Back to Journals » Clinical, Cosmetic and Investigational Dermatology » Volume 15
Bioinformatic Analysis and Translational Validation of Psoriasis Candidate Genes for Precision Medicine
Authors Li AH, Li WW, Yu XQ, Zhang DM, Liu YR, Li D
Received 13 June 2022
Accepted for publication 22 July 2022
Published 28 July 2022 Volume 2022:15 Pages 1447—1458
DOI https://doi.org/10.2147/CCID.S378143
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Jeffrey Weinberg
An-Hai Li,1 Wen-Wen Li,2 Xiao-Qian Yu,3 Dai-Ming Zhang,4 Yi-Ran Liu,5 Ding Li1
1Department of Dermatology, Qingdao Huangdao District Central Hospital, Qingdao, People’s Republic of China; 2Department of Hematology, Qingdao Women and Children’s Hospital, Qingdao, People’s Republic of China; 3Department of Dermatology, Qingdao Haici Hospital (Qingdao Traditional Chinese Medicine Hospital), Qingdao, People’s Republic of China; 4Department of Pharmacy, Affiliated Hospital of Qingdao University, Qingdao, People’s Republic of China; 5College of Traditional Chinese Medicine, Weifang Medical College, Weifang, People’s Republic of China
Correspondence: Ding Li, Department of Dermatology, Qingdao Huangdao District Central Hospital, Qingdao, People’s Republic of China, Email [email protected]
Background: Psoriasis is a recurrent, chronic, inflammation- and immune-mediated skin disease with multiple causative factors. However, the genetic markers associated with recurrence have not yet been fully elucidated. Accordingly, in this study, we aimed to identify markers associated with the recurrence of psoriasis.
Methods: We analyzed differentially expressed genes to determine which targets were associated with the recurrence of psoriasis and used these data to construct a protein-protein interaction network using Cytoscape software. The results were then validated by analysis of core targets using Gene Expression Omnibus (GEO) datasets and clinical samples. Functional enrichment analysis was used to explore the potential mechanisms mediating the recurrence of psoriasis.
Results: We screened out six core targets that played important roles in recurrence of psoriasis, and validation of GEO datasets and clinical samples showed that the expression levels of five core targets were higher in patients with psoriasis than in healthy individuals. Functional enrichment analysis revealed that the cell cycle and oocyte meiosis signaling pathways were involved in the recurrence of psoriasis.
Conclusion: Our findings provided insights into the mechanisms mediating the onset and recurrence of psoriasis.
Keywords: psoriasis, recurrence, cell cycle, gene expression profiling, pathogenic mechanism
Introduction
Psoriasis is a chronic, recurrent, inflammation- and immune-mediated skin disease that usually presents with varying degrees of skin itching and sharply demarcated erythematous plaques with whitish scales.1,52 In this complex immune-mediated skin disease, T lymphocytes, keratinocytes, dendritic cells, and cytokines (eg, interleukin [IL]-23, IL-17, and tumor necrosis factor) play central roles in the pathophysiological mechanism of psoriasis.2,3 Moreover, the skin flora has been shown to differ between patients with psoriasis and healthy individuals.4 Psoriasis can occur in both adults and children; however, the prevalence varies among populations, regions, and ages. A global systematic review showed that the incidence of psoriasis in adults ranged from 0.51% to 11.43%, whereas that in children ranged from 0% to 1.37%; the highest incidence rates of psoriasis were between 30 to 39 years old and between 50 to 69 years old.5,6 Studies by Icen7 and Tollefson8 showed that the incidence of psoriasis in children and adults is generally increasing.
The occurrence, development, and recurrence of psoriasis are caused by multiple factors, including mechanical stress, drugs, infection, lifestyle, obesity, diabetes mellitus, dyslipidemia, hypertension, and mental stress.9 Importantly, Cheng and colleagues showed that patients with psoriasis had a higher hospital readmission rate due to psoriasis and that readmission rates were increasing, resulting in a heavy economic burden.10 In recent years, biological agents have become important treatments for patients with moderate to severe psoriasis.11 Several clinical studies have shown that biological agents (eg, ustekinumab, secukinumab, ixekizumab, brodalumab, guselkumab, and tildrakizumab) can effectively alleviate the clinical symptoms of patients with moderate to severe psoriasis, reducing both the psoriasis area and severity index (PASI) score and incidence of side effects in patients with psoriasis.12–14 However, Johnson-Huang and colleagues15 showed that many patients with moderate to severe psoriasis exhibit relapse after stopping treatment with biological agents. While multiple studies by Giovanni Damiani have demonstrated that bioinhibitor combination therapy may improve the relapse of psoriasis, there are specific issues of drug resistance and loss of response.53–55 Thus, there is an urgent need to understand the biomarkers and mechanism of psoriasis recurrence, which will help us to better understand the pathogenesis of psoriasis.
Accordingly, in this study, we aimed to identify novel biomarkers and mechanism of recurrence in patients with psoriasis using bioinformatics analysis, which enables screening of targets of psoriasis recurrence and exploration of the mechanisms of psoriasis recurrence.16
Methods
Psoriasis Recurrence GEO Dataset Processing
From the Gene Expression Omnibus (GEO) database (https://www.ncbi.nlm.nih.gov/geo/), we chose one dataset related to psoriasis recurrence for analysis and selected three datasets related to psoriasis for validation. Among these datasets, GSE30768,15 which is based on the GPL571 platform, included skin samples from four samples with recurrence of psoriasis and four post-treatment psoriasis samples. This dataset was used to screen for differentially expressed genes via the “limma” package in R studio software 1.2 (https://www.rstudio.com/; Boston, United States), |logFC|≥1 and adjusted P value < 0.05 were set as filter values.
Psoriasis Validation GEO Dataset Processing
In addition, GSE41664 (based on GPL570)17 included skin tissues from 53 patients with psoriasis and 53 healthy individuals, GSE53552 (based on GPL570)18 included skin tissues from 25 patients with psoriasis and 24 healthy individuals, and GSE6710 (based on GPL96)19 included skin tissues from 13 patients with psoriasis and 13 healthy individuals, GSE13355 (based on GPL570)43 included skin tissues from 58 patients with psoriasis and 64 healthy individuals, GSE14905 (based on GPL570)44 included skin tissues from 33 patients with psoriasis and 21 healthy individuals, GSE30999 (based on GPL570)45 included skin tissues from 85 patients with psoriasis and 85 healthy individuals, GSE34248 (based on GPL570)17 included skin tissues from 14 patients with psoriasis and 14 healthy individuals, GSE42632 (based on GPL13497)46 included skin tissues from 6 patients with psoriasis and 6 healthy individuals, GSE50790 (based on GPL570)47 included skin tissues from 4 patients with psoriasis and 4 healthy individuals, GSE61281 (based on GPL6480)48 included skin tissues from 20 patients with psoriasis and 12 healthy individuals, GSE75343 (based on GPL571)49 included skin tissues from 22 patients with psoriasis and 10 healthy individuals; these datasets were used to verify the differential expression of recurrence-related targets in patients with psoriasis. The above datasets were normalized before formal analysis.
Construction of a Protein-Protein Interaction (PPI) Network and Screening of Core Targets
In the STRING database (https://www.string-db.org/), we constructed a PPI network based on the differentially expressed genes and downloaded PPI data with the organism set as Homo sapiens. In Cytoscape 3.7.2 software (http://www.cytoscape.org/; Bethesda, United States), we analyzed PPI data to rank differentially expressed genes via the “Cytohubba” package, with the calculation method set to MCC.
Validation Using GEO Datasets
To remove batch effects between the different platforms, we merged and normalized eleven GEO datasets (GSE6710, GSE13355, GSE14905, GSE30999, GSE34248, GSE41664, GSE42632, GSE50790, GSE53552, GSE61281 and GSE75343) using the “limma” and “sva” packages in R studio software and extracted and compared the expression levels of core targets in patients with psoriasis and healthy individuals (A total of 306 healthy volunteers and 333 psoriasis patients were included). We then performed correlation analysis based on the expression of core targets in order to elucidate the correlations between core targets.
Validation Using Clinical Samples
Validation of clinical samples was performed at the Qingdao Huangdao District Central Hospital. Twenty patients with psoriasis with a course of more than 5 years (seven men and 13 women) and five healthy controls (two men and three women) were recruited from the Qingdao Huangdao District Central Hospital. Two professional dermatologists scored the PASI and dermatology life quality index (DLQI) of patients with psoriasis (Table 1). Following centrifugation of the blood samples of healthy controls and psoriasis patients at 1500 g for 15 min, serum was collected and kept at −80°C until use. Serum samples were diluted, and the contents of core targets in the serum of patients and healthy individuals were detected using an enzyme-linked immunosorbent assay (ELISA) kit (Meimain, Nanjing, Jiangsu, China) according to the manufacturer’s instructions. The study protocol was approved by the ethics committees of Qingdao Huangdao District Central Hospital, and written informed consent was obtained from all participants or relatives before blood sampling.
 |
Table 1 Details of Psoriasis Patients and Healthy Individuals |
Functional Enrichment Analysis
To further explore the potential mechanisms of the recurrence and onset of psoriasis, we performed Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of differentially expressed genes using the “clusterProfiler” package and “org.Hs.eg.db” package in R studio software. The “ggplot2” package in R studio software was used to visualize the enrichment analysis results (P<0.05).
Artificial Neural Network (ANN) Model
In this section, we constructed a binary ANN model based on the GEO datasets and clinically validated core target expressions, to understand whether machine learning can effectively distinguish the psoriasis group and the control group through the expression of core targets. The psoriasis patients and control persons were completely randomly allocated; 70% of the total sample were selected as the training sample and 30% as the testing sample. The StandardScaler class from the sklearn function was used for data standardization. The sigmoid function was used as the activation function of the input and hidden layers, and the softmax function was used as the activation function of the output layer. The RMSProp was the optimizer for this model construction and the loss function was set as categorical cross entropy. The learning rate was 0.1, the rho was 0.9, the epsilon was 0, the decay was 0, the training period was 200, and the model training was completed when the training period reached 200.
Statistical Analysis
Statistical analyses were performed using GraphPad Prism 9.0 software (https://www.graphpad.com/; San Diego, United States). All data are presented as means ± standard deviations. The clinical samples data and the GEO datasets data were analyzed by student’s t test or Mann–Whitney U-test. P values less than 0.05 were considered statistically significant.
Results
Markers of Psoriasis Recurrence
First, we analyzed differentially expressed genes using the GSE30768 dataset and obtained 35 targets related to psoriasis recurrence, including 6 downregulated genes and 29 upregulated genes (Figure 1A, Table 2).
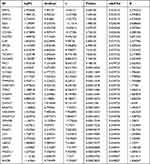 |
Table 2 Detailed Information of Genes Related to Psoriasis Recurrence in Differential Gene Expression Analysis |
PPI Network and Core Targets
We constructed PPI network based on the STRING database (Figure 1B), this PPI network contained 35 nodes and 85 edges, with an average node degree of 4.86, average local clustering coefficient of 0.588, expected number of edges of 12 and PPI enrichment p-value< 1.0e-16. And we screened out the six core targets (CDC20, CCNB1, CCNB2, MAD2L1, ZWINT, and SPC25) that may play important roles in the onset and recurrence of psoriasis (Figure 1C, Table 3).
 |
Table 3 Details of Core Genes in PPI Network |
Validation of Data from the GEO Database
After normalization and merging of the eleven GEO datasets, we extracted and compared the expression of core targets (Figure 2A and B). We found that the expression levels of CDC20, CCNB1, CCNB2, MAD2L1, ZWINT, and SPC25 were significantly higher in patients with psoriasis than in healthy individuals (Figure 2D). Correlation analysis showed that the expression levels of the six core targets were positively correlated (Figure 2C).
Validation of Data from Clinical Samples
Next, we used ELISA to detect the expression levels of the six core targets. The results of ELISA confirmed that the expression levels of CDC20, CCNB2, MAD2L1, ZWINT, and SPC25 were significantly higher in patients with psoriasis than in healthy individuals; however, the expression of CCNB1 was not significantly altered (Figure 3). Therefore, we hypothesized that the high expression of CCNB2, SPC25, CDC20, ZWINT, and MAD2L1 may be involved in the onset and recurrence of psoriasis.
Functional Enrichment Analyses
We then performed enrichment analyses of the six core targets using R studio software. Biological process analysis suggested that these six core targets were associated with mitotic nuclear division, chromosome segregation, nuclear division, and organelle fission (Figure 4A). Moreover, cellular component analysis suggested that the six core targets were also involved in condensed chromosome kinetochore, condensed chromosome, centromeric region, kinetochore, chromosome, centromeric region, condensed chromosome, and chromosomal region, whereas molecular function analysis revealed that the six core targets were enriched in cyclin-dependent protein serine/threonine kinase regulator activity, protein kinase regulator activity, protein C-terminus binding, and kinase regulator activity (Figure 4A). KEGG analysis revealed that the six core targets were enriched in cell cycle, oocyte meiosis, progesterone-mediated oocyte maturation, and human T-cell leukemia virus 1 infection (Figure 4B). As shown in Figure 4C and D, CNB1, MAD2L1, CDC20, and CCNB2 may participate in the onset and recurrence of psoriasis through pathways related to the cell cycle and oocyte meiosis.
ANN Model
We constructed a binary ANN model through the expression of core targets, and found that machine learning can effectively distinguish psoriasis patients and control persons based on the expression of core targets (loss=0.2348 and accuracy=0.9248) (Figure 4E and F).
Discussion
Psoriasis is a chronic, recurrent, inflammation- and immune-mediated skin disease that is pathophysiologically characterized by keratinocyte hyperproliferation and shortened cell cycle.38 Psychological stress, genetic factors, smoking, obesity, hypertension, diabetes, and other factors can promote the onset and recurrence of psoriasis.20–24 Long-term recurrent psoriasis also promotes the occurrence and exacerbation of cardiovascular disease.56 Additionally, symptoms of the disease can be alleviated by regulation of the cell cycle and meiosis in keratinocytes,42 and induction of cell cycle arrest in keratinocytes and inhibition of keratinocyte hyperproliferation can alleviate skin lesions in mouse models of psoriasis.39,40 Therefore, treatment of psoriasis should also be continuous for recurrence prevention, even if symptoms seem to be completely relieved.25 Although efalizumab biologics are no longer used in the treatment of psoriasis, the new therapies cannot change the situation of psoriasis recurrence after the treatment is stopped. Therefore, this study focuses on the recurrence mechanism of psoriasis and provides new theories and support for the study of psoriasis recurrence.
In this study, we first screened for potential recurrence-related targets in psoriasis through analysis of differentially expressed genes based on clinical data from the GEO database, yielding 35 potential targets. To further identified the core targets, we constructed a PPI network of recurrence-related targets and found that CDC20, CCNB1, CCNB2, MAD2L1, ZWINT, and SPC25 were the core targets in the PPI network. Validation of GEO datasets confirmed our conclusions, and we found that the expression levels of the six core targets were positively correlated based on the data from the GEO datasets. Furthermore, using clinical samples, five of the six core targets (CDC20, CCNB2, MAD2L1, ZWINT, and SPC25) were confirmed to show significantly different expression in psoriasis, and functional enrichment analysis identified two main signaling pathways, ie, cell cycle and oocyte meiosis signaling pathways, participating in recurrence of psoriasis. Importantly, in a previous study, Pasquali and colleagues used transcriptomics analysis and showed that dysregulated genes were mainly enriched in pathways related to immune response, cell cycle, and keratinization.37 While another work of Weighted Gene Co-expression Network Analysis (WGCNA) held by Sundarrajan S. identified crucial cascades of psoriasis which include immune response, keratinisation and cytoskeleton remodeling, that made up the shortcoming of the current work.50 Additionally, the core targets identified in this study have been shown to be involved in cell cycle progression and meiosis in psoriasis recurrence.41 As showed in Figure 4D, We found that core genes are actively involved in meiosis I and II in the oocyte meiosis signaling pathways. The high expression of CCNB1, CCNB2 and CDC20 may inhibit the prevention of DNA replication, bipolar spindle formation and meiosis I progression to participate the progression of psoriasis. The high expression of CDC20, MAD2L1, CCNB1 and CCNB2 may inhibit the meiosis II arrest and promote meiosis II exit to promote the development of psoriasis. Therefore, we initially believe that this may be one of the reasons for the genetic predisposition of psoriasis. This comprehension of bioinformatic and patients sample analyze provides new insights on the molecular pathogenesis of recurrent psoriasis and the identification of potential therapeutic targets.
Few studies exist which have integrated such gene expression microarray data of recurrent lesional and non-lesional samples. The results shown in Figure 3 from our Elisa test samples illustrated the expression levels of SPC25, ZWINT, MAD2L1, CCNB1 and CDC20 in psoriasis patients were higher than healthy individuals. Kinetochore protein Spc25 (SPC25) is one of the components of the NDC80 kinetochore complex. SPC25 and NDC80 have been shown to exhibit significantly different expression in patients with psoriasis,28 and NDC80 has important roles in regulating the cell cycle in keratinocytes.29 ZW10 interactor (ZWINT), like the other core targets, has been shown to regulate the cell cycle, and high ZWINT expression can promote cell proliferation.36 Both mitotic arrest deficient 2 like 1 (MAD2L1) and cell division cycle 20 (CDC20) are components of the mitotic checkpoint complex.30 CDC20 plays important regulatory roles in promoting the cell cycle, cell differentiation, and cell proliferation in keratinocytes and other related cells.31–33 Moreover, MAD2L1 has also been shown to promote cell proliferation, and decreased MAD2L1 expression inhibits the proliferation, migration, and invasion of cancer cells.34,35 Cyclin B2 (CCNB2) is a member of the cyclin family, and its specific roles in psoriasis have not yet been clarified. Wei et al showed that decreased CCNB2 expression inhibits the proliferation of keratinocytes and alleviates symptoms of psoriasis in a mouse model.26 Additionally, Li and colleagues demonstrated that CCNB2 expression was decreased in patients with psoriasis who showed alleviation of symptoms.27 These findings are similar to our results in the recurrence of psoriasis. In contrast to the results of our reports, the expression of most hub genes associated with the cell cycle and proliferation in scalp psoriatic samples, revealed no significant differences from that of paired control samples.51
Taken together, this comprehensive bioinformatic and patients samples analysis provides new insights into the molecular pathogenesis and recurrence of psoriasis. We propose that CDC20, CCNB2, MAD2L1, ZWINT, and SPC25 regulated the cell cycle and meiosis to participate in the pathogenesis of psoriasis, which is different from the Johnson-Huang LM study. The latter data set found there were CD11c+CD1c-, inflammatory myeloid DCs identified by TNFSF10/TRAIL, TNF, and iNOS. CD11c+ cells in relapsed lesions co-expressed CD14 and CD16 in situ. During a relapse, the majority of these genes reversed back to a lesional state.15 Besides, previous microarray analysis focuses on the comparison between the normal and diseased states, but the results of the current study obtained from a bioinformatics analysis and validation were focused on the recurrent cohort microarray data. The ANN part involved in this study also partially proves that artificial intelligence can screen patients with special conditions through gene expression, such as screening patients who do not respond to medication.
Conclusion
In summary, we not only demonstrated for the first time that CDC20, CCNB2, MAD2L1, ZWINT and SPC25 are key genes and some important pathways involved in psoriasis recurrence, but also demonstrated that this may be a new perspective on the pathogenesis of psoriasis. Among the screened out six psoriasis recurrence-related targets (CDC20, CCNB1, CCNB2, MAD2L1, ZWINT, and SPC25), further validation results showed that progression of recurrence and pathogenesis in psoriasis with mechanism of regulating the cell cycle and meiosis. Since these signature genes may serve as potential candidates for biomarker discovery leading to new therapeutic targets. Further experiments are required to strengthen our findings and fully elucidate the related mechanisms mediating psoriasis, as well as to provide alternative paths to mine out key drivers of psoriasis, which is one of the priorities of our future research.57
Data Sharing Statement
Data availability for GEO data (GSE6710, GSE13355, GSE14905, GSE30999, GSE34248, GSE41664, GSE42632, GSE50790, GSE53552, GSE61281 and GSE75343) downloading, readers can visit the website https://www.ncbi.nlm.nih.gov/geo/and choose “GEO datasets” in the drop-down list. For gene enrichment analysis, for inferring the protein-protein interaction, we used web based tool named STRING (https://www.string-db.org/) according to the instruction, and the species is set to Homo sapiens. For drawing the network, Cytoscape software was used. All patients were enrolled from April 2021 to June 2021.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Funding
This research is not funded.
Disclosure
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Boehncke WH, Schön MP. Psoriasis. Lancet. 2015;386(9997):983–994.
2. Hawkes JE, Chan TC, Krueger JG. Psoriasis pathogenesis and the development of novel targeted immune therapies. J Allergy Clin Immunol. 2017;140(3):645–653.
3. Lou F, Sun Y, Xu Z, et al. Excessive Polyamine Generation in Keratinocytes Promotes Self-RNA Sensing by Dendritic Cells in Psoriasis. Immunity. 2020;53(1):204–216.e10.
4. Fyhrquist N, Muirhead G, Prast-Nielsen S, et al. Microbe-host interplay in atopic dermatitis and psoriasis. Nat Commun. 2019;10(1):4703.
5. Michalek IM, Loring B, John SM. A systematic review of worldwide epidemiology of psoriasis. J Eur Acad Dermatol Venereol. 2017;31(2):205–212.
6. Parisi R, Symmons DP, Griffiths CE, et al. Global epidemiology of psoriasis: a systematic review of incidence and prevalence. J Invest Dermatol. 2013;133(2):377–385.
7. Icen M, Crowson CS, McEvoy MT, et al. Trends in incidence of adult-onset psoriasis over three decades: a population-based study. J Am Acad Dermatol. 2009;60(3):394–401.
8. Tollefson MM, Crowson CS, McEvoy MT, et al. Incidence of psoriasis in children: a population-based study. J Am Acad Dermatol. 2010;62(6):979–987.
9. Kamiya K, Kishimoto M, Sugai J, et al. Risk Factors for the Development of Psoriasis. Int J Mol Sci. 2019;20(18):4347.
10. Cheng BT, Silverberg JI. Predictors of hospital readmission in United States adults with psoriasis. J Am Acad Dermatol. 2020;82(4):902–909.
11. Sbidian E, Chaimani A, Garcia-Doval I, et al. Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis. Cochrane Database Syst Rev. 2017;12(12):CD011535.
12. Al-Hammadi A, Ruszczak Z, Magariños G, et al. Intermittent use of biologic agents for the treatment of psoriasis in adults. J Eur Acad Dermatol Venereol. 2021;35(2):360–367.
13. Bilal J, Berlinberg A, Bhattacharjee S, et al. A systematic review and meta-analysis of the efficacy and safety of the interleukin (IL)-12/23 and IL-17 inhibitors ustekinumab, secukinumab, ixekizumab, brodalumab, guselkumab and tildrakizumab for the treatment of moderate to severe plaque psoriasis. J Dermatolog Treat. 2018;29(6):569–578.
14. Kamata M, Tada Y. Safety of biologics in psoriasis. J Dermatol. 2018;45(3):279–286.
15. Johnson-Huang LM, Pensabene CA, Shah KR, et al. Post-therapeutic relapse of psoriasis after CD11a blockade is associated with T cells and inflammatory myeloid DCs. PLoS One. 2012;7(2):e30308.
16. Singer J, Irmisch A, Ruscheweyh HJ, et al. Bioinformatics for precision oncology. Brief Bioinform. 2019;20(3):778–788.
17. Bigler J, Rand HA, Kerkof K, et al. Cross-study homogeneity of psoriasis gene expression in skin across a large expression range. PLoS One. 2013;8(1):e52242.
18. Russell CB, Rand H, Bigler J, et al. Gene expression profiles normalized in psoriatic skin by treatment with brodalumab, a human anti-IL-17 receptor monoclonal antibody. J Immunol. 2014;192(8):3828–3836.
19. Reischl J, Schwenke S, Beekman JM, et al. Increased expression of Wnt5a in psoriatic plaques. J Invest Dermatol. 2007;127(1):163–169.
20. Stewart TJ, Tong W, Whitfeld MJ. The associations between psychological stress and psoriasis: a systematic review. Int J Dermatol. 2018;57(11):1275–1282.
21. Armstrong AW, Harskamp CT, Dhillon JS, et al. Psoriasis and smoking: a systematic review and meta-analysis. Br J Dermatol. 2014;170(2):304–314.
22. Jensen P, Skov L. Psoriasis and Obesity. Dermatology. 2016;232(6):633–639.
23. Armstrong AW, Harskamp CT, Armstrong EJ. Psoriasis and the risk of diabetes mellitus: a systematic review and meta-analysis. JAMA Dermatol. 2013;149(1):84–91.
24. Armstrong AW, Harskamp CT, Armstrong EJ. The association between psoriasis and hypertension: a systematic review and meta-analysis of observational studies. J Hypertens. 2013;31(3):433–442.
25. Masson Regnault M, Konstantinou MP, Khemis A, et al. Early relapse of psoriasis after stopping brodalumab: a retrospective cohort study in 77 patients. J Eur Acad Dermatol Venereol. 2017;31(9):1491–1496.
26. Wei JA, Han L, Lu CJ, et al. Formula PSORI-CM01 eliminates psoriasis by inhibiting the expression of keratinocyte cyclin B2. BMC Complement Altern Med. 2016;16:255.
27. Li YJ, Zhou T, Zhang J, et al. Clinical trait-connected network analysis reveals transcriptional markers of active psoriasis treatment with Liangxue-Jiedu decoction. J Ethnopharmacol. 2021;25(268):113551.
28. Zhang YJ, Sun YZ, Gao XH, et al. Integrated bioinformatic analysis of differentially expressed genes and signaling pathways in plaque psoriasis. Mol Med Rep. 2019;20(1):225–235.
29. Zanet J, Freije A, Ruiz M, et al. A mitosis block links active cell cycle with human epidermal differentiation and results in endoreplication. PLoS One. 2010;5(12):e15701.
30. Sitry-Shevah D, Kaisari S, Teichner A, et al. Role of ubiquitylation of components of mitotic checkpoint complex in their dissociation from anaphase-promoting complex/cyclosome. Proc Natl Acad Sci U S A. 2018;115(8):1777–1782.
31. Chen Z, Yu Y, Fu D, et al. Functional roles of PC-PLC and Cdc20 in the cell cycle, proliferation, and apoptosis. Cell Biochem Funct. 2010;28(4):249–257.
32. Quek LS, Grasset N, Jasmen JB, et al. Dual Role of the Anaphase Promoting Complex/Cyclosome in Regulating Stemness and Differentiation in Human Primary Keratinocytes. J Invest Dermatol. 2018;138(8):1851–1861.
33. Zhang Q, Huang H, Liu A, et al. Cell division cycle 20 (CDC20) drives prostate cancer progression via stabilization of β-catenin in cancer stem-like cells. EBioMedicine. 2019;42:397–407.
34. Teixeira JH, Silva P, Faria J, et al. Clinicopathologic significance of BubR1 and Mad2 overexpression in oral cancer. Oral Dis. 2015;21(6):713–720.
35. Li Y, Bai W, Zhang J. MiR-200c-5p suppresses proliferation and metastasis of human hepatocellular carcinoma (HCC) via suppressing MAD2L1. Biomed Pharmacother. 2017;92:1038–1044.
36. Ying H, Xu Z, Chen M, et al. Overexpression of Zwint predicts poor prognosis and promotes the proliferation of hepatocellular carcinoma by regulating cell-cycle-related proteins. Onco Targets Ther. 2018;Feb(11):689–702.
37. Pasquali L, Srivastava A, Meisgen F, et al. The Keratinocyte Transcriptome in Psoriasis: pathways Related to Immune Responses, Cell Cycle and Keratinization. Acta Derm Venereol. 2019;99(2):196–205.
38. Jia J, Li C, Yang J, et al. Yes-associated protein promotes the abnormal proliferation of psoriatic keratinocytes via an amphiregulin dependent pathway. Sci Rep. 2018;8(1):14513.
39. Yang XG, Jiang BW, Jing QQ, et al. Nitidine chloride induces S phase cell cycle arrest and mitochondria-dependent apoptosis in HaCaT cells and ameliorates skin lesions in psoriasis-like mouse models. Eur J Pharmacol. 2019;15(863):172680.
40. Thatikonda S, Pooladanda V, Sigalapalli DK, et al. Piperlongumine regulates epigenetic modulation and alleviates psoriasis-like skin inflammation via inhibition of hyperproliferation and inflammation. Cell Death Dis. 2020;11(1):21.
41. Lo YH, Li CS, Chen HL, et al. Galectin-8 Is Upregulated in Keratinocytes by IL-17A and Promotes Proliferation by Regulating Mitosis in Psoriasis. J Invest Dermatol. 2021;141(3):503–511.e9.
42. Wang R, Zhao Z, Zheng L, et al. MicroRNA-520a suppresses the proliferation and mitosis of HaCaT cells by inactivating protein kinase B. Exp Ther Med. 2017;14(6):6207–6212.
43. Nair RP, Duffin KC, Helms C, et al. Collaborative Association Study of Psoriasis. Genome-wide scan reveals association of psoriasis with IL-23 and NF-kappaB pathways. Nat Genet. 2009;41(2):199–204.
44. Yao Y, Richman L, Morehouse C, et al. Type I interferon: potential therapeutic target for psoriasis? PLoS One. 2008;3(7):e2737.
45. Suárez-Fariñas M, Li K, Fuentes-Duculan J, et al. Expanding the psoriasis disease profile: interrogation of the skin and serum of patients with moderate-to-severe psoriasis. J Invest Dermatol. 2012;132(11):2552–2564.
46. Niu X, Zhang K. Dysregulated expression of inflammation-related genes in psoriatic dermis mesenchymal stem cells. Acta Biochim Biophys Sin (Shanghai). 2016;48(6):587–588.
47. Swindell WR, Xing X, Stuart PE, et al. Heterogeneity of inflammatory and cytokine networks in chronic plaque psoriasis. PLoS One. 2012;7(3):e34594.
48. Pollock RA, Abji F, Liang K, et al. Gene expression differences between psoriasis patients with and without inflammatory arthritis. J Invest Dermatol. 2015;135(2):620–623.
49. Ruano J, Suárez-Fariñas M, Shemer A, et al. Molecular and Cellular Profiling of Scalp Psoriasis Reveals Differences and Similarities Compared to Skin Psoriasis. PLoS One. 2016;11(2):e0148450.
50. Sundarrajan S, Arumugam M. Weighted gene co-expression based biomarker discovery for psoriasis detection. Gene. 2016;593(1):225–234.
51. Ruano J, Suárez-Fariñas M, Shemer A, Oliva M, Guttman-Yassky E, Krueger JG. Molecular and Cellular Profiling of Scalp Psoriasis Reveals Differences and Similarities Compared to Skin Psoriasis. PLoS One. 2017;11(2):548.
52. Damiani G, Bragazzi NL, Karimkhani Aksut C, et al. The Global, Regional, and National Burden of Psoriasis: results and Insights From the Global Burden of Disease 2019 Study. Front Med. 2021;8:743180.
53. Damiani G, Conic RRZ, de Vita V, et al. When IL-17 inhibitors fail: real-life evidence to switch from secukinumab to Adalimumab or ustekinumab. Dermatol Ther. 2019;32(2):e12793.
54. Damiani G, Odorici G, Pacifico A, et al. Secukinumab Loss of Efficacy Is Perfectly Counteracted by the Introduction of Combination Therapy (Rescue Therapy): data from a Multicenter Real-Life Study in a Cohort of Italian Psoriatic Patients That Avoided Secukinumab Switching. Pharmaceuticals. 2022;15(1):95.
55. Damiani G, Conic RRZ, Pigatto PDM, et al. From randomized clinical trials to real life data. An Italian clinical experience with ixekizumab and its management. Dermatol Ther. 2019;32(3):e12886.
56. Conic RR, Damiani G, Schrom KP, et al. Psoriasis and Psoriatic Arthritis Cardiovascular Disease Endotypes Identified by Red Blood Cell Distribution Width and Mean Platelet Volume. J Clin Med. 2020;9(1):186.
57. Damiani G, Conic RRZ, Pigatto PDM, et al. Predicting Secukinumab Fast-Responder Profile in Psoriatic Patients: advanced Application of Artificial-Neural-Networks (ANNs). J Drugs Dermatol. 2020;19(12):1241–1246.
 © 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2022 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.




