Back to Journals » International Journal of General Medicine » Volume 15
Bioinformatic Analysis of Prognostic Value of SNTG2 with Immune Implications in Lung Adenocarcinoma
Authors Zhou J, Wen Y, Chen X, Guo L
Received 20 January 2022
Accepted for publication 26 April 2022
Published 24 May 2022 Volume 2022:15 Pages 5181—5196
DOI https://doi.org/10.2147/IJGM.S355393
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Scott Fraser
Jian Zhou,* Yang Wen,* Xiangtian Chen,* Linlang Guo
Department of Pathology, Zhujiang Hospital, Southern Medical University, Guangzhou, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Linlang Guo, Department of Pathology, Zhujiang Hospital, Southern Medical University, Guangzhou, People’s Republic of China, Email [email protected]
Background: Lung cancer is the most morbid and fatal cancer in the world, and nearly 85% of lung cancer is non-small cell lung cancer (NSCLC). Besides traditional chemotherapies, molecular targeted therapies and immunotherapies are increasing rapidly, but the treatment is still unsatisfactory. The study is to identify a new diagnostic and prognostic biomarker.
Methods: Data including mRNA expression and clinical information of lung adenocarcinoma (LUAD) patients were downloaded from The Cancer Genome Atlas (TCGA) database. Receiver operating characteristic (ROC) curve was constructed to assess the diagnostic value of syntrophin-γ 2 (SNTG2) expression and Kaplan–Meier (KM) survival curves were used to compare the survival disparities. The protein–protein interaction (PPI) network was analysed by Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) and the connections between SNTG2 and immune cell infiltration were found with Tumor Immunoassay Resource (TIMER). Genetic mutation in SNTG2 and its association with overall survival (OS) were evaluated by cBioPortal. The relationship between SNTG2 and methylation and its association with overall survival were evaluated by MethSurv. Chi square and Mann–Whitney tests were used for statistical analyses. Xiantao Academic Online Website was used for online analysis.
Results: Our results revealed that SNTG2 mRNA expression was lower in LUAD tissues than in both adjacent and non-adjacent normal tissues and low SNTG2 mRNA expression was verified to be correlated with histological grade, clinical stage, first therapy outcome and poor overall survival of LUAD. Next, ROC curve revealed diagnostic and prognostic value of SNTG2 for LUAD patients. Moreover, SNTG2 presented correlation with immune cell infiltration and immune checkpoints. Then, we revealed CC chemokine ligand 14 (CCL14), a co-expression gene with SNTG2, which has consistent influence with SNTG2. Furthermore, hypomethylation was found to be associated with high SNTG2 expression.
Conclusion: We revealed a potential diagnostic and prognostic indicator in LUAD and analyzed its influence on immunotherapy.
Keywords: NSCLC, LUAD, ICI, ICP, biomarker
Introduction
Lung cancer has been the malignant tumor with the highest morbidity and mortality,1–3 meanwhile within lung cancer, non-small cell lung cancer (NSCLC) accounts for about 85%. LUAD is the largest subgroup of NSCLC, for which traditional chemotherapy, surgery and radiotherapy have limitations in efficacy. Targeted therapy is a newly developed method in LUAD aiming at genes including ALK, EGFR, BRAF and so on.4,5 For example, EGFR-TKI (tyrosine kinase inhibitors) treatment, resulting from discovery of EGFR mutation in LUAD patients, has been widely used and shows effective responses.6
However, drug targets currently are very limited in their ability to oppose the developing cancer and the status for clinical treatment is not promising even with many targeted therapies showing promising effects. So it is important to explore more genetic information, identify new potential prognostic biomarkers and reveal novel drug targets for LUAD.
Fortunately, tremendous molecular data from next generation sequencing (NGS) provide chances to identify novel molecular targets for therapeutic improvement.7,8 The Cancer Genome Atlas (TCGA) database, which provides comprehensive and multi-dimensional data covering genome, transcriptome, epigenetics and other omics data, can help us find biomarkers and targets for LUAD.
Syntrophin-γ2 (SNTG2), a potential biomarker in LUAD in our work, has been reported to be required for eye development in Drosophila by affecting hNav1.5 gating kinetics by protein–protein interaction via PDZ domain in the distal C-terminus of the SCN5A-encoded sodium channel pore-forming subunit.9 It has also been reported that overexpression of SNTG2 mRNA is related to osteoporotic vertebral fracture in elderly women.10 However, the relationship between SNTG2 and cancer including LUAD remains unclear.
In this study, we first performed survival analysis for differently expressed genes (DEGs) from TCGA database and explored their association with LUAD prognosis and we confirmed that SNTG2 mRNA expression was lower in LUAD tissues than in normal tissues, and low expression of SNTG2 mRNA was correlated with poor overall survival of LUAD. Next, the link between SNTG2 and immune cell infiltration of tumors was investigated using the TIMER database. Moreover, PPI network information, co-expression genes with SNTG2, genetic mutations in SNTG2 and relationship between SNTG2 and methylation were also evaluated. In summary, this study should be useful for identifying a new potential prognostic biomarker in clinical LUAD treatment.
Methods
Data Source
Data were downloaded from TCGA database (LUAD mRNA expression (IlluminaHiseq), containing 535 LUAD samples). Raw data underwent a log2 transformation. Differential expression genes (DEGs) were calculated according to the criterion: |logFC| >1, FDR <0.05 by limma package in R version 3.5.3.21
Xiantao Academic Online Website
Xiantao Academic (https://www.xiantao.love/products) contains the TCGA database, which conducts online analysis using sequencing data and clinical information.
Survival Analysis
Patients were divided in two groups according to cut-off value of SNTG2 by median expression. Kaplan–Meier (KM) survival curves was used to compare the survival difference.
Univariate and Multivariate Logistic Regression Analysis
Univariate Cox regression analysis was used to calculate the association between the expression level of SNTG2 and patients’ OS. We use multivariate analysis to assess the value of SNTG2 as an independent prognostic factor.
Protein-Protein Interaction Comprehensive Analysis
STRING website22 helped us to gain the protein–protein interaction (PPI) network information and the score >0.7 was regarded significant.
TIMER Database Analysis
The Tumor Immune Estimation Resource (TIMER) database aims to calculate the abundance of immune infiltrates.23 The correlation of SNTG2 expression with infiltrating immune cells in LUAD was evaluated via TIMER database.
Gene Correlation Analysis
The correlation of SNTG2 with markers for immune cells and immune checkpoints was investigated by TIMER. The relation of SNTG2 gene with CCL14 gene was also analysed by TIMER database.
SNTG2 Mutations and Prognosis
We analyzed SNTG2 mutation status and its relationship with prognosis by cBioPortal,24 which helped us through exploring, visualizing, and analyzing.
Correlation Between SNTG2 Expression and Methylation
We analyzed the relationship between SNTG2 and methylation by MethSurv,25 a tool for analyses based on DNA methylation.
Statistical Analysis
We used Univariate Cox regression, Cox regression analysis, Kaplan–Meier curve and the ROC curve by R software. Chi square and Mann–Whitney tests were used for statistical analyses. The statistical significance was established at P <0.05.
Results
Patient Characteristics
We described the clinical information of 535 LUAD samples from TCGA database including patients’ gender, age at diagnosis, histological grade, pathological stage (T, N or M), primary therapy outcomes and OS time in Table 1.
 |
Table 1 Clinical Characteristics of the LUAD Patients |
SNTG2 Was Lowly Expressed in Tumor Samples
To assess the relationship between SNTG2 and multiple cancers, we analyzed data containing unpaired cancer and non-cancer tissues in TCGA database and found that SNTG2 was had low expression in various cancers (Figure 1A), including LUAD (Figure 1B). Furthermore, we compared SNTG2 expression in paired cancer with non-cancer tissues and the result was consistent with unpaired tissues (Figure 1C).
SNTG2 Was Closely Correlated with Clinicopathological Features
We next explored the association of SNTG2 with clinicopathological features of patients with LUAD and found differences in SNTG2 mRNA expression exist in groups based on pathological stage and clinical stage. Specifically, patients in high pathological stage showed lower SNTG2 mRNA expression compared with low pathological stage (Figure 2A) and patients with T3/T4 classification had lower SNTG2 mRNA expression than those with T1/T2 classification (Figure 2B). Furthermore, the results based on N classification (Figure 2C) and M classification (Figure 2D) were consistent.
 |
Figure 2 Association with SNTG2 mRNA expression and clinicopathologic characteristics. (A) Pathologic stage, (B) T stage, (C) N stage, (D) M stage. *P <0.05 and ***P <0.001. |
Association Between SNTG2 and Survival
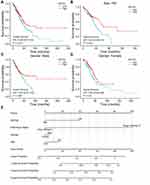
We then analyzed the prognostic relevance of SNTG2 expression on OS. The result shows that high SNTG2 expression prolonged OS for patients with HR value 0.69 (0.51–2.92) (Figure 3A) in both males with HR value 0.67 (0.44–1.03) (Figure 3C) and females with HR value 0.64 (0.42–0.96) (Figure 3D). Moreover, patients diagnosed with LUAD after 65 years of age with high SNTG2 expression had longer OS with HR value 0.53 (0.35–0.81) than those with low SNTG2 expression (Figure 3B). However, this phenomenon was not observed in patients diagnosed before 65 years of age, indicating that SNTG2 may play different roles in different age groups.
Nomogram chart is based on multivariate regression analysis, which integrates multiple predictors to express the relationship between variables in the prediction model. We made a nomogram here to predict the probability of 5-year, 3-year and 1-year overall survival (Figure 3E), on which a higher total number of points represents a worse prognosis, by summing up independent clinical risk factors and SNTG2 mRNA expression. The C-index value was 0.630 for SNTG2 based on 1000 bootstrap replicates, indicating the predictive model is useful.
Diagnostic and Prognostic Value of SNTG2 mRNA Expression in LUAD
The ROC curve is mainly used for the prediction accuracy of X to Y. According to the position of the curve, the entire graph is divided into two parts and the area under curve (AUC) is used to indicate the prediction accuracy. To evaluate the diagnostic value of SNTG2, we created an ROC curve and calculated AUC of SNTG2 was 0.802 (Figure 4A), suggesting that the expression level of SNTG2 has good efficiency to distinguish tumor patients from non-clinical individuals. We then assessed the diagnostic capability of SNTG2 for clinical stages and AUC for stage I, II, I II and IV were 0.769, 0.827, 0.893 and 0.746 (Figure 4E–H), showing that SNTG2 could also distinguish tumor-free individuals from patients of different clinical stages.
It is generally accepted that primary therapy outcomes include progressive disease (PD), stable disease (SD), partial response (PR), and complete response (CR). We then evaluated the diagnostic value of SNTG2 by the same method and our results indicated SNTG2 mRNA had potential prognostic ability to discriminate patients with different primary therapy outcomes according to AUC values of 0.551, 0.600 and 0.621 (Figure 4B–D).
Correlation Between SNTG2 and Clinical Characteristics of LUAD
Going further, we calculated the correlations between SNTG2 and clinical characteristics in LUAD by Cox regression. The results showed that SNTG2 mRNA expression was associated with primary therapy outcomes, confirming potential prognostic ability of SNTG2 (Table 2).
 |
Table 2 Univariate Analysis and Multivariate Analysis of SNTG2 |
 |
Table 3 Correlations Between SNTG2 and Biomakers of Immune Cells |
Correlation Analysis Between Infiltrating Immune Cells and SNTG2
Considering the significance of immune cell infiltration on survival of cancer patients,11 we then explored the relationship between SNTG2 expression and immune infiltration. Firstly, we assessed the difference in the infiltration of several types of immune cells between different SNTG2 expression and we observed that immune cells including T cells, CD8+ T cells, cytotoxic cells, B cells, DC cells and macrophages infiltrated more in patients with high SNTG2 expression (Figure 5A), indicating SNTG2 may suppress tumor by promoting specific immune cell infiltration. Secondly, we made a lollipop graph after calculating the correlation of infiltrating immune cells and tumor purity with SNTG2 and the result indicated SNTG2 expression was positively related with mast cells, pDC, eosinophils, B cells, T cells and NK cells, meanwhile being negatively related with Th2 cells and Tgd (Figure 5B). Thirdly, the analysis with TIMER database showed SNTG2 expression was positively correlated with infiltrating levels of B cells (r = 0.159, P = 2.52e-05), CD4+ T cells (r = 0.157, P = 5.42e-04) and macrophages (r = 0.094, P = 3.78e-02) in LUAD (Figure 5C).
In addition to the above, we also calculated the correlation between SNTG2 and immune markers accepted as symbols of immunocytes to intensely understand the function of SNTG2 in LUAD (Table 3).12
Correlation Analysis Between SNTG2 and Immune Checkpoints
Considering that immune checkpoints, aiming to help T cells to recognize and attack cancer cells, are extremely important molecular information in immunotherapy,13 better understanding of immune checkpoint regulation is expected to increase efficacy of immune checkpoint therapy (ICT). Hence, we analyzed the correlation of SNTG2 with immune checkpoints from TIMER database and the results showed SNTG2 expression was significantly positively correlated with KIR3DL1 (r = 0.15, P = 6.35e-04), CD27 (r = 0.18, P = 3.93e-05) and C10ORF54 (r = 0.243, P = 2.18e-085) (Figure 6), suggesting SNTG2 may affect the efficacy of ICT.
 |
Figure 6 Correlation of SNTG2 expression with immune checkpoints. (A–L) CD274, PDCD1, CTLA4, IDO1, KIR3DL1, LAG3, CD276, CD27, TNFRSF4, HAVCR2, TNFRSF18, C10ORF54. |
Networks of SNTG2 Protein Interaction
It is widely accepted that protein is the main bearer of biological function, hence deeper understanding of molecular interactions at the protein level is necessary. PPI network analyzed by STRING database showed DMD, SNTA1, SNTB1, DTNB, SNTB2, UTRN, DTNA, SGCE, SGCD, DAG1 were the top 10 correlational proteins (Figure 7). It is worth noting that DMD is one of the most common targets of de novo and consequential mutation in the genome, and Duchenne muscular dystrophy (DMD) is a severely debilitating and lethal genetic disease caused by DMD mutations.14 Notably, SNTA1, sharing structural similarity with SNTG2,9 has been regarded as being involved in cancer proliferation and carcinogenesis.15 Besides, UTRN is suspected as a tumor suppressor gene because nonsense and frameshift mutations in UTRN have been reported in several cancers.16 These all indicate that SNTG2 may play an important role not only in LUAD, but also in life activities.
 |
Figure 7 SNTG2-interaction proteins in LUAD tissue. Annotation of SNTG2-interacting proteins and their co-expression scores. |
SNTG2 mRNA and CCL14 mRNA Were Co-Expressed in LUAD Patients
For further gaining the knowledge of SNTG2 biological function in LUAD, co-expression genes of SNTG2 in TCGA-LUAD were detected by R software. The heat map showed the top 50 genes positively associated with SNTG2 (Supplementary Figure 1). Among these 50 genes, we selected CC chemokine ligand 14 (CCL14), the coding genes with the highest correlation with SNTG2, for follow-up studies after second verification of the association of SNTG2 with CCL14 by Timer database (Figure 8D).
There have been many reports of CCL14 in cancers. For example, CCL14 was reported to suppress HCC through modulating cell cycle and promoting apoptosis.17 In another article, CCL14 played an important in breast cancer18 and functioned in ovarian cancer.19 However, the relationship between CCL14 and LUAD remains ill-defined.
The Validation and Survival Analysis of CCL14 in LUAD
To explore the function of CCL14 on LUAD patients, we validated expression status and impact on survival of CCL14 in LUAD from TCGA. CCL14 was found significantly down-regulated both in unpaired and paired LUAD tissues than in normal tissues (Figure 8A and B) and lower CCL14 expression was related with shorter OS (Figure 8C). These results indicated that CCL14 correlates with LUAD malignancies and may serve as a prognostic indicator. Regrettably, CCL14 seems not to be a good prognostic indicator.
Genetic Mutation in SNTG2 and Its Associations with OS
Gene mutations are common in cancers, so we further explored SNTG2 mutations in LUAD. Among 305 patients with LUAD in TCGA database, SNTG2 genetic alteration was found in 8 patients, showing a mutation rate of 2.6% (Supplementary Figure 2). However, although the low mutation rate is low, our results suggest that SNTG2 alteration is associated with worse patient outcomes according to poor OS (Figure 9A). These results implied that mutation in SNTG2 could affect prognosis in LUAD.
 |
Figure 9 SNTG2 alteration and methylation are associated with worse OS in LUAD patients. Survival analysis for alteration of SNTG2 (A) and methylation of SNTG2 (B). |
Correlation Between SNTG2 Expression and Methylation
Finally, we assessed the relationship between SNTG2 expression and methylation level and its value for prognosis in LUAD by using MethSurv. Similar to gene alteration, SNTG2 behaves at a low methylation level (Supplementary Figure 3)20 and higher SNTG2 methylation was associated with poor prognosis (Figure 9B).
Discussion
LUAD is the largest subspecies of lung cancer, accounting for about 30%, which seriously threatens people’s health. We need deeper understanding of LUAD from more dimensions in addition to routine pathological grading, clinical staging and other clinicopathological features. At this time, the popularity of next-generation sequencing has provided us with knowledge at the molecular biology level. It is of broad and far-reaching significance to conduct in-depth exploration of genes in patients with LUAD, to clarify their relationship with the clinicopathological characteristics of patients, and to verify their evaluation value for diagnosis and prognosis.
Genetic testing and corresponding targeted therapy for patients have become routine methods in the process of clinical diagnosis and treatment of lung adenocarcinoma. At present, common therapeutic targets include EGFR, ALK, ROS1 and MET. However, these therapeutic targets do not cover all LUAD patients. Therefore, it is necessary to explore new therapeutic targets for LUAD.
Based on the above considerations, we deeply explored the relationship between SNTG2 and LUAD. The SNTG2 gene has been reported to be related to eye development in Drosophila and osteoporosis in elderly women, but there has been no report concerning its relationship with tumors, so the content of this study is sufficiently novel.
First of all, we explored the relationship between SNTG2 and LUAD patients, and obtained two main conclusions: 1. The expression level of SNTG2 in LUAD tissues was lower than that in normal tissues, and the level of SNTG2 was correlated with the prognosis of LUAD patients; 2. SNTG2 can be used as a diagnostic marker for LUAD, and SNTG2 is related to the clinical stage and pathological grade of LUAD patients. These results suggest that SNTG2 plays a role in LUAD, and importantly, it can be used as a potential biomarker for the diagnosis and prognosis of LUAD.
Immunotherapy has become a key treatment method in tumor treatment, such as anti-PD-1/PD-L1 immune checkpoint inhibitor therapy, which has brought considerable clinical benefits to tumor patients. Therefore, we further explored the relationship between SNTG2 and immune cell infiltration and immune checkpoints in LUAD, and obtained the following two main conclusions: 1. The infiltration of immune cells such as CD4+ T cells, B cells, DC cells and macrophages is positively related with SNTG2; 2. SNTG2 is positively correlated with immune checkpoints such as KIR3DL1, CD27 and C10ORF54. These results suggest that the expression level of SNTG2 may be related to the local immune micro-environment of LUAD patients and further influences the response to immunotherapy.
Considering that regulation of life activities is achieved through a very complex network, we further explored the genes co-expressed with SNTG2. Our results showed that, as the coding gene with the highest correlation with SNTG2, the relative expression of CCL14 in LUAD patients and its effect on the prognosis of LUAD patients are consistent with SNTG2. This results indicated that the molecular biological function of SNTG2 in LUAD is likely to be completed by cooperating with other molecules, which is worthy of further exploration.
Finally, for far more enriching understanding of SNTG2, we explored the interaction network of SNTG2 protein, the relationship between SNTG2 gene mutation and methylation and LUAD.
Our study had some limitations although it provides some value. Firstly, the conclusions of this study, resulting from bioinformatics analysis, have no molecular biology experimental support. Secondly, the specific role of SNTG2 in the development of LUAD should be comprehensively evaluated. Thirdly, the current study focuses on the transcription level of SNTG2 but not protein level, and the specific mechanism of SNTG2 in LUAD remains unknown.
In order to make the conclusion more meaningful, further biological experiments are needed to investigate the mechanism of SNTG2 and clinical trials are necessary for clinical translation.
Conclusions
Our study describes the relationship between SNTG2 and clinical features, immune cell infiltration, and immune checkpoints in LUAD patients, reveals the impact of SNTG2 mutation and methylation on LUAD patients, and explained the relationship between CCL14, an important gene in the SNTG2 interaction network, and LUAD. Most importantly, we established the status of SNTG2 as a diagnostic and prognostic indicator for LUAD.
Abbreviation
NSCLC, non-small cell lung cancer; LUAD, lung adenocarcinoma; NGS, next generation sequencing; ROC, receiver operating characteristic; AUC, Area Under the Curve; SNTG2, syntrophin-γ2; TIMER, Tumor Immunoassay Resource; PPI, protein–protein interaction; CCL14, CC chemokine ligand 14; ICI, Immune cell infiltration; ICP, Immune checkpoints; ICT, immune checkpoint therapy; OS, overall survival; DEGs, differential expression genes; STRING, Search Tool for the Retrieval of Interacting Genes/Proteins; CR, complete response; PR, partial response; SD, stable disease; PD, progressive disease; DMD, Duchenne muscular dystrophy.
Data Sharing Statement
Data in this research is from TCGA database and is available.
Ethic Approval
The research meets the review exemption condition for the collection or research of previously archived data, records which are public resources, by ethics committee of Zhujiang Hospital, Southern Medical University.
Consent for Publication
All authors in this research agree to publish the article.
Acknowledgments
We sincerely thank the public databases, including TCGA, TIMER, STRING, cBioPortal and MethSurv, for providing open access.
Funding
There is no funding to this research.
Disclosure
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. PMID: 25651787. doi:10.3322/caac.21262
2. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67(1):7–30. PMID: 28055103. doi:10.3322/caac.21387
3. Wakelee H, Kelly K, Edelman MJ. 50 years of progress in the systemic therapy of non-small cell lung cancer. Am Soc Clin Oncol Educ Book. 2014; (34):177–189. PMID: 24857075; PMCID: PMC5600272. doi:10.14694/EdBook_AM.2014.34.177
4. Stella GM, Luisetti M, Pozzi E, Comoglio PM. Oncogenes in non-small-cell lung cancer: emerging connections and novel therapeutic dynamics. Lancet Respir Med. 2013;1(3):251–261. PMID: 24429131. doi:10.1016/S2213-2600(13)70009-2
5. Paez JG, Jänne PA, Lee JC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304(5676):1497–1500. PMID: 15118125. doi:10.1126/science.1099314
6. Mitsudomi T, Yatabe Y. Mutations of the epidermal growth factor receptor gene and related genes as determinants of epidermal growth factor receptor tyrosine kinase inhibitors sensitivity in lung cancer. Cancer Sci. 2007;98(12):1817–1824. PMID: 17888036. doi:10.1111/j.1349-7006.2007.00607.x
7. Botling J, Edlund K, Lohr M, et al. Biomarker discovery in non-small cell lung cancer: integrating gene expression profiling, meta-analysis, and tissue microarray validation. Clin Cancer Res. 2013;19(1):194–204. PMID: 23032747. doi:10.1158/1078-0432.CCR-12-1139
8. Aibar S, Abaigar M, Campos-Laborie FJ, Sánchez-Santos JM, Hernandez-Rivas JM, De Las Rivas J. Identification of expression patterns in the progression of disease stages by integration of transcriptomic data. BMC Bioinform. 2016;17(Suppl 15):432. PMID: 28185568; PMCID: PMC5133487. doi:10.1186/s12859-016-1290-4
9. Wu G, Ai T, Kim JJ, et al. alpha-1-syntrophin mutation and the long-QT syndrome: a disease of sodium channel disruption. Circ Arrhythm Electrophysiol. 2008;1(3):193–201. PMID: 19684871; PMCID: PMC2726717. doi:10.1161/CIRCEP.108.769224
10. Jales Neto LH, Wicik Z, Torres GHF, et al. Overexpression of SNTG2, TRAF3IP2, and ITGA6 transcripts is associated with osteoporotic vertebral fracture in elderly women from community. Mol Genet Genomic Med. 2020;8(9):e1391. PMID: 32602654; PMCID: PMC7507059. doi:10.1002/mgg3.1391
11. Jiang P, Gu S, Pan D, et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat Med. 2018;24(10):1550–1558. PMID: 30127393; PMCID: PMC6487502. doi:10.1038/s41591-018-0136-1
12. Kadara H, Choi M, Zhang J, et al. Whole-exome sequencing and immune profiling of early-stage lung adenocarcinoma with fully annotated clinical follow-up. Ann Oncol. 2017;28(1):75–82. Erratum in: Ann Oncol. 2018 Apr 1;29(4):1072. PMID: 27687306; PMCID: PMC5982809. doi:10.1093/annonc/mdw436
13. Sade-Feldman M, Yizhak K, Bjorgaard SL, et al. Defining T cell states associated with response to checkpoint immunotherapy in melanoma. Cell. 2018;175(4):998–1013.e20. Erratum in: Cell. 2019 Jan 10;176(1–2):404.PMID: 30388456; PMCID: PMC6641984. doi:10.1016/j.cell.2018.10.038
14. Chamberlain JR, Chamberlain JS. Progress toward gene therapy for Duchenne muscular dystrophy. Mol Ther. 2017;25(5):1125–1131. PMID: 28416280; PMCID: PMC5417844. doi:10.1016/j.ymthe.2017.02.019
15. Bhat HF, Baba RA, Adams ME, Khanday FA. Role of SNTA1 in Rac1 activation, modulation of ROS generation, and migratory potential of human breast cancer cells. Br J Cancer. 2014;110(3):706–714. PMID: 24434436; PMCID: PMC3915110. doi:10.1038/bjc.2013.723
16. Zhou S, Ouyang W, Zhang X, et al. UTRN inhibits melanoma growth by suppressing p38 and JNK/c-Jun signaling pathways. Cancer Cell Int. 2021;21(1):88. PMID: 33632212; PMCID: PMC7905598. doi:10.1186/s12935-021-01768-4
17. Zhang X, Wan JX, Ke ZP, Wang F, Chai HX, Liu JQ. TMEM88, CCL14 and CLEC3B as prognostic biomarkers for prognosis and palindromia of human hepatocellular carcinoma. Tumour Biol. 2017;39(7):1010428317708900. PMID: 28718365. doi:10.1177/1010428317708900
18. Li Q, Shi L, Gui B, et al. Binding of the JmjC demethylase JARID1B to LSD1/NuRD suppresses angiogenesis and metastasis in breast cancer cells by repressing chemokine CCL14. Cancer Res. 2011;71(21):6899–6908. PMID: 21937684. doi:10.1158/0008-5472.CAN-11-1523
19. Feng L, Houck JR, Lohavanichbutr P, Chen C. Transcriptome analysis reveals differentially expressed lncRNAs between oral squamous cell carcinoma and healthy oral mucosa. Oncotarget. 2017;8(19):31521–31531. PMID: 28415559; PMCID: PMC5458226. doi:10.18632/oncotarget.16358
20. Yu Y, Wang Z, Zheng Q, Li J. FAM72 serves as a biomarker of poor prognosis in human lung adenocarcinoma. Aging. 2021;13(6):8155–8176. PMID: 33686947; PMCID: PMC8034972. doi:10.18632/aging.202625
21. Diboun I, Wernisch L, Orengo CA, Koltzenburg M. Microarray analysis after RNA amplification can detect pronounced differences in gene expression using limma. BMC Genomics. 2006;7(1):252. PMID: 17029630; PMCID: PMC1618401. doi:10.1186/1471-2164-7-252
22. Szklarczyk D, Morris JH, Cook H, et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45(D1):D362–D368. PMID: 27924014; PMCID: PMC5210637. doi:10.1093/nar/gkw937
23. Li T, Fan J, Wang B, et al. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77(21):e108–e110. PMID: 29092952; PMCID: PMC6042652. doi:10.1158/0008-5472.CAN-17-0307
24. Gao J, Aksoy BA, Dogrusoz U, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal. 2013;6(269):pl1. PMID: 23550210; PMCID: PMC4160307. doi:10.1126/scisignal.2004088
25. Modhukur V, Iljasenko T, Metsalu T, Lokk K, Laisk-Podar T, Vilo J. MethSurv: a web tool to perform multivariable survival analysis using DNA methylation data. Epigenomics. 2018;10(3):277–288. PMID: 29264942. doi:10.2217/epi-2017-0118
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.