 2012;36(7):993–999. | |
Wang WL, Patel NR, Caragea M, et al. Expression of ERG, an Ets family transcription factor, identifies ERG-rearranged Ewing sarcoma. Mod Pathol. 2012;25(10):1378–1383. | |
Miettinen M. Immunohistochemistry of soft tissue tumours – review with emphasis on 10 markers. Histopathology. 2014;64(1):101–118. | |
Miettinen M, Wang Z, Sarlomo-Rikala M, Abdullaev Z, Pack SD, Fetsch JF. ERG expression in epithelioid sarcoma: a diagnostic pitfall. Am J Surg Pathol. 2013;37(10):1580–1585. | |
Miettinen M, Wang ZF, Paetau A, et al. ERG transcription factor as an immunohistochemical marker for vascular endothelial tumors and prostatic carcinoma. Am J Surg Pathol. 2011;35(3):432–441. | |
Llombart-Bosch A, Navarro S. Immunohistochemical detection of EWS and FLI-1 proteinss in Ewing sarcoma and primitive neuroectodermal tumors: comparative analysis with CD99 (MIC-2) expression. Appl Immunohistochem Mol Morphol. 2001;9(3):255–260. | |
Terrier P, Llombart-Bosch A, Contesso G. Small round blue cell tumors in bone: prognostic factors correlated to Ewing’s sarcoma and neuroectodermal tumors. Semin Diagn Pathol. 1996;13(3):250–257. | |
Pagani A, Fischer-Colbrie R, Eder U, Pellin A, Llombart-Bosch A, Bussolati G. Neural and mesenchymal differentiations in Ewing’s sarcoma cell lines. Morphological, immunophenotypic, molecular biological and cytogenetic evidence. Int J Cancer. 1995;63(5):738–743. | |
Terrier P, Henry-Amar M, Triche TJ, et al. Is neuro-ectodermal differentiation of Ewing’s sarcoma of bone associated with an unfavourable prognosis? Eur J Cancer. 1995;31A(3):307–314. | |
Gu M, Antonescu CR, Guiter G, Huvos AG, Ladanyi M, Zakowski MF. Cytokeratin immunoreactivity in Ewing’s sarcoma: prevalence in 50 cases confirmed by molecular diagnostic studies. Am J Surg Pathol. 2000;24(3):410–416. | |
Elbashier SH, Nazarina AR, Looi LM. Cytokeratin immunoreactivity in Ewing sarcoma/ primitive neuroectodermal tumour. Malays J Pathol. 2013;35(2):139–145. | |
Schuetz AN, Rubin BP, Goldblum JR, et al. Intercellular junctions in Ewing sarcoma/primitive neuroectodermal tumor: additional evidence of epithelial differentiation. Mod Pathol. 2005;18(11):1403–1410. | |
Barisella M, Collini P, Orsenigo M, et al. Unusual myogenic and melanocytic differentiation of soft tissue pPNETs: an immunohistochemical and molecular study of 3 cases. Am J Surg Pathol. 2010;34(7):1002–1006. | |
Sorensen PH, Shimada H, Liu XF, Lim JF, Thomas G, Triche TJ. Biphenotypic sarcomas with myogenic and neural differentiation express the Ewing’s sarcoma EWS/FLI1 fusion gene. Cancer Res. 1995;55(6):1385–1392. | |
Tan SY, Burchill S, Brownhill SC, et al. Small round cell tumor with biphenotypic differentiation and variant of t(21;22)(q22;q12). Pediatr Dev Pathol. 2001;4(4):391–396. | |
Delattre O, Zucman J, Plougastel B, et al. Gene fusion with an ETS DNA-binding domain caused by chromosome translocation in human tumours. Nature. 1992;359(6391):162–165. | |
Sankar S, Lessnick SL. Promiscuous partnerships in Ewing’s sarcoma. Cancer Genet. 2011;204(7):351–365. | |
de Alava E, Kawai A, Healey JH, et al. EWS-FLI1 fusion transcript structure is an independent determinant of prognosis in Ewing’s sarcoma. J Clin Oncol. 1998;16(4):1248–1255. | |
Le Deley MC, Delattre O, Schaefer KL, et al. Impact of EWS-ETS fusion type on disease progression in Ewing’s sarcoma/peripheral primitive neuroectodermal tumor: prospective results from the cooperative Euro-E.W.I.N.G. 99 trial. J Clin Oncol. 2010;28(12):1982–1988. | |
Italiano A, Sung YS, Zhang L, et al. High prevalence of CIC fusion with double-homeobox (DUX4) transcription factors in EWSR1-negative undifferentiated small blue round cell sarcomas. Genes Chromosomes Cancer. 2012;51(3):207–218. | |
Pierron G, Tirode F, Lucchesi C, et al. A new subtype of bone sarcoma defined by BCOR-CCNB3 gene fusion. Nat Genet. 2012;44(4):461–466. | |
Kovar H, Jug G, Aryee DN, et al. Among genes involved in the RB dependent cell cycle regulatory cascade, the p16 tumor suppressor gene is frequently lost in the Ewing family of tumors. Oncogene. 1997;15(18):2225–2232. | |
Wei G, Antonescu CR, de Alava E, et al. Prognostic impact of INK4A deletion in Ewing sarcoma. Cancer. 2000;89(4):793–799. | |
Tsuchiya T, Sekine K, Hinohara S, Namiki T, Nobori T, Kaneko Y. Analysis of the p16INK4, p14ARF, p15, TP53, and MDM2 genes and their prognostic implications in osteosarcoma and Ewing sarcoma. Cancer Genet Cytogenet. 2000;120(2):91–98. | |
López-Guerrero JA, Pellín A, Noguera R, Carda C, Llombart-Bosch A. Molecular analysis of the 9p21 locus and p53 genes in Ewing family tumors. Lab Invest. 2001;81(6):803–814. | |
Maitra A, Roberts H, Weinberg AG, Geradts J. Aberrant expression of tumor suppressor proteins in the Ewing family of tumors. Arch Pathol Lab Med. 2001;125(9):1207–1212. | |
Honoki K, Stojanovski E, McEvoy M, et al. Prognostic significance of p16 INK4a alteration for Ewing sarcoma: a meta-analysis. Cancer. 2007;110(6):1351–1360. | |
Abudu A, Mangham DC, Reynolds GM, et al. Overexpression of p53 protein in primary Ewing’s sarcoma of bone: relationship to tumour stage, response and prognosis. Br J Cancer. 1999;79(7–8):1185–1189. | |
de Alava E, Antonescu CR, Panizo A, et al. Prognostic impact of P53 status in Ewing sarcoma. Cancer. 2000;89(4):783–792. | |
Huang HY, Illei PB, Zhao Z, et al. Ewing sarcomas with p53 mutation or p16/p14ARF homozygous deletion: a highly lethal subset associated with poor chemoresponse. J Clin Oncol. 2005;23(3):548–558. | |
López-Guerrero JA, Machado I, Scotlandi K, et al. Clinicopathological significance of cell cycle regulation markers in a large series of genetically confirmed Ewing’s sarcoma family of tumors. Int J Cancer. 2011;128(5):1139–1150. | |
Mora J, Rodríguez E, de Torres C, et al. Activated growth signaling pathway expression in Ewing sarcoma and clinical outcome. Pediatr Blood Cancer. 2012;58(4):532–538. | |
Ahmed AA, Sherman AK, Pawel BR. Expression of therapeutic targets in Ewing sarcoma family tumors. Hum Pathol. 2012;43(7):1077–1083. | |
Do I, Araujo ES, Kalil RK, et al. Protein expression of KIT and gene mutation of c-kit and PDGFRs in Ewing sarcomas. Pathol Res Pract. 2007;203(3):127–134. | |
Kelleher FC, Thomas DM. Molecular pathogenesis and targeted therapeutics in Ewing sarcoma/primitive neuroectodermal tumours. Clin Sarcoma Res. 2012;2(1):6. | |
Ban J, Bennani-Baiti IM, Kauer M, et al. EWS-FLI1 suppresses NOTCH-activated p53 in Ewing’s sarcoma. Cancer Res. 2008;68(17):7100–7109. | |
Bennani-Baiti IM, Aryee DN, Ban J, et al. Notch signalling is off and is uncoupled from HES1 expression in Ewing’s sarcoma. J Pathol. 2011;225(3):353–363. | |
Mendoza-Naranjo A, El-Naggar A, Wai DH, et al. ERBB4 confers metastatic capacity in Ewing sarcoma. EMBO Mol Med. 2013;5(7):1019–1034. | |
Strauss SJ, Ng T, Mendoza-Naranjo A, Whelan J, Sorensen PH. Understanding micrometastatic disease and Anoikis resistance in ewing family of tumors and osteosarcoma. Oncologist. 2010;15(6):627–635. | |
Kailayangiri S, Altvater B, Meltzer et al. The ganglioside antigen G(D2) is surface-expressed in Ewing sarcoma and allows for MHC-independent immune targeting. Br J Cancer. 2012;106(6):1123–1133. | |
Zhu L, McManus MM, Hughes DP. Understanding the biology of bone sarcoma from early initiating events through late events in metastasis and disease progression. Front Oncol. 2013;3:230. | |
DuBois SG, Marina N, Glade-Bender J. Angiogenesis and vascular targeting in Ewing sarcoma: a review of preclinical and clinical data. Cancer. 2010;116(3):749–757. | |
DuBois S, Demetri G. Markers of angiogenesis and clinical features in patients with sarcoma. Cancer. 2007;109(5):813–819. | |
Fuchs B, Inwards CY, Janknecht R. Vascular endothelial growth factor expression is up-regulated by EWS-ETS oncoproteins and Sp1 and may represent an independent predictor of survival in Ewing’s sarcoma. Clin Cancer Res. 2004;10(4):1344–1353. | |
Wiles ET, Bell R, Thomas D, Beckerle M, Lessnick SL. ZEB2 represses the epithelial phenotype and facilitates metastasis in Ewing sarcoma. Genes Cancer. 2013;4(11–12):486–500. | |
Machado I, Navarro S, Giner F, Alberghini M, Bertoni F, Llombart-Bosch A. Ezrin immunohistochemical expression in chondrosarcomas, osteosarcomas and Ewing sarcoma family of tumors. Virchows Arch. 2010;457(1):87–89. | |
Tennstedt P, Bölch C, Strobel G, et al. Patterns of TPD52 overexpression in multiple human solid tumor types analyzed by quantitative PCR. Int J Oncol. 2014;44(2):609–615. | |
Machado I, López-Guerrero JA, Calabuig-Fariñas S, et al. Clinical significance of tumor protein D52 immunostaining in a large series of Ewing’s sarcoma family of tumors. Pediatr Dev Pathol. 2011;14(3):255–256. | |
Berghuis D, Schilham MW, Santos SJ, et al. The CXCR4-CXCL12 axis in Ewing sarcoma: promotion of tumor growth rather than metastatic disease. Clin Sarcoma Res. 2012;2(1):24. | |
Berghuis D, Santos SJ, Baelde HJ, et al. Pro-inflammatory chemokine-chemokine receptor interactions within the Ewing sarcoma microenvironment determine CD8(+) T-lymphocyte infiltration and affect tumour progression. J Pathol. 2011;223(3):347–357. | |
Berghuis D, de Hooge AS, Santos SJ, et al. Reduced human leukocyte antigen expression in advanced-stage Ewing sarcoma: implications for immune recognition. J Pathol. 2009;218(2):222–231. | |
Fujiwara T, Fukushi J, Yamamoto S, et al. Macrophage infiltration predicts a poor prognosis for human ewing sarcoma. Am J Pathol. 2011;179(3):1157–1170. | |
Yabe H, Tsukahara T, Kawaguchi S, et al. Overexpression of papillomavirus binding factor in Ewing’s sarcoma family of tumors conferring poor prognosis. Oncol Rep. 2008;19(1):129–134. | |
Zhang T, Zhao C, Luo L, et al. The clinical and prognostic significance of CCN3 expression in patients with cervical cancer. Adv Clin Exp Med. 2013;22(6):839–845. | |
Perbal B, Lazar N, Zambelli D, et al. Prognostic relevance of CCN3 in Ewing sarcoma. Hum Pathol. 2009;40(10):1479–1486. | |
Zambelli D, Zuntini M, Nardi F, et al. Biological indicators of prognosis in Ewing’s sarcoma: an emerging role for lectin galactoside-binding soluble 3 binding protein (LGALS3BP). Int J Cancer. 2010;126(1):41–52. | |
Meynet O, Scotlandi K, Pradelli E, et al. Xg expression in Ewing’s sarcoma is of prognostic value and contributes to tumor invasiveness. Cancer Res. 2010;70(9):3730–3738. | |
Teleki I, Krenacs T, Szasz MA, et al. The potential prognostic value of connexin 26 and 46 expression in neoadjuvant-treated breast cancer. BMC Cancer. 2013;13:50. | |
Bui MM, Han G, Acs G, et al. Connexin 43 is a potential prognostic biomarker for ewing sarcoma/primitive neuroectodermal tumor. Sarcoma. 2011;2011:971050. | |
Bennani-Baiti IM, Machado I, Llombart-Bosch A, Kovar H. Lysine-specific demethylase 1 (LSD1/KDM1A/AOF2/BHC110) is expressed and is an epigenetic drug target in chondrosarcoma, Ewing’s sarcoma, osteosarcoma, and rhabdomyosarcoma. Hum Pathol. 2012;43(8):1300–1307. | |
Moreaux J, Kassambara A, Hose D, Klein B. STEAP1 is overexpressed in cancers: a promising therapeutic target. Biochem Biophys Res Commun. 2012;429(3–4):148–155. | |
Grunewald TG, Ranft A, Esposito I, et al. High STEAP1 expression is associated with improved outcome of Ewing’s sarcoma patients. Ann Oncol. 2012;23(8):2185–2190. | |
Grunewald TG, Diebold I, Esposito I, et al. STEAP1 is associated with the invasive and oxidative stress phenotype of Ewing tumors. Mol Cancer Res. 2012;10(1):52–65. | |
Jahromi MS, Putnam AR, Druzgal C, et al. Molecular inversion probe analysis detects novel copy number alterations in Ewing sarcoma. Cancer Genet. 2012;205(7–8):391–404. | |
Schleiermacher G, Peter M, Oberlin O, et al; Société Française d’Oncologie Pédiatrique. Increased risk of systemic relapses associated with bone marrow micrometastasis and circulating tumor cells in localized ewing tumor. J Clin Oncol. 2003;21(1):85–91. | |
Dubois SG, Epling CL, Teague J, Matthay KK, Sinclair E. Flow cytometric detection of Ewing sarcoma cells in peripheral blood and bone marrow. Pediatr Blood Cancer. 2010;54(1):13–18. | |
Ash S, Luria D, Cohen IJ, et al. Excellent prognosis in a subset of patients with Ewing sarcoma identified at diagnosis by CD56 using flow cytometry. Clin Cancer Res. 2011;17(9):2900–2907. | |
Jiang X, Gwye Y, Russell D, et al. CD133 expression in chemo-resistant Ewing sarcoma cells. BMC Cancer. 2010;10:116. | |
Kovar H, Alonso J, Aman P, et al. The first European interdisciplinary ewing sarcoma research summit. Front Oncol. 2012;2:54. | |
Scotlandi K, Benini S, Nanni P, et al. Blockage of insulin-like growth factor-I receptor inhibits the growth of Ewing’s sarcoma in athymic mice. Cancer Res. 1998;58(18):4127–4131. | |
Scotlandi K, Benini S, Sarti M, et al. Insulin-like growth factor I receptor-mediated circuit in Ewing’s sarcoma/peripheral neuroectodermal tumor: a possible therapeutic target. Cancer Res. 1996;56(20):4570–4574. | |
Kolb EA, Gorlick R, Houghton PJ, et al. Initial testing (stage 1) of a monoclonal antibody (SCH 717454) against the IGF-1 receptor by the pediatric preclinical testing program. Pediatr Blood Cancer. 2008;50(6):1190–1197. | |
Olmos D, Tan DS, Jones RL, Judson IR. Biological rationale and current clinical experience with anti-insulin-like growth factor 1 receptor monoclonal antibodies in treating sarcoma: twenty years from the bench to the bedside. Cancer J. 2010;16(3):183–194. | |
Malempati S, Weigel B, Ingle AM, et al. Phase I/II trial and pharmacokinetic study of cixutumumab in pediatric patients with refractory solid tumors and Ewing sarcoma: a report from the Children’s Oncology Group. J Clin Oncol. 2012;30(3):256–262. | |
Tap WD, Demetri G, Barnette P, et al. Phase II study of ganitumab, a fully human anti-type-1 insulin-like growth factor receptor antibody, in patients with metastatic Ewing family tumors or desmoplastic small round cell tumors. J Clin Oncol. 2012;30(15):1849–1856. | |
Asmane I, Watkin E, Alberti L, et al. Insulin-like growth factor type 1 receptor (IGF-1R) exclusive nuclear staining: a predictive biomarker for IGF-1R monoclonal antibody (Ab) therapy in sarcomas. Eur J Cancer. 2012;48(16):3027–3035. | |
Scotlandi K, Manara MC, Serra M, et al. Expression of insulin-like growth factor system components in Ewing’s sarcoma and their association with survival. Eur J Cancer. 2011;47(8):1258–1266. | |
Garofalo C, Manara MC, Nicoletti G, et al. Efficacy of and resistance to anti-IGF-1R therapies in Ewing’s sarcoma is dependent on insulin receptor signaling. Oncogene. 2011;30(24):2730–2740. | |
Wang Z, Wang F, Tang T, Guo C. The role of PARP1 in the DNA damage response and its application in tumor therapy. Front Med. 2012;6(2):156–164. | |
Soldatenkov VA, Albor A, Patel BK, Dreszer R, Dritschilo A, Notario V. Regulation of the human poly(ADP-ribose) polymerase promoter by the ETS transcription factor. Oncogene. 1999;18(27):3954–3962. | |
Brenner JC, Feng FY, Han S, et al. PARP-1 inhibition as a targeted strategy to treat Ewing’s sarcoma. Cancer Res. 2012;72(7):1608–1613. |
 © 2014 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2014 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.