Back to Journals » International Journal of General Medicine » Volume 15
Bioinformatics Analysis of Prognostic Value of SPC24 in ccRCC and Pan-Cancer
Authors Zhang J, Chen X, Wang J , Zhang P, Han X, Zhang Y, Wang Y , Yang X
Received 20 November 2021
Accepted for publication 31 December 2021
Published 23 January 2022 Volume 2022:15 Pages 817—836
DOI https://doi.org/10.2147/IJGM.S348859
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Scott Fraser
Jipeng Zhang,1 Xinlei Chen,1 Jirong Wang,2 Pengfei Zhang,1 Xue Han,3 Youzhi Zhang,1 Yonghua Wang,1 Xiaokun Yang1
1Department of Urology, The Affiliated Hospital of Qingdao University, Qingdao, Shandong, People’s Republic of China; 2Department of Urology, The Second Affiliated Hospital of Lanzhou University, Lanzhou, Gansu, People’s Republic of China; 3Department of Oncology, The Second Affiliated Hospital of Shandong First Medical University, Taian, Shandong, People’s Republic of China
Correspondence: Yonghua Wang; Xiaokun Yang, Department of Urology, The Affiliated Hospital of Qingdao University, No. 16, Jiangsu Road, Shinan District, Qingdao, Shandong, 266071, People’s Republic of China, Email [email protected]; [email protected]
Purpose: Clear cell renal cell carcinoma (ccRCC) is one of the most common diseases in the world, with high morbidity and mortality. Recent studies have revealed the important role of SPC24, a subunit of the Ndc80 complex, in the occurrence and development of carcinoma. However, the latent effect of SPC24 in the progress of ccRCC remains to be further explored. The intent of this research is to investigate whether SPC24 can be used as an index to predict the progression of ccRCC and to explore its relationship with the immune microenvironment and pan-cancer.
Materials and Methods: Based on data from public databases, we determined the expression level and clinical value of SPC24 in ccRCC and human pan-cancer. RT-qPCR analysis was carried out to detect the expression level of SPC24 in the OSRC/786O (human ccRCC cells) cell lines and HK2 (human normal kidney cells) cell line. The signal transduction pathways activated by different levels of SPC24 expression were explored by Gene Set Enrichment Analysis (GSEA), and the CIBERSORT algorithm was applied to analyze the relationship between infiltrating immune cells and SPC24 expression in ccRCC and pan-cancer tissues.
Results: SPC24 is overexpressed in ccRCC and several types of tumors, which is associated with poor prognosis. GSEA and CIBERSORT algorithms suggested that the high expression group of SPC24 enriched various pathways including immune-related pathways, and the several infiltrated immunocytes were related to the expression of SPC24.
Conclusion: Our study revealed that SPC24 is a prognostic factor in ccRCC related to immunomodulation and has generalized value in pan-cancer.
Keywords: gene expression, SPC24, clear cell renal cell carcinoma, pan-cancer, prognosis, bioinformatics
Introduction
Clear cell renal cell carcinoma (ccRCC) accounts for 70–85% of RCC and has the highest mortality.1 Surgery is the main treatment for localized ccRCC, while immunotherapy and targeted drugs are the first choice for advanced and metastatic ccRCC.2 Despite the continuous progress in cancer treatment, mortality in ccRCC is still rising.3 At present, there is still a lack of effective diagnostic and prognostic biomarkers.4,5 Thus, identifying new biomarkers is essential for the early detection and treatment of ccRCC patients.
SPC24 is part of the Ndc80 complex, regulating the dynamic interaction between nuclear spindle microtubules and centromeres, ensuring faithful and correct chromosome separation during mitosis.6 It has been reported that destruction of SPC24 gene can lead to spindle checkpoint defects, which allows cells to bypass mitosis without correct chromosome segregation,7,8 a process that resembles the characteristics of the onset of tumorigenesis. In addition, previous studies have shown that SPC24 is abnormally expressed in a variety of malignant tumors, such as breast, lung, liver, and thyroid cancers and osteosarcoma.9–13 However, the correlation between SPC24 and the prognosis of ccRCC has not been fully elucidated.
In recent years, researchers have developed a great interest in tumor microenvironment (TME). The latest evidence suggests that resting dendritic cells, activation of dendritic cells, resting mast cells, activation of mast cells, and eosinophils are associated with a favorable prognosis in ccRCC, while B-cell memory, T-cell follicular helper cells and regulatory T cells (Tregs) in ccRCC indicate a poor prognosis.14 Moreover, it has been reported that macrophages have a crucial function in both promoting and preventing tumor growth. The expression of CD163 and CD204 in M2 macrophages is highly associated with poor prognosis in renal cell carcinoma,15 whereas Hutterer et al16 have found that tumor-associated macrophages could independently reduce the risk of death in renal cell carcinoma. Therefore, it is of great significance to determine the dynamic immune components in TME through genetic analysis to guide the treatment and prognosis in ccRCC patients.
Therefore, the purpose of this study was to evaluate the prognostic value of SPC24 and its effect on the immune components of TME in patients with ccRCC and human pan-cancers. More specifically, we compared the expression of SPC24 between tumors and corresponding normal tissues. Then, we evaluated the clinical value of SPC24 in patients with ccRCC by survival and clinical correlation analysis. Afterward, we verified the expression of SPC24 in ccRCC cell lines by RT-qPCR analysis. In addition, we performed Gene Set Enrichment Analysis (GSEA) to identify the signal transduction pathways related to the regulatory mechanism of SPC24. To reveal the correlation between SPC24 expression and the immune microenvironment, the proportion of types of various infiltrated immunocytes in ccRCC was analyzed by the CIBERSORT algorithm. Last, to investigate the potential prognostic value of SPC24 in pan-cancer, we performed further research on SPC24 in 32 other cancer types. Our results show that SPC24 is closely related to the immune subsets in infiltrating tumors, can be used as a biomarker for the diagnosis and prognosis of ccRCC, and has extensive application value in pan-cancer.
Materials and Methods
Data Source
The ccRCC dataset, comprising genome data and corresponding clinical information of 539 tumor samples and 72 normal samples, was obtained from the TCGA database (https://portal.gdc.cancer.gov/repository). The ccRCC microarray data of GSE40435, comprising genome data and corresponding clinical information on 101 tumor samples and 101 para-cancerous samples, was obtained from the Gene Expression Omnibus (GEO) database (http://www.ncbi.nlm.nih.gov/geo/). The t-test was used to conduct the power analysis of the sample data obtained from TCGA and GEO databases based on the R software package “pwr.” The results showed that the power of both groups of sample data is greater than 0.8, which proved that our sample size is sufficient. The detailed clinical data include age at diagnosis, gender, histological grade, clinical stage, T classification, M classification, N classification and survival status (Table 1). In addition, the overall survival time (OS) of ccRCC patients was obtained from the TCGA database, and the disease-free survival time (DFS), progression-free survival time (PFS), and disease-specific survival time (DSS) of ccRCC patients were obtained from the UCSC Xena database. And pan-cancer data including genomic data and corresponding clinical data (32 cancer types, including ACC, BRCA, CESC, CHOL, COAD, DLBC, ESCA, GBM, HNSC, KICH, KIRC, KIRP, LAML, LGG, LIHC, LUAD, LUSC, MESO, OV, PAAD, PCPG, PRAD, READ, SARC, SKCM, STAD, TGCT, THCA, THYM, UCEC, UCS, UVM) were also downloaded from the UCSC Xena database. All the above analysis methods and the R package were implemented by R Foundation for Statistical Computing version 4.1.1, and a P value <0.05 was considered as statistically significant.
 |
Table 1 Clinical Characteristics of ccRCC Patients |
Expression of SPC24 in ccRCC
To explore the expression of SPC24 in ccRCC, the Wilcoxon signed-rank test was used to analyze the comparison of SPC24 expression between ccRCC and normal groups based on the R software package “ggpubr.” In order to further confirm our point, we used the same method to analyze the expression of SPC24 in ccRCC and para-cancerous tissues.
The Prognostic Prediction Value of SPC24 in ccRCC
To explore the relationship between the expression of SPC24 and the prognosis of patients with ccRCC, we divided the samples into a high-expression group and low- expression group according to the median value of SPC24 expression. Based on the R software packages “survival” and “survminer,” we plotted the Kaplan-Meier curves to reveal the differences of the OS, DFS, PFS, and DSS between the SPC24 high- expression group and low-expression group.
The Clinical Correlation Analysis of the SPC24 in ccRCC
Patients with ccRCC in TCGA were divided into groups according to the grade, stage, T stage, N stage, and M stage, and patients with ccRCC in the GEO database were divided into groups according to the grade. In addition, we applied Wilcoxon signed-rank test to determine the correlations between SPC24 expression and clinical characteristics of ccRCC based on the R software package “ggpubr.” Furthermore, to exclude the effects of confounders, univariate COX regression was performed to explore the prognostic value of SPC24 in ccRCC. In order to find independent prognostic factors for ccRCC, we combined SPC24 gene expression with other disease-related factors, including age, gender, grade, stage, T stage, N stage, and M stage to conduct multivariate COX regression analysis.
Establishment and Validation of the Nomogram
To predict the prognosis of patients with ccRCC more effectively, we used the R software package “rms” to establish a prognosis nomogram to evaluate the probability of 1-, 3-, and 5-years of OS in patients with ccRCC based on a number of clinical indicators (age, gender, grade, and stage) and SPC24 expression. Then, a calibration plot was used to evaluate the consistency between the predicted probability and the actual results.
RNA Isolation and Reverse Transcription Quantitative PCR
According to the manufacturer’s protocol, the total RNA in the cultured cells was extracted by using Trizol (Takara). OSRC/786O (human ccRCC cells) and HK2 (human kidney cell biochemistry Pillon) cell lines were provided by the cell bank of the Chinese Academy of Sciences. To detect the level of mRNA, the PrimeScriptTMRT reagent kit (Perfect Real Time; Takara) was used to transcribe the total RNA (500ng) into cDNA. All the primers were synthesized by Huada Gene (Beijing, China), and the sequence was presented as follows: F: 5’-CCCAGAGCCTTCTCAATGCGA-3’; R: 5’-GGCTCACACTCATAATCCCACT-3’. Reverse transcription quantitative PCR (RT-qPCR) was performed by Roche Light Cycler 480II real-time PCR detection system (Roche, Basel, Switzerland). β-actin was used as the internal control, and all operations were repeated three times. The relative expression levels of SPC24 were analyzed by the 2-ΔΔCt method.
GSEA
GSEA is a method of calculation that determines whether a set of predefined genes shows statistically significant and consistent differences between two biological states.17 In this study, GSEA first generated an ordered list of genes based on the correlation between all genes and SPC24 expression and then clarified the significant survival differences observed between high- and low- expression groups. For each analysis, a total of 1000 genome permutations were performed. Nominal P value (NOM p) < 0.05 and false discovery rate (FDR) < 0.05 are considered to be two necessary conditions for a significant enrichment of the gene sets.
Immune Cell Infiltration Analysis
As an immune-related gene, SPC24 has been shown to influence the prognosis of tumors by regulating the immune process and remodeling the immune microenvironment. CIBERSORT is a useful resource for comprehensive analysis of tumor-infiltrating immune cells.18 Therefore, to study the relationship between SPC24 expression and immune cell infiltration, we applied CIBERSORT to determine the proportion of 22 infiltrating immune cells in each ccRCC patient. After that, we analyzed the differences in immune infiltrating cells between high-expression and low-expression groups of SPC24 and used the Wilcoxon rank sum test for significance testing. The Spearman correlation test was performed to examine the correlations between the expression level of SPC24 and the infiltrating immune cells.
Pan-Cancer Analysis
To explore the potential prognostic value of SPC24 in pan-cancer, we conducted the further research on SPC24 in the remaining 32 cancers, similar to the above analysis method. In the pan-cancer data, the expression levels of SPC24 between tumor and normal tissue were compared. Afterward, we also evaluated the prognostic value of SPC24 in the remaining 32 cancers. Finally, we analyzed the correlations between the expression level of SPC24 and the immune cells infiltrated in the pan-cancer data.
Results
SPC24 Overexpression in TCGA and GEO ccRCC Patients
The power analysis that we conducted proved that the sample size obtained from the TCGA and GEO databases was sufficient. In order to explore the relationship between SPC24 and ccRCC, we conducted the analysis of differential gene expression of SPC24. The expression levels of SPC24 in ccRCC and normal kidney tissues were compared, and the results indicated that SPC24 was overexpressed in ccRCC compared with normal kidney tissues (Figure 1A and B). In addition, we analyzed the expression of SPC24 in ccRCC and adjacent tissues and found that the expression of SPC24 in adjacent tissues was also lower than that in ccRCC tissues (Figure 1C and D). The above data suggested that SPC24 may be the carcinogenic gene in ccRCC.
The Expression Level of SPC24 is Correlated with the Survival Outcome of ccRCC Patients
To assess the predictive ability of SPC24, we used survival analysis to detect differences in survival outcomes between groups defined by SPC24 expression. The results demonstrated that compared with patients with low SPC24 expression, patients with high SPC24 expression have significantly poor OS (Figure 2A), DFS (Figure 2B), PFS (Figure 2C), and DSS (Figure 2D).
The Relationship Between the Expression of SPC24 and the Clinical Characteristics of TCGA and GEO ccRCC Patients
The clinicopathological characteristics of ccRCC patients largely determine the treatment strategy. Therefore, we analyzed the relationship between SPC24 expression and clinical characteristics. The results indicated that in TCGA ccRCC patients (Figure 3A–E), the increased expression of SPC24 was closely related to high histological grade, pathological stage, and TNM stage. In the GEO ccRCC patients (Figure 3F), we found that increased SPC24 expression was also dramatically related to high histological grade.
SPC24 is an Independent Prognostic Factor for ccRCC
Univariate and multivariate Cox regression analysis were performed according to the TCGA data to evaluate the prognostic significance of SPC24 and various clinical factors. Univariate Cox regression analysis suggested that age (HR=1.023; P = 0.012), grade (HR=2.242; P < 0.001), stage (HR=1.862; P< 0.001), T stage (HR =1.943; P < 0.001), M stage (HR = 4.073; P < 0.001), N stage (HR = 2.932; P = 0.001), SPC24 (HR = 1.186; P < 0.001) are related to the overall survival rate in ccRCC patients (Figure 4A). Multivariate Cox regression analysis indicated that in the TCGA ccRCC patients, the expression of SPC24 was an independent risk factor for their prognosis (Figure 4B). Based on the above results, we have reason to believe that SPC24 is a risk-related gene and may participate in the occurrence and progress of carcinoma.
 |
Figure 4 Univariate Cox regression and multivariate Cox regression analysis. Forest plot based on univariate (A) and multivariate (B) Cox regression analysis of OS in TCGA database. |
Establishing a Nomogram to Predict OS in ccRCC Patients
To establish a clinically practical method for monitoring the prognosis of ccRCC patients, we constructed a prognostic nomogram to predict the probability of 1-year, 3-year, and 5-year OS (Figure 5A). The predictors in the nomogram include clinical characteristics (age, gender, grade, and stage) and SPC24. Compared with the reference line, the analysis of the calibration curve indicated that the actual and predicted 1-year, 3-year, and 5-year survival times were consistent (Figure 5B-D). These data suggested that the nomogram was trustworthy and accurate.
Increased Expression of SPC24 Was Detected in ccRCC Cells
To clarify the potential role of SPC24 in ccRCC progression, we further measured the expression level of SPC24 with RT-qPCR. We applied the paired t-test to evaluate the differences of SPC24 expression between ccRCC and normal kidney tissues. The results demonstrated that the expression level of SPC24 in ccRCC cell lines (OSRC, 786O) was significantly enhanced compared to that in normal HK2 cell line (Figure 6). Therefore, we believe that SPC24 is a risk-related gene in the progression of ccRCC.
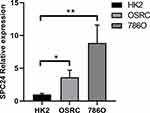 |
Figure 6 SPC24 is highly up-expressed in ccRCC cell lines. Relative SPC24 expression in ccRCC cell lines (OSRC, 786O) compared with normal cell line (HK2) by RT-qPCR analysis. *P<0.05, **P<0.01. |
GSEA
The above studies have indicated that the expression of SPC24 plays a key role in the development of ccRCC. To clarify the underlying mechanism of SPC24 expression affecting the prognosis of ccRCC, we conducted GSEA to explore the differentially activated signaling pathways between the low and high SPC24 expression groups. GSEA revealed significant differences (FDR < 0:05, NOM p < 0.05) in enrichment of the MSigDB Collection (c2.cp.kegg.v6.2.symbols.gmt). According to their normalized enrichment score (NES), the most significantly enriched signaling pathways were selected. The results indicated that five gene sets (colorectal cancer, endometrial cancer, prostate cancer, renal cell carcinoma, and the WNT signaling pathway) may be significantly related to the progression of ccRCC (Figure 7A), and four metabolic-related genomes (base excision repair, DNA replication, homologous recombination, and proteasome pathway) were enriched in the SPC24 low-expression group (Figure 7B). In addition, to explore the relationship between SPC24 and the tumor immune microenvironment, we conducted a more targeted GSEA. The results indicated that in the high SPC24 expression group, immune-related phenotypes (immune response and immune system process pathways) were significantly enriched19 (Figure 7C and D). The results demonstrated that SPC24 may have a certain relationship with the tumor immune microenvironment.
Correlation Analysis Between SPC24 Expression and Immune Cell Infiltration in ccRCC
To further explore the relationship between SPC24 expression and the immune microenvironment, the CIBERSORT calculation method was used to determine the infiltration ratio of 21 immune cells in each ccRCC patient (Figure 8). Then, the differences in the level of infiltrating immune cells between the SPC24 high- expression group and the SPC24 low-expression group, the correlation between the SPC24 expression level and infiltrating immune cells was analyzed (Figure 9). As shown in Figure 10, the results suggested that five kinds of immune cells were positively correlated with the expression of SPC24 (Macrophages M0, T cells follicular helper, T cells CD8, Tregs, T cells CD4 memory activated), and seven kinds of immune cells were negatively correlated with the expression of SPC24 (B cells naive, dendritic cells activated, dendritic cells resting, macrophages M2, mast cells resting, monocytes, and T cells CD4 memory resting). The above data further proved that the levels of SPC24 expression can affect the immune activity in the tumor immune microenvironment.
 |
Figure 9 The plot showed the ratio differentiation of 21 kinds of immune cells between ccRCC samples with low or high SPC24 expression relative to the median of SPC24 expression level. |
 |
Figure 10 Scatter plot showed the correlation of 12 kinds of immunocytes infiltrated proportion with the SPC24 expression. |
The Promotion Value of SPC24 in Pan-Cancer
To explore the potential prognostic value of SPC24 in cancers, we conducted further research on SPC24 in the pan-cancer data. As shown in Figure 11, compared with the corresponding normal tissues, SPC24 was overexpressed in BLCA, BRCA, CESC, CHOL, COAD, ESCA, GBM, HNSC, KICH, KIRC, KIRP, LIHC, LUAD, LUSC, PCPG, PRAD, READ, SARC, STAD, THCA, and UCEC. In addition, survival analysis suggested that patients with high SPC24 expression had significantly poor OS in ACC, KIRP, LIHC, LUAD, MESO, and PAAD (Figure 12A–F). Then, we analyzed the difference in PFS between groups defined by SPC24 expression through survival analysis to rule out the possible impact of cancer follow-up treatment on OS. We found that in ACC, KIRP, LIHC, LUAD, MESO, and PAAD, the PFS of the high SPC24 expression group was also worse than in the low SPC24 expression group (Figure 13A–F). Afterward, based on pan-cancer data, we analyzed the relationship between SPC24 expression and pathological stage. We got the same results: increased SPC24 expression was significantly correlated with high pathological stages in ACC, KIRP, LIHC, LUAD, MESO, and PAAD (Figure 14A–F). Tumor-infiltrating immune cells are considered to be an independent predictor for prognosis in cancer patients.20 Therefore, we further explored the relationship between the expression of SPC24 and the immune microenvironment in 32 cancer types and used the CIBERSORT calculation method to determine the proportion of 21 immune cells infiltrated in each patient. The results showed that the level of infiltration of the five immune cells (CD4 memory resting T cells, CD4 memory activated T cells, follicular helper T cells, NK cells, and macrophages) was correlated with the expression of SPC24 in KIRP, LIHC, and LUAD (Figure 15A–C). The above findings suggested that SPC24 is a predictor of prognosis and has universal value in pan-cancer.
 |
Figure 11 The difference of SPC24 expression between tumor and corresponding normal tissues in 32 cancer types. *P<0.05, **P<0.01, ***P<0.001. |
 |
Figure 12 High SPC24 expression patients were significant related to short OS in ACC (A), KIRP (B), LIHC (C), LUAD (D), MESO (E), and PAAD (F). |
 |
Figure 13 High SPC24 expression groups PFS were worse than that of low SPC24 expression groups in ACC (A), KIRP (B), LIHC (C), LUAD (D), MESO (E), and PAAD (F). |
 |
Figure 14 The relationship between the expression SPC24 and pathological stage in pan-cancer data. ACC (A), KIRP (B), LIHC (C), LUAD (D), MESO (E), and PAAD (F). |
Discussion
CCRCC is one of the tumors with the highest incidence in the urinary system and is known for its high invasiveness and recurrence. Unfortunately, although partial or radical nephrectomy is often performed for early or localized renal cell carcinoma, about 30% of renal cell carcinoma patients are diagnosed in the metastatic stage, and one-third of patients relapse after surgery.21–23 Therefore, further research on ccRCC is urgent, especially in the identification of biomarkers that contribute to early detection, risk stratification, and identification of appropriate interventions.
Recently, SPC24 has been reported to be associated with poor patient prognosis and overexpressed in many cancer entities. For example, Yin et al9 believe that in anaplastic thyroid cancer, SPC24 activates the EMT pathway by down-regulating E-cadherin to cause carcinogenesis. Zhou et al10 proved that in breast cancer, SPC24 caused carcinogenesis by activating the PI3K/Akt pathway. Moreover, they also demonstrated that SPC24 is overexpressed in NSCLC. Knockdown of SPC24 in NSCLC cells can significantly inhibit tumor cell proliferation and promote tumor cell apoptosis.11 Zhu et al12 believe that the levels of SPC24 mRNA and protein in human HCC tissues are significantly higher than in adjacent non-cancerous tissues and normal tissues, indicating that the high expression of SPC24 may be involved in the development of HCC. Sheng et al13 proved that SPC24 promotes osteosarcoma progression by enhancing EGFR/MAPK signal transduction. However, the specific clinical role of SPC24 in ccRCC is not clear as well as whether it can be an effective therapeutic target for patients with ccRCC.
In this research, our results showed that compared with neighboring normal tissues or normal tissues, the expression of SPC24 in ccRCC tissues was up-regulated, which was also confirmed by the results of our next RT-qPCR analysis. Second, in TCGA and GEO data, the up-regulation of SPC24 in ccRCC was associated with late clinical features (grade, stage, T stage, N stage, and M stage) and poor survival outcomes. However, we found there is no significant difference in the expression of SPC24 in patients with G1 and G2/G3, stage I and stage II, and T1 and T2 in the TCGA database. We believe that this is caused by a too small sample size of low-grade and low-stage patients in the TCGA database. Third, based on univariate and multivariate Cox regression, we determined that SPC24 is an independent prognostic factor in TCGA ccRCC patients. Based on these results, we concluded that SPC24 may be a valuable prognostic biomarker in ccRCC.
Afterward, in order to clarify the potential mechanism of SPC24 expression affecting the prognosis of ccRCC, GSEA was carried out to explore the differentially activated signal pathways between the low and high SPC24 expression groups. Our results revealed that SPC24 high-expression phenotype was related to colorectal cancer, endometrial cancer, prostate cancer, renal cell carcinoma, and the WNT signaling pathway. Among them, the WNT signaling pathway plays an important role in the occurrence and progression of a variety of cancers. Wang et al24 found that the WNT signal axis plays a key role in regulating the proliferation of ccRCC cells. Abnormal activation of the WNT signaling pathway in liver cancer can promote the malignant transformation of normal liver cells.25 Abnormal activation of the WNT signaling pathway in colorectal cancer can also inhibit the proliferation of colon cancer cells by inhibiting the activity of the WNT signaling pathway.26–28 However, further mechanistic studies are needed to verify the regulatory mechanism of SPC24 in ccRCC, which may provide a novel treatment method for ccRCC.
It is worth noting that the results of GSEA research also showed that immune-related phenotypes tend to be enriched in the high SPC24 expression group. Increasing evidence shows that immune cells and stromal cells in TME play a role in tumorigenesis and development. In ccRCC, previous studies have shown that the subtypes of tumor-infiltrating immune cells are related to the prognosis.29,30 In order to explore the relationship between the expression of SPC24 and the immune microenvironment of ccRCC, we used the CIBERSORT algorithm to further analyze it. The results showed that 12 kinds of infiltrating immune cells are associated with the expression of SPC24, including those closely related to tumors, such as macrophage M0, Tregs, and so on. Previous studies have reported the relationship between these immune cells and tumor proliferation and progression. Yi et al31 found that there was a significant positive correlation between high macrophage M0 and poor OS in patients with lung adenocarcinoma. Zhang et al 32 reported that M0 macrophages can be transformed into M2 macrophages under certain conditions and promote the proliferation and metastasis of ovarian cancer. De Martino et al33 proved that Tregs are closely related to breast cancer. Wang et al34 found that elevated Tregs play a key negative regulatory role in tumor-associated immunity and contribute to tumor growth and progression. In our research, we found that SPC24 is a gene indicating increased risk, and its expression was positively correlated with the infiltration of macrophages M0 and Tregs. This showed that our research is consistent with previous reports. Overall, we have reason to believe that SPC24 is related to the immune microenvironment.
In order to evaluate the prognostic value of SPC24 in cancer, we conducted a pan-cancer analysis through similar analytical methods. Based on our results, we found that the expression of SPC24 in tumor and normal tissues was significantly different in many cancers. We also found that the expression level of SPC24 was related to clinical stage, so we can speculate that abnormal SPC24 expression is a prognostic factor in some types of cancer. More importantly, the CIBERSORT calculation method proved that the expression of SPC24 has a significant correlation with immune cells in many cancers, so we have reason to believe that SPC24 is related to the tumor immune microenvironment.
It is important to identify the abnormal gene expression in different tumor types, and it is even more important to identify the specific targets or characteristics of the tumor for individualized treatment to increase the chances of successful treatment or cure of cancer cases.35 Pan‐cancer analysis of SPC24 is valuable for identifying differential expression and the role of SPC24 in many cancer types. Pan‐cancer cellular analysis can be performed to assess the expression of genes, which may shed light on future cellular experiments. On the other hand, TCGA genomic and survival analysis may provide guidance for clinical implications and future studies.36 It is worth noting that another key finding of this research is that SPC24 is associated with the level of immune cell infiltration in cancer. This proves that SPC24 may affect the degree of immune infiltration and the activation of various immune cells and provides an immuno-oncological perspective on SPC24 as a prognostic marker of tumor.
Inevitably, our research also has some limitations that need to be pointed out. Because of the limitations of the data, SPC24ʹs ability to predict prognosis has only been verified only in the TCGA and GEO data sets, and it is expected that more patient data sets can be used to verify the significance of SPC24. In addition, in order to accelerate clinical application, these findings need to be verified by more data, such as immunohistochemistry or flow cytometry. Therefore, we will verify the feasibility of SPC24 as a therapeutic target of ccRCC in future studies.
Conclusion
Our study identifies a novel prognostic molecular marker associated with immune cell infiltration in ccRCC. In addition, we also revealed the promoter role of SPC24 in pan-cancer. These findings may provide new insights into prognosis in ccRCC and provide a reference for further study of tumor immunotherapy in ccRCC.
Abbreviations
SPC24, SPC24 component of NDC80 kinetochore complex; ccRCC, clear cell renal cell carcinoma; OS, overall survival; DFS, disease-free survival; PFS, progression-free survival; DSS, disease-specific survival; FDR, false discovery rate; ACC, Adrenocortical carcinoma; BRCA, Breast invasive carcinoma; CESC, Cervical squamous cell carcinoma and endocervical adenocarcinoma; CHOL, Cholangiocarcinoma; COAD, Colon adenocarcinoma; DLBC, Lymphoid Neoplasm Diffuse Large B-cell Lymphoma; ESCA, Esophageal carcinoma; GBM, Glioblastoma multiforme; HNSC, Head and Neck squamous cell carcinoma; KICH, Kidney Chromophobe; KIRC, Kidney renal clear cell carcinoma; KIRP, Kidney renal papillary cell carcinoma; LAML, Acute Myeloid Leukemia; LGG, Brain Lower Grade Glioma; LIHC, Liver hepatocellular carcinoma; LUAD, Lung adenocarcinoma; LUSC, Lung squamous cell carcinoma; MESO, Mesothelioma; OV, Ovarian serous cystadenocarcinoma; PAAD, Pancreatic adenocarcinoma; PCPG, Pheochromocytoma and Paraganglioma; PRAD, Prostate adenocarcinoma; READ, Rectum adenocarcinoma; SARC, Sarcoma; SKCM, Skin Cutaneous Melanoma; STAD, Stomach adenocarcinoma; TGCT, Testicular Germ Cell Tumors; THCA, Thyroid carcinoma; THYM, Thymoma; UCEC, Uterine Corpus Endometrial Carcinoma; UCS, Uterine Carcinosarcoma; UVM, Uveal Melanoma.
Ethical Approval and Consent to Participate
Ethics approval and written consent were not needed for this study exploring the public data. The medical ethics committee of The Affiliated Hospital of Qingdao University agreed to the ethical waiver.
Acknowledgments
The author sincerely thanks all participants who participated in this study.
Disclosure
The authors declare that they have no known competing financial interests or personal relationships that may affect the work reported in this article.
References
1. Chen L, Peng T, Luo Y, et al. ACAT1 and metabolism-related pathways are essential for the progression of Clear Cell Renal Cell Carcinoma (ccRCC), as determined by co-expression network analysis. Front Oncol. 2019;9:957. doi:10.3389/fonc.2019.00957
2. Chen VJ, Hernandez-Meza G, Agrawal P, et al. Time on therapy for at least three months correlates with overall survival in metastatic renal cell carcinoma. Cancers (Basel). 2019;11:7. doi:10.3390/cancers11071000
3. Wu Z, Zhang Y, Chen X, et al. Characterization of the prognostic values of the CXCR1-7 in Clear Cell Renal Cell Carcinoma (ccRCC) microenvironment. Front Mol Biosci. 2020;7:601206. doi:10.3389/fmolb.2020.601206
4. Song E, Song W, Ren M, et al. Identification of potential crucial genes associated with carcinogenesis of clear cell renal cell carcinoma. J Cell Biochem. 2018;119(7):5163–5174. doi:10.1002/jcb.26543
5. Yang JF, Shi SN, Xu WH, et al. Screening, identification and validation of CCND1 and PECAM1/CD31 for predicting prognosis in renal cell carcinoma patients. Aging (Albany NY). 2019;11(24):12057–12079. doi:10.18632/aging.102540
6. Cheeseman IM, Desai A. Molecular architecture of the kinetochore-microtubule interface. Nat Rev Mol Cell Biol. 2008;9(1):33–46. doi:10.1038/nrm2310
7. Bharadwaj R, Yu H. The spindle checkpoint, aneuploidy, and cancer. Oncogene. 2004;23(11):2016–2027. doi:10.1038/sj.onc.1207374
8. Wigge PA, Kilmartin JV. The Ndc80p complex from Saccharomyces cerevisiae contains conserved centromere components and has a function in chromosome segregation. J Cell Biol. 2001;152(2):349–360. doi:10.1083/jcb.152.2.349
9. Yin H, Meng T, Zhou L, et al. SPC24 is critical for anaplastic thyroid cancer progression. Oncotarget. 2017;8(13):21884–21891. doi:10.18632/oncotarget.15670
10. Zhou J, Pei Y, Chen G, et al. SPC24 regulates breast cancer progression by PI3K/AKT signaling. Gene. 2018;675:272–277. doi:10.1016/j.gene.2018.07.017
11. Zhou J, Yu Y, Pei Y, et al. A potential prognostic biomarker SPC24 promotes tumorigenesis and metastasis in lung cancer. Oncotarget. 2017;8(39):65469–65480. doi:10.18632/oncotarget.18971
12. Zhu P, Jin J, Liao Y, et al. A novel prognostic biomarker SPC24 up-regulated in hepatocellular carcinoma. Oncotarget. 2015;6(38):41383–41397. doi:10.18632/oncotarget.5510
13. Sheng J, Yin M, Sun Z, et al. SPC24 promotes osteosarcoma progression by increasing EGFR/MAPK signaling. Oncotarget. 2017;8(62):105276–105283. doi:10.18632/oncotarget.22167
14. Pan Q, Wang L, Chai S, et al. The immune infiltration in clear cell renal cell carcinoma and their clinical implications: a study based on TCGA and GEO databases. J Cancer. 2020;11(11):3207–3215. doi:10.7150/jca.37285
15. Motoshima T, Miura Y, Wakigami N, et al. Phenotypical change of tumor-associated macrophages in metastatic lesions of clear cell renal cell carcinoma. Med Mol Morphol. 2018;51(1):57–63. doi:10.1007/s00795-017-0174-7
16. Hutterer GC, Pichler M, Chromecki TF, et al. Tumour-associated macrophages might represent a favourable prognostic indicator in patients with papillary renal cell carcinoma. Histopathology. 2013;63(3):309–315. doi:10.1111/his.12163
17. Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. doi:10.1073/pnas.0506580102
18. Chen B, Khodadoust MS, Liu CL, et al. Profiling tumor infiltrating immune cells with CIBERSORT. Methods Mol Biol. 2018;1711:243–259.
19. Wang W, Zhao Z, Yang F, et al. An immune-related lncRNA signature for patients with anaplastic gliomas. J Neurooncol. 2018;136(2):263–271. doi:10.1007/s11060-017-2667-6
20. Azimi F, Scolyer RA, Rumcheva P, et al. Tumor-infiltrating lymphocyte grade is an independent predictor of sentinel lymph node status and survival in patients with cutaneous melanoma. J Clin Oncol. 2012;30(21):2678–2683. doi:10.1200/JCO.2011.37.8539
21. Patard JJ, Pignot G, Escudier B, et al. ICUD-EAU International Consultation on Kidney Cancer 2010: treatment of metastatic disease. Eur Urol. 2011;60(4):684–690. doi:10.1016/j.eururo.2011.06.017
22. Capitanio U, Montorsi F. Renal cancer. Lancet. 2016;387(10021):894–906. doi:10.1016/S0140-6736(15)00046-X
23. Rini BI, Campbell SC, Escudier B. Renal cell carcinoma. Lancet. 2009;373(9669):1119–1132. doi:10.1016/S0140-6736(09)60229-4
24. Wang G, Zhang ZJ, Jian WG, et al. Novel long noncoding RNA OTUD6B-AS1 indicates poor prognosis and inhibits clear cell renal cell carcinoma proliferation via the Wnt/β-catenin signaling pathway. Mol Cancer. 2019;18(1):15. doi:10.1186/s12943-019-0942-1
25. Liu Z, Zhang Y, Xu Z. UNC119 promotes the growth and migration of hepatocellular carcinoma via Wnt/β-catenin and TGF-β/EMT signaling pathways. J Buon. 2018;23(1):185–187.
26. Chen H, Wang S, Chen L, et al. MicroRNA-344 inhibits 3T3-L1 cell differentiation via targeting GSK3β of Wnt/β-catenin signaling pathway. FEBS Lett. 2014;588(3):429–435. doi:10.1016/j.febslet.2013.12.002
27. Li T, Zhu J, Wang X, et al. Long non-coding RNA lncTCF7 activates the Wnt/β-catenin pathway to promote metastasis and invasion in colorectal cancer. Oncol Lett. 2017;14(6):7384–7390. doi:10.3892/ol.2017.7154
28. Mokkapati S, Niopek K, Huang L, et al. β-catenin activation in a novel liver progenitor cell type is sufficient to cause hepatocellular carcinoma and hepatoblastoma. Cancer Res. 2014;74(16):4515–4525. doi:10.1158/0008-5472.CAN-13-3275
29. Fu H, Zhu Y, Wang Y, et al. Tumor Infiltrating Mast Cells (TIMs) confers a marked survival advantage in nonmetastatic clear-cell renal cell carcinoma. Ann Surg Oncol. 2017;24(5):1435–1442. doi:10.1245/s10434-016-5702-5
30. Jensen HK, Donskov F, Marcussen N, et al. Presence of intratumoral neutrophils is an independent prognostic factor in localized renal cell carcinoma. J Clin Oncol. 2009;27(28):4709–4717. doi:10.1200/JCO.2008.18.9498
31. Yi M, Li A, Zhou L, et al. Immune signature-based risk stratification and prediction of immune checkpoint inhibitor’s efficacy for lung adenocarcinoma. Cancer Immunol Immunother. 2021;70(6):1705–1719. doi:10.1007/s00262-020-02817-z
32. Zhang Q, Li H, Mao Y, et al. Apoptotic SKOV3 cells stimulate M0 macrophages to differentiate into M2 macrophages and promote the proliferation and migration of ovarian cancer cells by activating the ERK signaling pathway. Int J Mol Med. 2020;45(1):10–22. doi:10.3892/ijmm.2019.4408
33. De Martino M, Daviaud C, Diamond JM, et al. Activin A promotes regulatory T-cell-mediated immunosuppression in irradiated breast cancer. Cancer Immunol Res. 2021;9(1):89–102. doi:10.1158/2326-6066.CIR-19-0305
34. Wang Q, Gao X, Yuan Z, et al. Methionine enkephalin (MENK) improves lymphocyte subpopulations in human peripheral blood of 50 cancer patients by inhibiting regulatory T cells (Tregs). Hum Vaccin Immunother. 2014;10(7):1836–1840. doi:10.4161/hv.28804
35. Andre F, Mardis E, Salm M, et al. Prioritizing targets for precision cancer medicine. Ann Oncol. 2014;25(12):2295–2303. doi:10.1093/annonc/mdu478
36. Liu JN, Kong XS, Sun P, et al. An integrated pan-cancer analysis of TFAP4 aberrations and the potential clinical implications for cancer immunity. J Cell Mol Med. 2021;25(4):2082–2097. doi:10.1111/jcmm.16147
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.