Back to Journals » OncoTargets and Therapy » Volume 14
Bioinformatics Analysis of the SIRT Family Members and Assessment of Their Potential Clinical Value
Authors Liu M, Yu J, Jin H, Wang S, Ding J, Xing H, He S , Zeng Y
Received 31 December 2020
Accepted for publication 11 March 2021
Published 15 April 2021 Volume 2021:14 Pages 2635—2649
DOI https://doi.org/10.2147/OTT.S298616
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Prof. Dr. Nicola Silvestris
Mingjiang Liu,* Jingjing Yu,* Hu Jin, Sifan Wang, Jin Ding, Hao Xing, Songqing He, Yonglian Zeng
Department of Hepatobiliary Surgery, The First Affiliated Hospital of Guangxi Medical University, Nanning, 530021, Guangxi, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Songqing He; Yonglian Zeng
Department of Hepatobiliary Surgery, The First Affiliated Hospital of Guangxi Medical University, Nanning, 530021, Guangxi, People’s Republic of China
Tel/Fax +86 771-5352512
Email [email protected]; [email protected]
Background: Hepatocellular carcinoma (HCC) is a highly malignant and common tumor. Many biomarkers have been identified for HCC. However, the available ones are not accurate enough in term of prognostic value and new markers are needed for the prognosis of this disease. Sirtuins are NAD(+)-dependent histone deacetylases involved in many biological processes of cancers, consisting of family members SIRT1-SIRT7. However, the prognostic value of the SIRTs in HCC remains largely unknown.
Methods: Differential expression of SIRTs and survival analysis were assessed in patients with HCC using Oncomine and UALCAN databases. Gene set enrichment analysis (GSEA) was used for pathway analysis. Metascape software was used to construct gene ontologies, metabolic pathways and protein-protein interaction networks. Moreover, a HCC murine model was used to validate the expression levels of SIRT3/6/7 expression.
Results: Differential expression analysis suggested that SIRT2-7, not SIRT1, were expressed at higher levels in HCC tissues compared to adjacent normal tissues. These SIRTs showed some similarities, as revealed by GO and KEGG pathway. Higher SIRT3/6/7 mRNA expression levels were found to be significantly associated with shorter overall survival (OS) in HCC patients. Both SIRT3/6/7 mRNA and protein levels were highly expressed in HCC. In addition, over-expression of SIRT3/6/7 was associated with tumor stage and grade in HCC patients. Univariate analysis showed that SIRT 6/7 expressions were linked to a shorter OS of HCC patients. Multivariate analysis showed that SIRT7 levels were independently associated with a significantly shorter OS in HCC patients.
Conclusion: Differentially expressed SIRT3/6/7 were significantly associated with tumor stage, grade and OS in HCC patients. In addition, SIRT7 were independently associated with a significantly shorter OS in HCC patients. Thus, SIRT3/6/7 can be used as prognostic biomarkers to predict the survival of HCC patients.
Keywords: SIRT3, SIRT6, SIRT7, biomarker, hepatocellular carcinoma
Introduction
Liver cancer is fatal and represent the second cause of cancer mortality in men worldwide.1 The incidence of liver cancer has rapidly increased in recent years.2 The main type of liver cancer is hepatocellular carcinoma (HCC). HCC causes vary between different countries. In more developed countries, HCC is mainly caused by the hepatitis C virus (HCV),3 whereas, in the less developed countries, the first risk factor is chronic HBV infection.4 Even though great efforts have put into elucidating the disease mechanisms of HCC, the prognosis of HCC remains poor. Early diagnosis and treatment are essential for a good prognosis for HCC. Efforts have made to study HCC biomarkers, but not many of them have the potential to be routinely used in clinical practice.
In mammals, there are seven different types of histone deacetylases known as sirtuins (SIRTs), including SIRT1-SIRT7.5,6 Their subcellular localization differs but they are mainly located in the nucleus, mitochondria and cytoplasm.7 All Sirtuins take part in cell metabolism and regulate many cellular functions.8,9 SIRT1 regulates inflammation and cellular senescence through deacetylation of transcription factors and histones.10 SIRT2 deacetylates histones and several non-histone proteins.10 SIRT3 is a mitochondrial protein that target the mitochondria for lysine deacetylation.11 SIRT4 and SIRT5 are very weak deacetylated but are critical for cellular metabolism and DNA damage responses.12,13 SIRT6 is a complex enzyme with multiple catalytic activities and has been associated with metabolic diseases and cancer.14 SIRT7 is the least studied member, but research has shown its involvement in various metabolic processes and cancer.15
Recent studies revealed that several members of SIRT are involved in progression of HCC.16,17 Therefore, the work presented here aimed to explore the potential clinical value of SIRTs in HCC.
Materials and Methods
Oncomine
The Oncomine database (https://www.oncomine.org)18 is a bioinformatics tool that was used to analyze the expression level of the seven SIRTs in common types of cancer, including cancers of the liver, bladder and breast. Differences in transcriptional expression levels were compared using a Student’s t-test. A p < 0.05, fold change >1.5 and gene rank of 10% were the criteria used to determine statistical significance.
UALCAN
UALCAN (http://ualcan.path.uab.edu) is a web resource with data obtained from The Cancer Genome Atlas (TCGA). UALCAN was used to analyze relative transcriptional expression levels of genes in tumor tissues and adjacent normal tissues. The prognostic value of mRNA expression of distinct SIRTs in liver cancers was analyzed by using UALCAN. In addition, the association of gene expression levels with pathologic data was determined.19 In this study, UALCAN was used to analyze mRNA expressions of levels of SIRTs in HCC tissues and their association with clinical stage and grade. A p <0.05 was determined as statistically significant using a Student’s t-test.
TCGA
Information about sequencing and clinical data of more than 30 kinds of human tumors can be analyzed in TCGA.20 In our analysis, the mRNA expression levels of SIRTs from 371 HCC patients were downloaded from the Firehose website (http://gdac.broadinstitute.org). 50 of 371 HCC patients were excluded because of the absence of follow-up data. Finally, 321 HCC patients were included in our analysis (Supplementary Table 1). Clinical data from HCC patients including gender, age, weight, PLT, albumin, creatinine, prothrombin time, AFP, Child-Pugh stage, neoplasm histologic grade and pathologic stage are shown in Supplementary Table 1.
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) Analysis
Enrichment analysis of GO terms and KEGG pathways was performed for SIRT1-7. Three GO terms included biological processes (BPs), cellular components (CCs) and molecular functions (MFs). Single gene enrichment was enriched and visualized using LinkedOmics (http://www.linkedomics.org/).21 GSEA enrichment of KEGG pathways was also enriched using LinkedOmics. The multiple gene enrichment of SIRTs were performed and visualized using Metascape.22 The STRING website (https://string-db.org/)23 was used to construct a PPI network to explore SIRT interactions.
Animals
Male C57BL/6J mice 8 weeks of age were purchased from Hunan SJA Laboratory Animal Co. Ltd. Mice were maintained in 12-hour dark or light cycles and housed in the Animal Facilities of Guangxi Medical University. For in vivo studies, mice were fed a normal chow diet (ND), (TrophicDiets, LAD0011, Nantong, China) and water or West Diet (WD) (TrophicDiets, TP23100, Nantong, China) containing 42% fat, 21% sucrose 1.25% cholesterol and a high sugar solution (23.1 g/L fructose (TrophicDiets, China) and 18.9 g/L glucose (TrophicDiets, China). CCl4 (Aladdin, C112044-500ML,China) at the dose of 0.2 ul (0.32 ug)/g of body weight once per week. Corn oil (Aladdin, C116025-500ML,China) was injected intraperitoneally once a week as a control. Corn oil and CCl4 injections began with dietary management and continued for 24 weeks.24 There were four experimental groups (ND/Oil, ND/CCl4, WD/Oil, WD/CCl4 24 weeks) each containing 6–8 mice each. The mice were euthanized, and samples were collected after 24 weeks. Liver and serum samples were collected and stored at −80 °C until analyzed further. The protocol for animal experiment was approved by the Institutional Animal Care and Use Committee of Guangxi Medical University, and manipulations were performed in accordance with the ethical regulations laid down by the Committee of Guangxi Medical University.
Quantitative Real-Time PCR
Total RNA from tissues was isolated using TRIzol reagent (15596026, Invitrogen, USA). RNA was reverse transcribed into cDNA using a cDNA synthesis kit (Thermo Fisher Scientific, Waltham, USA) following instructions provided by the manufacturer. RT-PCR was performed using SYBR green mix (Bio-Rad, California, USA) on a Real-Time system (CFX96 Bio-Rad, California, USA).
Patient Samples
Samples (HCC tissue and paired adjacent tissue) from a total of 30 HCC patients (22 males and 8 females) treated at The First Affiliated Hospital of Guangxi Medical University from March 2017 to May 2018 were included in this analysis. All patients provided written, informed consent for participation in the study. The Ethics Committee of the First Affiliated Hospital of Guangxi Medical University (Nanning, China) approved the use of these samples for total RNA isolation, quantitative reverse transcription-polymerase chain reaction (qRT-PCR) analysis, immunohistochemical and hematoxylin and eosin (H&E) staining (Solarbio, China). Patient inclusion criteria included patients diagnosed with HCC based on a pathologic biopsy and TNM staging criteria for HCC proposed by the AJCC in 2009,25 patients that were not previously treated using surgery, radiotherapy or chemotherapy.
Histopathology and Immunohistochemistry
Liver tissues were resected (2cm*2cm) and fixed in 10% formalin for 12 hours. Paraffin-embedded sections (4-μm thick) were prepared for immunohistochemical and hematoxylin and eosin (H&E) staining. Sections were incubated with the following primary antibody at 4°C overnight, anti-SIRT3 (Abcam, UK,1:300), anti-SIRT6 (Proteintech.,China,1:100), anti-SIRT7 (Abcam, UK,1:1000). After incubation with labeled the following day using an anti-rabbit universal two-step detection kit (Beijing Zhongshan Jinqiao Biotechnology Co. LTD, China). Finally, the signal was detected by DAB method (Beijing Zhongshan Jinqiao Biotechnology Co. LTD, China). Images were collected using NanoZoomer S60 C13210 series (S60 C13210, Hamamatsu Photonics K.K., Japan).
Biochemical Analysis
Serum ALT and AST levels were measured using an autoanalyzer (Antech Diagnostics, CA, USA)
Statistical Analysis
One-way analysis of variance was used to analyze gene expression. The Cox test was used to analyze survival with the help of SPSS23.0 software. Survival data include the Cox proportional risk ratio and 95% confidence interval. SPSS 23.0 software was used to perform univariate and multivariate analyses. A p < 0.05 was considered statistically significant.
Results
Gene Expression Analysis in HCC Patients
Studies investigating SIRTs in various cancers are presented in the figure (Figure 1). The expression was analyzed using Oncomine and UALCAN. The roles of SIRTs were previously explored in many cancer types. Differences in transcriptional expression of these SIRTs were analyzed using a Student’s t-test. SIRT expression levels in HCC and normal liver tissues (Figure 2). All SIRT family members were highly expressed in HCC tissues compared with adjacent normal tissues, except for SIRT1 (Figure 2).
Enrichment Analysis of GO and KEGG Pathway
After analyzing the expression levels of SIRTs in HCC patients, metabolic pathways and GO terms for these SIRTs were next explored. Functional enrichment and KEGG pathway analyses of SIRTs in HCC were forced at a limitation of p <0.05. Single gene enrichment of SIRTs showed similar changes in BPs, CCs and MFs, (Figure 3). Changes in SIRT1-7 in GO BPs mainly included biological regulation, metabolic processes and stimulus-response as the top three. Changes in GO CCs were focused on the enrichment of the membrane, nucleus and membrane-enclosed lumen. Meanwhile, in MFs, changes were found to be significant in protein binding, ion binding and nucleic acid-binding. GSEA enrichment analysis of KEGG pathway was enriched in Type II diabetes mellitus (SIRT1, Figure 4A), oxidative phosphorylation (SIRT2, Figure 4B), glycine and serine and threonine metabolism (SIRT5, Figure 4E). Others (SIRT3/4/6/7) were enriched in the ribosome (Figure 4C and D, 4F and G). SIRTs were enriched in fatty liver disease (no-alcoholic susceptibility to two) and other liver diseases as well as an alcoholic susceptibility to one, as shown by multiple gene enrichment analysis (Figure 5). These findings imply that SIRTs are closely related to liver diseases.
 |
Figure 4 GSEA enrichment analysis of KEGG pathway SIRT1-7. (A–G) The results of pathways of SIRT1-7 cut-off of criteria were as follows: NSE>1, FDR <0.05, p <0.05. |
 |
Figure 5 Summary of SIRT enrichment analyses. SIRTs were enriched in fatty liver disease (no-alcoholic susceptibility to two) and liver diseases as well as an alcoholic susceptibility to one. |
PPI Network Analysis
To distinguish the relationship between SIRTs, a PPI network was constructed using String. Their adjacent genes were also analyzed and found to be remarkably related to SIRT functions (Figure 6). Liver disease-related genes, including Foxo3, Foxo1, TP53, EP300 and HDAC1, were significantly linked to SIRT functions (Figure 6). Some of these genes were related to tumor formation. These findings suggest that SIRTs may play important roles in liver cancer.
 |
Figure 6 PPI network construction. Protein-protein interaction networks of SIRT1-7. Network nodes: proteins, edges: protein-protein associations. |
Prognostic Value of SIRT mRNA Expression Levels in HCC Patients
UALCAN was used to analyze the prognostic value of SIRT expression in HCC patients. Higher mRNA expression of SIRT3 (p=0.00074), SIRT6 (p=0.0027) and SIRT7 (p< 0.0001) were notably associated with shorter overall survival (OS) of HCC patients (Figure 7C, F and G), while there was no correlation between SIRT1/2/4/5 and HCC patient prognosis (Figure 7A, B, D and E). These results imply that SIRT3/6/7 might be selected as useful biomarkers to predict HCC patient survival.
Gene and Protein Expression Levels of SIRT 3/6/7 in HCC
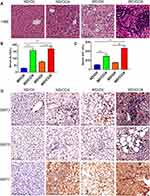
To confirm changes in SIRT3/6/7 in HCC, a liver cancer mouse model was generated to explore the differential expression of SIRT3/6/7. The WD/CCl4 group showed changes in liver pathology, similar to what is observed in liver cancer (Figure 8A). In addition, ALT and AST levels were also significantly increased in WD/CCl4 mice (Figure 8B and C). We performed immunohistochemical staining to verify SIRT3/6/7 expression levels at the protein level. SIRT3/6/7 accumulation increased in the WD/CCl4 mouse group (Figure 8D).
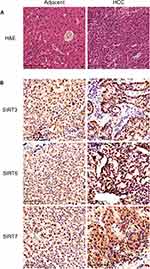
Furthermore, we repeated the validation experiment on human HCC and normal tissues. 30 paired patient tissues were selected from The First Affiliated Hospital of Guangxi Medical University. Results showed that SIRT3/6/7 levels were obviously upregulated in HCC compared to adjacent normal tissues (Figure 9A, B and C). In the 30 paired tissues, SIRT3/6/7 expression levels in HCC were distinctly higher than in adjacent normal tissues, with average fold changes being 3.103, 4.025 and 11.814, respectively (Figure 9D–I). Next, we performed H&E and immunohistochemical (IHC) staining to further identify the positions and expression levels of SIRT3/6/7 protein in cells (Figure 10A and B). Protein expression levels in HCC tissues were higher than in normal tissues (Figure 10B). These results implied that SIRT3/6/7 levels were significantly upregulated at both the mRNA and protein levels.
Relationship Between SIRT3/6/7 Expression Levels and Tumor Stages and Grades in HCC Patients
SIRT3/6/7 expression levels were high in HCC patients. Therefore, we analyzed the relationship between SIRT3/6/7 expression levels and tumor stages and grades in HCC patients by UALCAN. SIRT3/6/7 expression levels correlated with tumor stage (Figure 11). The more advanced the disease, the higher the SIRT3/6/7 mRNA expression levels and the highest levels were observed in stage 3 (Figure 11). Similarly, SIRT3/6/7 expression was distinctly correlated with tumor grade. An increase in grade was significantly associated with higher SIRT3/6/7 expression levels (Figure 12). The highest SIRT3/6 mRNA expression levels were observed in grade 4 tumors (Figure 12A and B), while SIRT7 mRNA expression was the highest in grade 3 (Figure 12C). These results show that SIRT 3/6/7 expression levels are associated with tumor stage and grade in HCC patients.
Independent Prognostic Value of SIRT 3/6/7 mRNA Expression and OS of HCC Patients
After confirming that SIRT 3/6/7 mRNA expression levels are significantly associated with HCC patient prognosis, we explored the prognostic value of SIRT 3/6/7 expression in relation to the OS of HCC patients. Next, base data (Supplementary Table 1) and mRNA expression data for SIRT 3/6/7 in 321 HCC patients from TCGA database were obtained from Firehorse (http://gdac.broadinstitute.org) for Cox survival analysis. Using univariate analysis, high pathologic stage (HR=1.649, 95% CI: 1.315–2.067 and p<0.001) and high mRNA expression levels of SIRT 6/7 (HR=1.001, 95% CI: 1.000–1.001 and p=0.021)/(HR= 1.001, 95% CI: 1.000–1.001 and p=0.005) were related to poor OS for HCC patients (Supplementary Table 2). Multivariate analysis revealed that high expression levels of SIRT3/6 were no significant (Supplementary Tables 3 and 4). On the other hand, SIRT7 (HR=1.001, 95% CI: 1.000–1.001 and p=0.015) was independently related to poor OS in HCC patients (Supplementary Table 5). These results shows that the transcriptional expression of SIRT7 is an independent prognostic factor affecting OS in HCC patients.
Discussion
It was previously reported that SIRTs are involved in the development of a variety of cancers, including HCC.26,27 No one has done a study on bioinformatics analysis of the entire SIRT family in HCC. In this study, the expression levels and prognostic value of different SIRTs in HCC were analyzed. We uncovered that SIRTs are highly expressed in HCC tissues compared to adjacent normal tissues, except for SIRT1. In addition, SIRT mRNA expression levels were remarkably associated with tumor stage and grade in HCC patients. Furthermore, SIRT3/6/7 expression was associated with OS.
Studies have shown that SIRT3, a major deacetylase in mitochondria, may play a dual role in cancer.28 Over-expression of SIRT3 is observed in colorectal cancer, gastric cancer and non-small cell lung cancer.29–31 In gastric cancer tissues, higher SIRT3 expression correlates with gastric cancer subtypes. SIRT3 may increase glycolysis and cell proliferation in cancer cells that have high SIRT3 expression.30 Similarly, over-expression of SIRT3 was also related to poor prognosis in colorectal cancer and non-small cell lung cancer.29,31 In contrast, down-regulation of SIRT3 was observed in breast cancer, kidney cancer and prostate cancer.32–34 Therefore, SIRT3 clearly has differential expression in different cancers.
In this study, SIRT3 levels were elevated at both the mRNA and protein level in tumor tissues. SIRT3 mRNA expression was significantly associated with tumor stage and grade, in agreement with previously published research.29,30 Higher SIRT3 mRNA expression was a related prognostic factor affecting poor OS of HCC patients.
SIRT6 is a member of the SIRT family and has multiple roles in different diseases. Studies showed that SIRT6 promotes specific oncogenic pathways.35 However, the role of SIRT6 in different human cancers is still unclear.36 To better elucidate SIRT6 function, Kugel et al generated SIRT6 deficient mice to determine pancreatic cancer progression to find that its loss did facilitate the progression of pancreatic cancer.37 Others have shown opposite outcomes.38 Some studies reported that SIRT 6 is a tumor suppressor in colorectal cancer, non-small cell lung cancer and renal cell carcinoma.39–41 However, SIRT6 also acts as an oncogene in other cancers.38,42 Jun Sang Bae et al conducted a study investigating SIRT6 in breast carcinoma and demonstrated that CSNK2A1 and SIRT6 were markers for poor prognosis.42
In our study, SIRT6 showed higher expression levels in HCC compared with adjacent normal tissues. The expression of SIRT6 was related to tumor stage and grade, which was similar to the findings published by Ming et al.38 Higher expression of SIRT6 was also linked to shorter OS in HCC patients, acting as an effective prognostic marker.
Similar to SIRT3, upregulation of SIRT7 was observed in numerous malignancies, such as colorectal, gastric, thyroid, node-positive breast, bladder and ovarian cancers.15 In breast cancer patients, SIRT7 is linked to poor prognosis, and low SIRT7 expression is associated with increased metastasis occurrence over time.43 High expression of SIRT7 indicates poor prognosis in breast cancer.44 In gastric cancer patients, the mRNA and protein levels of SIRT7 were both higher and associated with the cancer stage.45
Likewise, in this study, SIRT7 was elevated at both the mRNA and protein levels in tumor tissues. mRNA expression of SIRT7 was remarkably associated with tumor stage and grade. Higher mRNA expression of SIRT7 was also related to a shorter OS in HCC patients. Furthermore, SIRT7 was an independent prognostic factor affecting OS in HCC patients. Our results imply that SIRT7 acts as an oncogene in HCC.
Our study had several limitations. First, a larger population would be needed to validate these results. Second, the potential therapeutic roles of SIRT3/6/7 in HCC were not studied and would require further work. Lastly, the mechanisms underlying SIRT3/6/7 in HCC need more exploration.
Conclusion
In general, the study presented here showed that SIRT3/6/7 significantly correlated with tumor stage, grade and OS in HCC patients. In addition, SIRT7 were independently associated with a significantly shorter OS in HCC patients. These findings suggest that SIRT3/6/7 may serve as prognostic biomarkers for HCC patient survival.
Ethics Approval
This study was approved by the Ethics Committee of the First Affiliated Hospital of Guangxi Medical University. The study was performed in accordance with the Declaration of Helsinki.
Acknowledgments
The authors are grateful for the data support provided by the TCGA databases. The authors are grateful for the contributions of professor Chuang Qin in this research.
Author Contributions
All authors made substantial contributions to conception and design, acquisition of data, implementation of the experiment, establishment of animal models, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; agreed to submit to the current journal; gave final approval of the version to be published; and agreed to be accountable for all aspects of the work.
Funding
This work was supported by National Natural Science Foundation of China (81771674 and 91949122, to Songqing He); The 111 Projects (D17011, to Songqing He).
Disclosure
The authors have no conflicts of interest to disclose.
References
1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi:10.3322/caac.21262
2. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492
3. Testino G, Leone S, Patussi V, Scafato E, Borro P. Hepatocellular carcinoma: diagnosis and proposal of treatment. Minerva Med. 2016;107(6):413–426.
4. de Martel C, Ferlay J, Franceschi S, et al. Global burden of cancers attributable to infections in 2008: a review and synthetic analysis. Lancet Oncol. 2012;13(6):607–615. doi:10.1016/s1470-2045(12)70137-7
5. Carafa V, Rotili D, Forgione M, et al. Sirtuin functions and modulation: from chemistry to the clinic. Clin Epigenetics. 2016;8(1):61. doi:10.1186/s13148-016-0224-3
6. Mendes KL, Lelis DF, Santos SHS. Nuclear sirtuins and inflammatory signaling pathways. Cytokine Growth Factor Rev. 2017;38:98–105. doi:10.1016/j.cytogfr.2017.11.001
7. Houtkooper RH, Pirinen E, Auwerx J. Sirtuins as regulators of metabolism and healthspan. Nat Rev Mol Cell Biol. 2012;13(4):225–238. doi:10.1038/nrm3293
8. Wątroba M, Dudek I, Skoda M, Stangret A, Rzodkiewicz P, Szukiewicz D. Sirtuins, epigenetics and longevity. Ageing Res Rev. 2017;40:11–19. doi:10.1016/j.arr.2017.08.001
9. Vargas-Ortiz K, Pérez-Vázquez V, Macías-Cervantes MH. Exercise and sirtuins: a way to mitochondrial health in skeletal muscle. Int J Mol Sci. 2019;20(11):2717. doi:10.3390/ijms20112717
10. Hwang JW, Yao H, Caito S, Sundar IK, Rahman I. Redox regulation of SIRT1 in inflammation and cellular senescence. Free Radic Biol Med. 2013;61:95–110. doi:10.1016/j.freeradbiomed.2013.03.015
11. Chen Y, Fu LL, Wen X, et al. Sirtuin-3 (SIRT3), a therapeutic target with oncogenic and tumor-suppressive function in cancer. Cell Death Dis. 2014;5(2):e1047. doi:10.1038/cddis.2014.14
12. Min Z, Gao J, Yu Y. The roles of mitochondrial SIRT4 in cellular metabolism. Front Endocrinol (Lausanne). 2018;9:783. doi:10.3389/fendo.2018.00783
13. Bringman-Rodenbarger LR, Guo AH, Lyssiotis CA, Lombard DB. Emerging roles for SIRT5 in metabolism and cancer. Antioxid Redox Signal. 2018;28(8):677–690. doi:10.1089/ars.2017.7264
14. Tasselli L, Zheng W, Chua KF. SIRT6: novel mechanisms and links to aging and disease. Trends Endocrinol Metab. 2017;28(3):168–185. doi:10.1016/j.tem.2016.10.002
15. Wu D, Li Y, Zhu KS, Wang H, Zhu WG. Advances in cellular characterization of the sirtuin isoform, SIRT7. Front Endocrinol (Lausanne). 2018;9:652. doi:10.3389/fendo.2018.00652
16. Wen X, Ling S, Wu W, et al. Ubiquitin-specific protease 22/silent information regulator 1 axis plays a pivotal role in the prognosis and 5-fluorouracil resistance in hepatocellular carcinoma. Dig Dis Sci. 2020;65(4):1064–1073. doi:10.1007/s10620-019-05844-8
17. Wang JX, Yi Y, Li YW, et al. Down-regulation of sirtuin 3 is associated with poor prognosis in hepatocellular carcinoma after resection. BMC Cancer. 2014;14(1):297. doi:10.1186/1471-2407-14-297
18. Rhodes DR, Kalyana-Sundaram S, Mahavisno V, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9(2):166–180. doi:10.1593/neo.07112
19. Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19(8):649–658. doi:10.1016/j.neo.2017.05.002
20. Blum A, Wang P, Zenklusen JC. SnapShot: TCGA-analyzed tumors. Cell. 2018;173(2):530. doi:10.1016/j.cell.2018.03.059
21. Vasaikar SV, Straub P, Wang J, Zhang B. LinkedOmics: analyzing multi-omics data within and across 32 cancer types. Nucleic Acids Res. 2018;46(D1):D956–D963. doi:10.1093/nar/gkx1090
22. Zhou Y, Zhou B, Pache L, Chang M. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10(1):1523. doi:10.1038/s41467-019-09234-6
23. Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–D613. doi:10.1093/nar/gky1131
24. Tsuchida T, Lee YA, Fujiwara N, et al. A simple diet- and chemical-induced murine NASH model with rapid progression of steatohepatitis, fibrosis and liver cancer. J Hepatol. 2018;69(2):385–395. doi:10.1016/j.jhep.2018.03.011
25. Chun YH, Kim SU, Park JY, et al. Prognostic value of the 7th edition of the AJCC staging system as a clinical staging system in patients with hepatocellular carcinoma. Eur J Cancer. 2011;47(17):2568–2575. doi:10.1016/j.ejca.2011.07.002
26. George J, Ahmad N. Mitochondrial sirtuins in cancer: emerging roles and therapeutic potential. Cancer Res. 2016;76(9):2500–2506. doi:10.1158/0008-5472.can-15-2733
27. Olmos Y, Brosens JJ, Lam EW. Interplay between SIRT proteins and tumour suppressor transcription factors in chemotherapeutic resistance of cancer. Drug Resist Updat. 2011;14(1):35–44. doi:10.1016/j.drup.2010.12.001
28. Torrens-Mas M, Oliver J, Roca P, Sastre-Serra J. SIRT3: oncogene and tumor suppressor in cancer. Cancers (Basel). 2017;9(7):90. doi:10.3390/cancers9070090
29. Wei Z, Song J, Wang G, et al. Deacetylation of serine hydroxymethyl-transferase 2 by SIRT3 promotes colorectal carcinogenesis. Nat Commun. 2018;9(1):4468. doi:10.1038/s41467-018-06812-y
30. Cui Y, Qin L, Wu J, et al. SIRT3 enhances glycolysis and proliferation in SIRT3-expressing gastric cancer cells. PLoS One. 2015;10(6):e0129834. doi:10.1371/journal.pone.0129834
31. Yang GC, Fu BC, Zhang DY, et al. The expression and related clinical significance of SIRT3 in non-small-cell lung cancer. Dis Markers. 2017;2017:8241953. doi:10.1155/2017/8241953
32. Liu H, Li S, Liu X, Chen Y, Deng H. SIRT3 overexpression inhibits growth of kidney tumor cells and enhances mitochondrial biogenesis. J Proteome Res. 2018;17(9):3143–3152. doi:10.1021/acs.jproteome.8b00260
33. Li R, Quan Y, Xia W. SIRT3 inhibits prostate cancer metastasis through regulation of FOXO3A by suppressing Wnt/β-catenin pathway. Exp Cell Res. 2018;364(2):143–151. doi:10.1016/j.yexcr.2018.01.036
34. Finley LW, Carracedo A, Lee J, et al. SIRT3 opposes reprogramming of cancer cell metabolism through HIF1α destabilization. Cancer Cell. 2011;19(3):416–428. doi:10.1016/j.ccr.2011.02.014
35. Vitiello M, Zullo A, Servillo L, et al. Multiple pathways of SIRT6 at the crossroads in the control of longevity, cancer, and cardiovascular diseases. Ageing Res Rev. 2017;35:301–311. doi:10.1016/j.arr.2016.10.008
36. Ioris RM, Galié M, Ramadori G, et al. SIRT6 suppresses cancer stem-like capacity in tumors with PI3K activation independently of its deacetylase activity. Cell Rep. 2017;18(8):1858–1868. doi:10.1016/j.celrep.2017.01.065
37. Kugel S, Sebastián C, Fitamant J, et al. SIRT6 suppresses pancreatic cancer through control of Lin28b. Cell. 2016;165(6):1401–1415. doi:10.1016/j.cell.2016.04.033
38. Ming M, Han W, Zhao B, et al. SIRT6 promotes COX-2 expression and acts as an oncogene in skin cancer. Cancer Res. 2014;74(20):5925–5933. doi:10.1158/0008-5472.can-14-1308
39. Qi J, Cui C, Deng Q, et al. Downregulated SIRT6 and upregulated NMNAT2 are associated with the presence, depth and stage of colorectal cancer. Oncol Lett. 2018;16(5):5829–5837. doi:10.3892/ol.2018.9400
40. Chen T, Sun Z, Liu F, Wang Q. RASSF1A and SIRT6 in non-small cell lung cancer: relationship with clinical outcome. Oncol Lett. 2017;14(5):5759–5764. doi:10.3892/ol.2017.6172
41. Jeh SU, Park JJ, Lee JS, et al. Differential expression of the sirtuin family in renal cell carcinoma: aspects of carcinogenesis and prognostic significance. Urol Oncol. 2017;35(12):
42. Bae JS, Park SH, Jamiyandorj U, et al. CK2α/CSNK2A1 phosphorylates SIRT6 and is involved in the progression of breast carcinoma and predicts shorter survival of diagnosed patients. Am J Pathol. 2016;186(12):3297–3315. doi:10.1016/j.ajpath.2016.08.007
43. Tang X, Shi L, Xie N, et al. SIRT7 antagonizes TGF-β signaling and inhibits breast cancer metastasis. Nat Commun. 2017;8(1):318. doi:10.1038/s41467-017-00396-9
44. Huo Q, Li Z, Cheng L, Yang F, Xie N. SIRT7 is a prognostic biomarker associated with immune infiltration in luminal breast cancer. Front Oncol. 2020;10:621. doi:10.3389/fonc.2020.00621
45. Zhang S, Chen P, Huang Z, et al. Sirt7 promotes gastric cancer growth and inhibits apoptosis by epigenetically inhibiting miR-34a. Sci Rep. 2015;5(1):9787. doi:10.1038/srep09787
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.