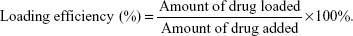Back to Journals » International Journal of Nanomedicine » Volume 13
Bioengineering the spider silk sequence to modify its affinity for drugs
Authors Kucharczyk K, Weiss M , Jastrzebska K, Luczak M, Ptak A , Kozak M, Mackiewicz A, Dams-Kozlowska H
Received 13 March 2018
Accepted for publication 15 April 2018
Published 20 July 2018 Volume 2018:13 Pages 4247—4261
DOI https://doi.org/10.2147/IJN.S168081
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Dr Thomas Webster

Kamil Kucharczyk,1,2 Marek Weiss,3 Katarzyna Jastrzebska,1,2 Magdalena Luczak,4,5 Arkadiusz Ptak,3 Maciej Kozak,6,7 Andrzej Mackiewicz,1,2 Hanna Dams-Kozlowska1,2
1Department of Medical Biotechnology, Poznan University of Medical Sciences, Poznan, Poland; 2Department of Diagnostics and Cancer Immunology, Greater Poland Cancer Centre, Poznan, Poland; 3Division of Computational Physics and Nanomechanics, Institute of Physics, Faculty of Technical Physics, Poznan University of Technology, Poznan, Poland; 4Department of Biomedical Proteomics, Institute of Bioorganic Chemistry, Polish Academy of Sciences, Poznan, Poland; 5Department of Organic Chemistry, Institute of Chemical Technology and Engineering, Poznan University of Technology, Poznan, Poland; 6Department of Macromolecular Physics, Adam Mickiewicz University, Poznan, Poland; 7Joint Laboratory for SAXS Studies, Adam Mickiewicz University, Poznan, Poland
Background: Silk is a biocompatible and biodegradable material, able to self-assemble into different morphological structures. Silk structures may be used for many biomedical applications, including carriers for drug delivery. The authors designed a new bioengineered spider silk protein, EMS2, and examined its property as a carrier of chemotherapeutics.
Materials and methods: To obtain EMS protein, the MS2 silk monomer (that was based on the MaSp2 spidroin of Nephila clavipes) was modified by the addition of a glutamic acid residue. Both bioengineered silks were produced in an Escherichia coli expression system and purified by thermal method. The silk spheres were produced by mixing with potassium phosphate buffer. The physical properties of the particles were characterized using scanning electron microscopy, atomic force microscopy, Fourier-transform infrared spectroscopy, and zeta potential measurements. The MTT assay was used to examine the cytotoxicity of spheres. The loading and release profiles of drugs were studied spectrophotometrically.
Results: The bioengineered silk variant, EMS2, was constructed, produced, and purified. The EMS2 silk retained the self-assembly property and formed spheres. The spheres made of EMS2 and MS2 silks were not cytotoxic and had a similar secondary structure content but differed in morphology and zeta potential values; EMS2 particles were more negatively charged than MS2 particles. Independently of the loading method (pre- or post-loading), the loading of drugs into EMS2 spheres was more efficient than the loading into MS2 spheres. The advantageous loading efficiency and release rate made EMS2 spheres a good choice to deliver neutral etoposide (ETP). Despite the high loading efficiency of positively charged mitoxantrone (MTX) into EMS2 particles, the fast release rate made EMS2 unsuitable for the delivery of this drug. A faster release rate from EMS2 particles compared to MS2 particles was observed for positively charged doxorubicin (DOX).
Conclusion: By modifying its sequence, silk affinity for drugs can be controlled.
Keywords: silk, bioengineering, spheres, drug delivery, chemotherapeutics, cancer therapy
Introduction
Chemotherapy is one of the methods used to treat cancer. However, drugs administered intravenously may cause toxicity toward both tumor cells and healthy cells, which can lead to severe adverse side effects. A delivery system capable of binding, transporting, and releasing the drug at the target site is needed. A drug carrier should be biocompatible and biodegradable. Its physical and chemical properties should be well defined, and its manufacturing process should be reproducible and controllable.
Most drug delivery systems that are investigated are based on synthetic polymers such as poly(lactic-co-glycolic acid) (PLGA).1 This material presents good pharmacokinetic properties and has been approved by the US Food and Drug Administration (FDA). However, factors such as processing in organic solvents or the acidic degradation products of this polymer may be restrictive for some therapeutic applications. Therefore, increasing effort and studies have recently been devoted to naturally derived materials. The protein-based biomaterials, such as collagen, gelatin, silk, sericin, and the polysaccharide-based biomaterials including cellulose, chitin/chitosan, glucose, agarose, alginate, and hyaluronic acid, attracted the interest of scientists.2–5 These materials were extensively investigated as drug delivery platforms.6–9 Moreover, the blends of natural and synthetic polymers can be explored as a new class of materials that combine the mechanical properties and biocompatibility.10,11 Silk may be one of the most promising candidates for an alternative delivery system.12 Biocompatibility, biodegradability, and superb mechanical properties make silk an ideal material for a wide range of biomedical applications.13 Silkworm silk is obtained from cocoons, but the harvesting of spider silk from nature is limited, due to the difficulties in breeding spiders on a large scale. Moreover, spiders produce several types of silk fibers that consist of different types of silk proteins.14 However, the recent development of bioengineered spider silk production and purification has solved the accessibility problem and provided new opportunities for silk materials.15
Although spiders produce only silk fibers, the silk protein can be processed under laboratory conditions into various morphological forms, such as films, fibers, hydrogels, scaffolds, capsules, microspheres, and nanospheres.16 These structures can find application in the study of tumor biology and therapy or as carriers of chemotherapeutics or other drugs.17,18 The potential of spheres made of silkworm silk as an effective delivery system has been demonstrated.19,20 The results of various studies indicated that fibroin nanoparticles exhibited both good encapsulation efficiency and good release profiles for model drugs.21–25 Moreover, silk fibroin nanoparticles were relatively nontoxic and were able to accumulate in the cytoplasm of carcinoma cells.26 Drug delivery applications for bioengineered spider silk proteins have also been explored.12 Lammel et al27 demonstrated that spheres made of bioengineered spider silk protein eADF4(C16) (mimicking the ADF4 protein from the European garden spider Araneus diadematus) were stable and could efficiently bind and release low-molecular-weight model drugs. The eADF4(C16) particles were also successfully evaluated as appropriate drug carriers for the delivery of high-molecular-weight protein drugs.28 The study by Blüm and Scheibel29 showed that the loading and release of a model drug can be controlled by the processing conditions of eADF4(C16) silk during the sphere formation process, indicating the influence of crosslinking and preparation route as major factors. The potential application of spheres as drug delivery vehicles was also reported for bioengineered silks based on Nephila clavipes proteins MaSp1 and MaSp2.30–34 The bioengineered MS1 silk (based on MaSp1) was functionalized with H2.1 and H2.2 binding peptides that specifically recognize the Her2 receptor. Spheres made of the functionalized bioengineered silks were loaded with doxorubicin (DOX) and found to efficiently target and reduce the viability of Her2-positive cancer cells.30 The analysis of bioengineered MS2 silk spheres (based on MaSp2) showed that their properties and their interactions with drugs depended on the method of protein purification.31 The application of two bioengineered proteins, MS1 and MS2, functionalized with H2.1 peptide and blended together, resulted in spheres with the same targeting potential and greatly improved the physical and chemical properties compared with functionalized MS1 particles.32 The direct comparison of MS1 and MS2 particles indicated that the difference in amino acid composition of silks determined the processing conditions and physical properties of the silk spheres.34 MS2 spheres were smaller, of solid core, of higher β-sheet structure content, more dispersed, and of opposite (negative) charge than MS1 particles.34 Our previous results indicated that MS2 spheres were a better choice for mitoxantrone (MTX) and etoposide (ETP) delivery; however, MS1 spheres showed greater applicability for DOX delivery.34 Genetic modifications changing even a singular amino acid residue to produce bioengineered spider silk made it possible to obtain silk with modified properties.33,35 Doblhofer and Scheibel33 replaced a glutamic acid residue with lysine in the amino acid sequence of the eADF4(C16) protein. The newly derived silk protein, eADF4(κ16), was positively charged, opposite to the original eADF4(C16). This modification enabled the encapsulation of negatively charged model drugs.33 The study by Szela et al35 demonstrated the introduction of methionines in positions flanking the polyalanine regions that were responsible for β-sheet formation. Chemical agents targeting the added methionines allowed the control of β-sheet formation, resulting in a protein with a different solubility.35
The aim of this study was to control the properties of bioengineered spider silk by amino acid sequence alteration to modify its affinity for drugs. We designed and produced a bioengineered silk variant, ie, EMS2, based on the sequence of MS2 silk. MS2 consists of the 15 units of the consensus motif of the MaSp2 protein from the spider N. clavipes.32 For EMS2 construction, the repeat unit of MS2 was modified by the addition of one glutamic acid residue. The obtained protein differed in the theoretical isoelectric point (pI) value: 3.15 vs 5.27 for EMS2 vs MS2, respectively. The new bioengineered silk EMS2 was analyzed for its ability to form spheres under different preparation conditions. Moreover, EMS2 spheres were investigated in terms of their morphology, size, zeta potential, secondary structure, and toxicity against cells in comparison with the control MS2 spheres. The silk sphere variants were loaded with drugs using two methods: pre- and post-loading. The loading and release of three chemotherapeutic drugs (ETP, MTX, and DOX) were investigated. The addition of one amino acid residue to the repeat unit of silk made it possible to obtain protein with different properties and to control the loading and release of drugs used for cancer therapy.
Materials and methods
Construction of expression plasmids pETNX-EMS2 and pETNX-MS2
For protein expression, the pET30(a)+ vector was modified with the linker NX (pETNX) to provide the restriction sites NheI and SpeI for cloning silk genes.36 The construction of a synthetic gene encoding 15 repeats of MS2 was performed as described previously.32 In constructing the EMS2 gene, the following monomer oligonucleotide sequences were used: EMS2F-CTAGCGAAGGTCCAGGCGGCTATGGTCCGGGCCAGCAAGGGCCGAGCGGTCCGGGCTCGGCGGCC
GCGGCTGCGGCAGCGGCCGGACCTGGCGGCTATGGTCCGGGCCAGCAGA and EMS2R-CTAGTCTGCTGGCCCGGACCATAGCCGCCAGGTCCGGCCGCTGCCGCAGCCGCGGCCGCCGAGCC
CGGACCGCTCGGCCCTTGCTGGCCCGGACCATAGCCGCCTGGACCTTCG. After annealing, the synthetic silk monomer EMS2, containing cohesive ends complementary to NheI and SpeI, was inserted into the NheI and SpeI restriction sites of the pETNX expression vector. Plasmid pETNX-EMS2, consisting of 15 repeats of EMS2, was obtained by multiple ligation of the EMS2 monomer into the NheI restriction site of the obtained pETNX plasmid. The sequence of the construct was confirmed by sequencing at the Core Facility of the Adam Mickiewicz University in Poznan, Poland. The enzymes for digestion and ligation were supplied by Fermentas (Thermo Fisher Scientific Inc., Waltham, MA, USA) and A&A Biotechnology (Gdynia, Poland), respectively.
Protein expression and purification
The expression plasmids pETNX-EMS2 and pETNX-MS2 were transformed into E. coli strain BLR bacteria (DE3) (Novagen, Madison, WI, USA). For large-scale production, a Bioflo 3000 (New Brunswick Scientific, Edison, NJ, USA) fermentor and protocol were used as previously reported.30,31,36
Silk proteins were purified by using a thermal denaturation method (named 80/20), as described previously.34,36 The protein concentration was calculated by measuring the absorbance by UV spectroscopy at 280 nm and using the molar extinction coefficient of 44,700 cm−1M−1 for EMS2 and MS2 and their respective molecular masses. The protein quality was analyzed by 12.5% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) and staining of the proteins with colloidal blue (Roti-Blue) (Carl Roth, Karlsruhe, Germany).
MALDI-TOF/TOF mass spectrometry
The molecular masses of proteins were determined using a MALDI-TOF/TOF mass spectrometer. The MALDI spectra were acquired on an UltrafleXtreme (Bruker Daltonics, Bremen, Germany) mass spectrometer operated in the positive ion mode with a reflector, using delayed ion extraction. For each sample, 0.5 μL was co-crystallized with a sinapinic acid matrix and spotted directly onto the MALDI AnchorChip target plate (Bruker Daltonics). The MS spectra were externally calibrated using the Protein Calibration Standard mixture (Bruker Daltonics). FlexControl v. 3.3 was used for the acquisition of spectra, and all further data processing was performed using flexAnalysis v. 3.3.
Sphere preparation
Spheres were formed using EMS2 and MS2 proteins in several processing variants: (1) variable initial concentration of potassium phosphate buffer, (2) variable pH of potassium phosphate buffer, and (3) variable initial concentration of silk proteins. For the first set of variants, spheres were prepared using 2.5 mg/mL silk solution and potassium phosphate in the following concentrations: 0.5, 1, and 2 M, pH 7. The second set of processing conditions combined 2.5 mg/mL silk solution and 1.75 M potassium phosphate at pH 4, 7, and 10. The third set of sphere variants were formed by using 2 M potassium phosphate at pH 7 and the following concentrations of silk: 0.5, 2.5, and 5 mg/mL. For each case, 100 μL of silk solution was rapidly mixed by pipette with 1 mL of potassium phosphate buffer and incubated for 2 h at room temperature. Next, the sphere suspension was dialyzed against distilled water overnight, centrifuged at 10,000 × g for 15 min, and suspended in ultrapure water. Samples were stored at 4°C.
Spheres were prepared for the secondary structure and drug post-loading analysis by using silk proteins at an initial concentration of 2.5 mg/mL and 2 M potassium phosphate at pH 7 and for the cytotoxicity study by using 0.5 mg/mL silk proteins and 2 M potassium phosphate at pH 7. For the stability analysis, spheres were prepared using an initial concentration of 2.5 mg/mL silk proteins and 2 M potassium phosphate at pH 7 and stored in water at 4°C for 6 months.
Zeta potential measurement
The zeta potential (ZP) of spheres was measured using Zetasizer Nano XS (Malvern Instruments. Ltd, Worcestershire, UK). Spheres were suspended in 1 mL of ultrapure water and sonicated for 5 min in an ultrasonic water bath. The ZP was measured three times in triplicate at room temperature.
Scanning electron microscopy (SEM)
The morphology of the spheres was examined using SEM. The sphere suspension was placed on a cover slip and dried. Next, the samples were sputtered with an Au/Pd layer in a Quorum Sputter Coater Q150T ES (Quorum Technologies, Ringmer, UK) and analyzed with a JEOL JSM-7001F (JEOL Ltd, Tokyo, Japan) field emission scanning electron microscope at 10 kV accelerating voltage. The size was determined by measuring the diameter of spheres in three images captured at 10,000× magnification using the SmileView software program (JEOL Ltd). The experiment was repeated three times.
Atomic force microscopy (AFM)
Spheres were placed on a cover slip, air-dried, and then analyzed by using an AFM Veeco Innova (Veeco Instruments Inc., New York, NY, USA). AFM studies were performed in soft intermittent contact mode, directly on spheres, using Nanosensors PPP-NCLR μ-cantilevers (Nanosensors, Neuchatel, Switzerland) with the radius at the nominal end of the tip of <10 nm. Data were analyzed by using the Gwyddion software (Czech Metrology Institute, Brno, Czech Republic).
Fourier-transform infrared spectroscopy (FTIR)
The sphere secondary structure analysis was performed as described previously.34 The amide I band components were assigned to the following elements of the secondary structure: 1,605–1,615 cm−1 tyrosine side chains, 1,616–1,637 cm−1 and 1,697–1,705 cm−1 β-sheets, 1,638–1,655 cm−1 random coils, 1,665–1,662 cm−1 helices, and 1,663–1,696 cm−1 turns, as previously described.37 The experiment was repeated three times.
Cytotoxicity study
NIH 3T3 fibroblasts (ATCC, Manassas, VA, USA) were cultured in DMEM (Sigma-Aldrich, St Louis, MO, USA) medium supplemented with 10% fetal bovine serum (PAA Laboratories GmbH, Pasching, Austria) and 80 μg/mL gentamycin (KRKA, Novo Mesto, Slovenia). Cells were grown at 37°C in a humidified atmosphere containing 5% CO2. Cells were seeded at 2.5×104/well into a 96-well plate and incubated overnight. On the next day, sphere suspensions were added to the cell cultures at different concentrations and then incubated for 72 h. Cells without spheres were used as a negative control. After incubation, 50 μL (5 mg/mL) of MTT reagent (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide; Sigma-Aldrich) was added to each well. After 4 h, the medium was removed, and insoluble purple formazan was dissolved in 200 μL DMSO (dimethyl sulfoxide; Sigma-Aldrich). The absorbance of the solution was measured at a wavelength of 560 nm using a ELX808IV (Bio-Tek Instruments, Winooski, VT, USA) microplate reader. The experiment was repeated three times in triplicate.
Drug loading and release
EMS2 and MS2 spheres were loaded with anticancer drugs using two methods: (i) pre-loading (for the drugs MTX and ETP) and (ii) post-loading (for MTX, ETP, and DOX). In the pre-loading method, 50 μL of protein solution at 5 mg/mL was mixed with 50 μL of 2 mg/mL MTX (Oncotron, Mumbai, India) or 50 μL of 2 mg/mL ETP (Ebewe, Unterach, Austria) and then mixed with 1,000 μL of 2 M potassium phosphate solution, pH 8 using a pipette. The spheres were incubated overnight at room temperature. Next, samples were centrifuged for 15 min at 10,000 × g, and the amount of drug loaded was determined spectrophotometrically by measuring the absorbance of the supernatant at a wavelength of 610 nm for MTX and 230 nm for ETP. The drug quantification was based on the concentration curves for the individual drugs.
For the post-loading method, 250 μg of spheres (the amount of silk corresponded to the amount used for the pre-loading method) was suspended in 250 μL of PBS, mixed with 50 μL of 2 mg/mL MTX, 50 μL of 2 mg/mL ETP, or 50 μL of 2 mg/mL DOX (Adriamycin; Pfizer Inc., New York City, NY, USA) and incubated overnight at room temperature with agitation. The spheres were centrifuged for 15 min at 10,000 × g, and the drug concentration in supernatant was measured spectrophotometrically by measuring the absorbance at a wavelength of 508 nm for DOX, and for MTX and ETP, as described above. The drug loading efficiency was calculated using the following equation:
|
For the drug release study, the drug-loaded spheres were suspended in 1 mL of phosphate-buffered saline (PBS) at pH 4.5, 6, and 7.4, and incubated at 37°C with agitation. The PBS buffer was replaced at the indicated time points with fresh PBS at the appropriate pH. The drug concentration in the collected samples was determined by measuring the absorbance at appropriate wavelengths for each drug, as specified above. The experiment was repeated three times in triplicate.
Statistics
The statistical significance of the differences between sphere groups was calculated using analysis of variance (ANOVA). Post hoc tests with the Bonferroni correction were performed. The differences between groups were considered significant if the P-value was <0.05.
Results
Production and purification of recombinant spider silk proteins
Two bioengineered silk proteins were produced: EMS2 and MS2. Figure 1 shows the amino acid sequences of EMS2. EMS2 was constructed by the addition of a glutamic acid residue to the repeat unit of MS2, and the repeat unit of MS2 was derived from the protein MaSp2 from N. clavipes.31,34 Both proteins comprised 15 repeats of the corresponding unit.
Bioengineered silk proteins were expressed in E. coli and purified using a thermal denaturation method. The average amount of protein from 1 g of bacterial pellet after purification was 3 and 2 mg for EMS2 and MS2, respectively. SDS-PAGE indicated good quality and purity of both proteins (Figure 1B), and MALDI-TOF analysis showed that the molecular mass of EMS2 protein corresponded with the theoretical calculations: 48.657 kDa (Figure 1C). The molecular mass of MS2 was analyzed previously.34
Silk sphere formation and sphere morphology
Silk spheres were produced in several processing variants, determining the influence of three variables on sphere formation: (1) initial concentration of potassium phosphate, (2) pH of potassium phosphate, and (3) initial concentration of silk proteins. For comparison, spheres made of MS2 silk were shown.
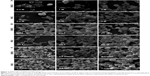
The SEM analysis showed that 2 M for EMS2 and 1 M for MS2 were the respective concentrations of potassium phosphate buffer that enabled sphere formation (Figure 2A). The mixing of the protein solution with potassium phosphate at lower concentrations resulted in silk structures of undefined, irregular shapes. MS2 spheres were slightly better separated and more spherical in shape compared with EMS2 particles (Figure 2A). Increasing the pH of potassium phosphate resulted in a more spherical morphology, which was particularly observed for EMS2 particles (Figure 2B); spheres were more separated as the pH of phosphate buffer increased. The detailed analysis indicated that the initial amount of silk proteins was crucial for the size of the spheres: a higher silk concentration resulted in larger spheres (Figure 2C).
Regardless of the preparation conditions, the EMS2 spheres were larger than the MS2 particles (Table 1). A trend toward reduction in size was observed with increasing pH of potassium phosphate for both sphere types. The concentration of silk had the greatest influence on the EMS2 and MS2 particle size, and the differences between sphere variants were significant. The mean size of the EMS2 spheres was 0.53 μm (±0.09) and 1.25 μm (±0.2) for spheres formed at concentrations of 0.5 and 5 mg/mL silk protein, respectively. The average size of the control MS2 spheres was ~2.5-fold smaller for spheres prepared at 0.5 mg/mL protein concentration (0.47 μm, ±0.1) compared with spheres prepared at 5 mg/mL (1.22 μm, ±0.14).
AFM investigation confirmed that both EMS2 and MS2 spheres were round, and their surfaces were smooth (Figure 3). The spheres revealed good structural integrity after being transferred to the top of the glass substrate, even if they formed clusters. As shown in the set of 2D topographical images, the topography of the EMS2 and MS2 spheres was similar (Figure 3).
Both EMS2 and MS2 particles were stable and preserved their spherical morphology during 6 months of storage at 4°C in water (Figure 4).
Zeta potential
Both EMS2 and MS2 particles demonstrated negative ZP (Table 1). Independently of the conditions of sphere production, the ZP values of EMS2 spheres were several times lower than the ZP values of MS2 particles (Table 1). EMS2 spheres exhibited similar values of ZP for all sphere variants (~−33 mV). A significant difference was observed between the ZP values of spheres prepared using potassium phosphate buffer at pH 4 and at pH 10 (Table 1).
The concentration of phosphate buffer and concentration of silk protein did not affect the ZP of the control MS2 spheres. Increasing the pH of the phosphate buffer used for MS2 sphere formation resulted in significant decreases in the ZP values of the obtained spheres. The ZP was >8-fold lower for MS2 spheres prepared at pH 10 than for those formed at pH 4 (Table 1).
Secondary structure analysis
Secondary structure estimation of EMS2 spheres was performed using the Fourier deconvoluted infrared spectra of amide I band analysis (Figure 5A). For comparison, the structure of MS2 spheres was indicated. The EMS2 and MS2 spheres did not differ significantly in the secondary structure content, and β-sheet structure accounted for ~40% in both sphere types (Figure 5B).
Cytotoxicity
The cytotoxicity study showed that both EMS2 and MS2 spheres were not toxic (Figure 6). A slight decrease in the viability of cells was observed at the highest tested sphere concentrations. The differences between EMS2 and MS2 particles were not significant.
Drug loading
Drug loading was performed using two methods: pre-loading and post-loading, as described in the Materials and methods section. Equivalent total amounts of silk protein (0.25 mg) and spheres (0.25 mg) were used for the pre- and post-loading experiments, respectively. Spheres for the pre- and post-loading methods were prepared under the same conditions in terms of pH and the concentrations of silk and phosphate buffer. For comparison and statistical analysis, the drug loading was also performed into MS2 spheres as described previously.34 Independently of the studied drug, the post-loading method was more efficient for the loading of drugs into EMS2 spheres (Table 2). Moreover, the loading efficiency of drugs into EMS2 spheres was significantly higher than that into MS2 particles, independently of the loading method. The only exception was DOX, where a similar efficiency of post-loading was observed for both types of spheres. The highest drug incorporation was reported for ETP loaded into EMS2 spheres using the post-loading method.
Cumulative drug release
The release of incorporated drugs was performed at 37°C, over 15 days, against PBS at pH 4.5, 6, or 7.4 (Figure 7). Regardless of the drug loading method, MTX, ETP, and DOX demonstrated pH-dependent release profiles from both EMS2 and MS2 spheres, with the fastest and most efficient release at pH 4.5, and the slowest and least efficient release at pH 7.4 (Figure 7).
EMS2 spheres loaded with MTX exhibited enhanced release of the drug in comparison with MS2 particles, independently of the loading method (Figure 7A–D). The post-loading method resulted in ~2 times higher MTX incorporation into EMS2 spheres than into MS2 spheres, but the release of MTX was also faster from EMS2 than from MS2 sphere (Figure 7C and D). MS2 spheres pre-loaded with MTX exhibited slower drug release than post-loaded MS2 particles (Figure 7B and D).
In contrast to MTX, the pre-loaded EMS2 spheres released ETP with considerably slower kinetics than MS2 particles (Figure 7E and F). After the first day, with equal levels of release, the post-loaded ETP was released slightly faster from EMS2 particles than from MS2 spheres (Figure 7G and H).
Moreover, the EMS2 particles demonstrated faster release of DOX than MS2 particles, with similar loading efficiency for both types of spheres (Figure 7I and J).
Discussion
We previously designed the bioengineered silk named MS2, based on the consensus motif of MaSp2 spidroin from the spider N. clavipes.31,32,34 For this study, we designed a new variant of MS2 called EMS2. For the construction of EMS2, the repeat unit of MS2 was modified by the addition of a glutamic acid residue (Glu/E). The MS2 and EMS2 proteins consisted of 15 repeat units; thus, in one molecule of EMS2, the E residue occurred 15 times. Glutamic acid contains a side-chain carboxylic acid, which will be deprotonated and, thus, negatively charged in its anionic form at physiological pH. We added E to the MS2 repeat unit to modify its theoretical isoelectric point and, as a result, to obtain a new bioengineered silk with different characteristics. By comparing the new silk with the control (MS2 silk), we determined the influence of this modification on the process of sphere formation and on the sphere properties. The modification of the amino acid composition could be one of the key factors for controlling the loading and release profile of the drug, thus making it possible to obtain the most effective drug delivery system.
Spider silk spheres were produced by protein precipitation caused by rapid mixing with potassium phosphate buffer. The salting out method was used for the production of microspheres from bioengineered spider silks that were based on the dragline silk of N. clavipes or A. diadematus or silk fibroin.27,30,31,33,38,39 Potassium phosphate, when mixed with silk solution, increases the hydrophobic interactions between silk molecules. As a consequence, alanine residues form hydrogen bonds and β-sheet structures to eventually assemble into spherical microparticles. It was shown that the process parameters, such as the protein and potassium phosphate concentration, method of silk purification, pH, or mixing method, affected the size and morphology of the spheres.21,22,31,38,39 Both the EMS2 and MS2 proteins formed stable spherical microparticles after the application of a high molar concentration of potassium phosphate. For the EMS2 protein, a higher phosphate concentration was required for sphere formation (2 M) than for the MS2 protein (1 M). This phenomenon could be caused by the presence of hydrophilic glutamic acid residues in the EMS2 sequence, which made EMS2 more hydrophilic than MS2. According to the Grand Average of Hydropathicity, EMS2 silk is slightly less hydrophobic than MS2 (−0.613 and −0.535 for EMS2 and MS2, respectively).
The major difference between the two silks was their theoretical isoelectric point: 3.15 and 5.27 for EMS2 and MS2, respectively. Accordingly, in ddH2O, both EMS2 and MS2 particles showed negative zeta potential; however, EMS2 spheres exhibited >2-fold lower values of ZP, which indicated that, independently of the sphere preparation conditions, the E residues were exposed on the surface of the EMS2 particles. Moreover, for sphere formation, potassium phosphate pH values of 4, 7, and 10 were used to test the influence of pH on the silk assembly process and the final charge of the silk particles. Although the zeta potential of EMS2 particles was in the range of -31 to -34 for all prepared variants, indicating the presence of the E residue (pKa ~4.3), a trend of decreasing ZP with increasing pH was observed. The difference in ZP values for EMS2 prepared at pH 4 and 10 was even significant. For MS2 spheres, significant differences in ZP values were observed for all particle variants. The MS2 sequence provides one N- and one C-terminus, with one amino and one carboxyl group, respectively. Moreover, it contains two tyrosine residues (Y, pKa 10.07) per repeat of the protein sequence. The tyrosine residue will be of importance only at very high pH. One possible explanation of the higher zeta potential value (closer to 0) for MS2 when measured in ddH2O is a more abundant presence of the N-terminus on the sphere surface. Thus, lower pH could favor a mode of MS2 assembly that exposes more N-termini on the sphere surface. The same may apply for EMS2 spheres. However, the possibility needs further study.
Interestingly, despite the difference in zeta potential values for EMS2 and MS2 spheres (−32.3 and −10.23, respectively), EMS2 particles exhibited a slightly stickier morphology. Most likely, the repulsive interaction was not strong enough to separate the EMS2 spheres. The observed behavior of the EMS2 spheres could be explained by differences in the assembly process compared with the assembly of MS2 particles. The bioengineered eADF3 spheres (protein derived from the spider A. diadematus) were proposed to possess hydrophilic side chains that stuck out of the spheres and formed dangling ends, which could mediate interactions between neighboring aggregates.40 Whether this concept applies to EMS2 spheres needs evaluation.
The stability of different morphological forms of silk is the consequence of the presence of polyalanine motifs that form β-sheet structures, which are responsible for the material strength.41 The EMS2 and MS2 proteins share the sequence and number of repetitive motifs consisting mainly of GPGXX, GPGGX, and GGX groups, and polyalanine chains. FTIR analysis of spheres made of both silk variants indicated that their secondary structure content was very similar. The addition of an E residue, at least not outside the vicinity of the polyalanine motif, did not influence the overall structure content, and the structure content was not the major factor determining the differences in the morphology and properties of the two sphere types.
To determine the potential of bioengineered spider silk spheres as drug carriers, the loading efficiency and release profiles of three model substances were evaluated: ETP (physiological charge=0, MW=588.557, experimental logP=0.6), DOX (physiological charge=+1, MW=543.52 g/mol, logP=1.27), and MTX (physiological charge=+2, MW=444.481 g/mol, experimental logP=−3.1). As mentioned previously, the properties of the silks were also different: EMS2 was slightly more hydrophilic and had a 3 times more negative zeta potential than control MS2. The drugs were incorporated into the EMS2 and MS2 spheres using two methods: pre- and post-loading. The varying affinities to drugs observed during the pre-loading method could be caused by the influence of potassium phosphate, a salt with high ionic strength that could have an impact on the electrostatic interactions between the drug and the protein. To obtain more data regarding the interaction of drug–silk, the EMS2 spheres were prepared and then loaded with drugs using the same methodology as described previously to study the properties of spheres made of bioengineered silks that were based on N. clavipes proteins MaSp1 and MaSp2 (ie, MS1 and MS2, respectively).34
MTX was the most hydrophilic and most positively charged drug studied. The high concentration of ions could disturb the electrostatic interactions and binding of this drug to both silk spheres, resulting in poor loading efficiencies when tested in the pre-loading assay. However, we observed a higher affinity of MTX to EMS2 spheres during post-loading, with almost a 2-fold higher efficiency compared to MS2 spheres. This result was consistent with the higher negative charge of EMS2 compared with MS2 particles. The difference was probably based on the stronger electrostatic interplay. The important role of electrostatic interactions was described by Lammel et al,27 who found that only positively charged model molecules could be incorporated into eADF4(C16) particles of the opposite charge. The release of MTX was faster from EMS2 spheres than from MS2 particles independently of the loading method. This result indicated that the initial fast release of drug was caused by washing away its molecules bound to the surface of the spheres. Most likely, the drug molecules did not penetrate into the core of the sphere, or the interactions between the drug and the silk were not strong enough to withstand the influx of protons that led to the displacement of drug molecules from the sphere. Although the EMS2 spheres enabled higher MTX loading, due to the fast drug release, they are not the best choice for in vivo application. The MS2 spheres, due to the low release rate at pH 7.4, are more suitable for carrying the drug through blood plasma. The MS2 spheres were also superior for MTX delivery compared with spheres made of bioengineered silk MS1.34
For both EMS2 and MS2 spheres, the loading efficiency of neutral ETP exhibited a loading method-dependent relationship. In our studies, the post-loading method demonstrated higher loading efficiency for ETP, suggesting the possibility of drug diffusion into the sphere matrix. Despite having the highest molecular weight, moderate hydrophobicity, and neutral charge, the ETP molecules were able to penetrate the sphere matrix. This conclusion can be supported by the observations of the drug release processes. The burst drug release was observed only for MS2 pre-loaded spheres, indicating that the drug molecules were mostly on the surface of the particles. For post-loaded MS2 and for EMS2 spheres produced by both loading methods, the release of ETP was gradual, and for pre-loaded EMS2 spheres, the drug release process was the slowest. After a moderate burst release of ETP from the sphere surface, the prolonged and slower release from pre-loaded EMS2 spheres was presumably caused by the diffusion of the drug from the protein matrix to the surface of the particles. It was indicated previously that post-loaded MS2 spheres showed greater applicability for ETP delivery than MS1 silk spheres.34 However, a new variant of silk, ie, EMS2 examined in this study, was superior compared with MS2 particles. Higher loading efficiency and slower drug release make EMS2 spheres a better choice to deliver ETP.
DOX is one of the most frequently examined drugs in silk-based delivery systems.24,30–32,42–45 It was shown that, in the case of DOX, one of the main loading factors is hydrophobic interactions. In our previous study, we showed that particles made of different variants of positively charged bioengineered MS1 silks were successfully loaded with DOX.30 Moreover, the studies of Hofer et al28 demonstrated the effective loading of positively charged eADF4(C16) bioengineered silk spheres with positively charged FITC-lysozyme and FITC-BSA, suggesting that electrostatic forces are not the only interactions between spider silk proteins and drugs. In this study, we examined only the post-loading method of incorporation of DOX, since, in a high salt concentration at pH 8, the drug co-precipitated with the silk, making the spectrometric measurement of the unbound drug in the supernatant unreliable (data not shown). Despite the high difference in zeta potential between the two types of silk spheres, the loading of positive DOX was similar, confirming that electrostatic interaction played a minor role in the loading of this drug. DOX was the most hydrophobic drug examined in this study, and hydrophobic interactions had the greatest influence on the loading of this drug into silk spheres.
For both sphere variants, a pH-dependent release kinetics of DOX was observed. The similar release profile of DOX was as reported previously for silk fibroin particles24 and spider silk spheres.30,32 For silk fibroin particles, the pH dependency of DOX release was probably because the low pH value caused a loss of the negative net charge of silk fibroin and weakened the electrostatic interaction between the silk and the drug.24 It was also suggested that lower pH increased the hydrophilicity of DOX, resulting in a faster release rate.46 In this study, the release of DOX was higher for EMS2 than for MS2 spheres. This difference could be the result of the slightly more hydrophilic character of the EMS2 protein, due to the presence of the glutamic acid residues. Moreover, it needs to be mentioned that, according to our results described previously, the most promising bioengineered silk for DOX delivery, mainly due to higher loading efficiency, was MS1 variant.34
Conclusion
We designed a new variant of bioengineered silk called EMS2. EMS2 was constructed based on the sequence of MS2 silk with additional glutamic acid residues. As a result, we obtained spheres with different characteristics. Despite a very similar secondary structure content, EMS2 spheres were bigger and had a higher tendency to aggregate than MS2 particles. Moreover, the spheres differed in their zeta potential values, and EMS2 particles were more negatively charged than MS2 particles. Interestingly, the more negative zeta potential was not advantageous for interactions with the drug with the greatest positive charge. Although the MTX loading could be increased, a burst release of this drug was observed. A faster release rate of positive DOX from EMS2 spheres than from MS2 particles was also observed. The reasonable loading efficiency and advantageous release rate make the EMS2 spheres the most suitable to deliver the drug with neutral charge – ETP. The loading and release of drug molecules into carriers made of bioengineered silks are dependent on many properties of both components (silk and drug), such as the hydrophobicity, charge, size, molecular weight, or shape of drug molecules. For spheres made of eADF4(C16), similar conclusions were drawn.33
Similar to the other bioengineered silk variants,30–32,36 the EMS2 silk was not cytotoxic. Moreover, long storage in ddH2O did not modify the morphology of either sphere type. These factors, as well as additional factors such as processing in an aqueous environment and biodegradability, make bioengineered silk one of the most promising candidates for an alternative drug delivery system, and the new silk variant EMS2 can be used to deliver a drug with neutral charge, such as ETP.
Abbreviations
MaSp2, major ampullate spidroin 2; MaSp1, major ampullate spidroin 1; MALDI-TOF, matrix-assisted laser desorption ionization-time-of-flight mass spectrometer; FITC, fluorescein isothiocyanate; BSA, bovine serum albumin.
Acknowledgments
We would like to thank NanoBioMedical Centre (NBMC) at Adam Mickiewicz University in Poznan, Poland, for the availability of the use of the microscope (JEOL JSM-7001F). The NBMC was supported from the National Centre for Research and Development under research grant “Nanomaterials and their application to biomedicine”, contract number PBS1/A9/13/2012. The project was partially supported by grant from the National Science Centre, Poland (2014/15/B/NZ7/00903). MK acknowledge partial support by HARMONIA3 grant (DEC-2012/06/M/ST4/00036) from the National Science Centre, Poland. MW and AP thank the Ministry of Science and Higher Education in Poland for financial support within Project No 06/62/DSPB/2183.
Author contributions
KK carried out the molecular construction, production and purification of EMS2, sphere preparation, SEM, zeta potential, drug loading/release and cytotoxicity study, and analysis of the data, and participated in writing the manuscript; MW and AP carried out the AFM study; KJ and MK carried out the FTIR study; ML performed MALDI-TOF study; AM participated in data analysis and writing the manuscript; HDK conceived, designed and coordinated the study, analyzed the data, and wrote the manuscript. All authors read and approved the final manuscript. All authors contributed toward data analysis, revising the paper and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
Zhang Y, Chan HF, Leong KW. Advanced materials and processing for drug delivery: the past and the future. Adv Drug Deliv Rev. 2013;65(1):104–120. | ||
Ha TLB, Quan TM, Vu DN, Si DM. Naturally derived biomaterials: preparation and application. In: Andrades JA, editor. Regenerative Medicine and Tissue Engineering. Rijeka: InTech; 2013:Ch. 11. | ||
Ige OO, Umoru LE, Aribo S. Natural products: a minefield of biomaterials. ISRN Mater Sci. 2012;2012:ID 983062. | ||
Lamboni L, Gauthier M, Yang G, Wang Q. Silk sericin: a versatile material for tissue engineering and drug delivery. Biotechnol Adv. 2015;33(8):1855–1867. | ||
Hu X, Tang Y, Wang Q, et al. Rheological behaviour of chitin in NaOH/urea aqueous solution. Carbohydr Polym. 2011;83(3):1128–1133. | ||
Crivelli B, Perteghella S, Bari E, et al. Silk nanoparticles: from inert supports to bioactive natural carriers for drug delivery. Soft Matter. 2018;14(4):546–557. | ||
Shi Z, Gao X, Ullah MW, Li S, Wang Q, Yang G. Electroconductive natural polymer-based hydrogels. Biomaterials. 2016;111:40–54. | ||
Jao D, Xue Y, Medina J, Hu X. Protein-based drug-delivery materials. Materials (Basel). 2017;10(5):pii: E517. | ||
Hong SJ, Ahn MH, Sangshetti J, Choung PH, Arote RB. Sugar-based gene delivery systems: current knowledge and new perspectives. Carbohydr Polym. 2018;181:1180–1193. | ||
Jahangirian H, Lemraski EG, Webster TJ, Rafiee-Moghaddam R, Abdollahi Y. A review of drug delivery systems based on nanotechnology and green chemistry: green nanomedicine. Int J Nanomedicine. 2017;12:2957–2978. | ||
Sionkowska A. Current research on the blends of natural and synthetic polymers as new biomaterials: Review. Progr Polym Sci. 2011;36(9):1254–1276. | ||
Numata K, Kaplan DL. Silk-based delivery systems of bioactive molecules. Adv Drug Deliv Rev. 2010;62(15):1497–1508. | ||
Rising A, Widhe M, Johansson J, Hedhammar M. Spider silk proteins: recent advances in recombinant production, structure-function relationships and biomedical applications. Cell Mol Life Sci. 2011;68(2):169–184. | ||
Heim M, Keerl D, Scheibel T. Spider silk: from soluble protein to extraordinary fiber. Angew Chem Int Ed Engl. 2009;48(20):3584–3596. | ||
Tokareva O, Michalczechen-Lacerda VA, Rech EL, Kaplan DL. Recombinant DNA production of spider silk proteins. Microb Biotechnol. 2013;6(6):651–663. | ||
Schacht K, Scheibel T. Processing of recombinant spider silk proteins into tailor-made materials for biomaterials applications. Curr Opin Biotechnol. 2014;29:62–69. | ||
Jastrzebska K, Kucharczyk K, Florczak A, Dondajewska E, Mackiewicz A, Dams-Kozlowska H. Silk as an innovative biomaterial for cancer therapy. Rep Pract Oncol Radiother. 2015;20(2):87–98. | ||
Wongpinyochit T, Johnston BF, Seib FP. Manufacture and drug delivery applications of silk nanoparticles. J Vis Exp. 2016;(116). | ||
Li L, Puhl S, Meinel L, Germershaus O. Silk fibroin layer-by-layer microcapsules for localized gene delivery. Biomaterials. 2014;35(27):7929–7939. | ||
Wongpinyochit T, Uhlmann P, Urquhart AJ, Seib FP. PEGylated silk nanoparticles for anticancer drug delivery. Biomacromolecules. 2015;16(11):3712–3722. | ||
Lammel AS, Hu X, Park SH, Kaplan DL, Scheibel TR. Controlling silk fibroin particle features for drug delivery. Biomaterials. 2010;31(16):4583–4591. | ||
Wenk E, Wandrey AJ, Merkle HP, Meinel L. Silk fibroin spheres as a platform for controlled drug delivery. J Control Release. 2008;132(1):26–34. | ||
Chen M, Shao Z, Chen X. Paclitaxel-loaded silk fibroin nanospheres. J Biomed Mater Res A. 2012;100(1):203–210. | ||
Seib FP, Jones GT, Rnjak-Kovacina J, Lin Y, Kaplan DL. pH-dependent anticancer drug release from silk nanoparticles. Adv Healthc Mater. 2013;2(12):1606–1611. | ||
Wang X, Wenk E, Matsumoto A, Meinel L, Li C, Kaplan DL. Silk microspheres for encapsulation and controlled release. J Control Release. 2007;117(3):360–370. | ||
Kundu J, Chung YI, Kim YH, Tae G, Kundu SC. Silk fibroin nanoparticles for cellular uptake and control release. Int J Pharm. 2010;388(1–2):242–250. | ||
Lammel A, Schwab M, Hofer M, Winter G, Scheibel T. Recombinant spider silk particles as drug delivery vehicles. Biomaterials. 2011;32(8):2233–2240. | ||
Hofer M, Winter G, Myschik J. Recombinant spider silk particles for controlled delivery of protein drugs. Biomaterials. 2012;33(5):1554–1562. | ||
Blüm C, Scheibel T. Control of drug loading and release properties of spider silk sub-microparticles. Bio Nano Science. 2012;2(2):67–74. | ||
Florczak A, Mackiewicz A, Dams-Kozlowska H. Functionalized spider silk spheres as drug carriers for targeted cancer therapy. Biomacromolecules. 2014;15(8):2971–2981. | ||
Jastrzebska K, Felcyn E, Kozak M, et al. The method of purifying bioengineered spider silk determines the silk sphere properties. Sci Rep. 2016;6:28106. | ||
Florczak A, Jastrzebska K, Mackiewicz A, Dams-Kozlowska H. Blending two bioengineered spider silks to develop cancer targeting spheres. J Mater Chem B. 2017;5(16):3000–3011. | ||
Doblhofer E, Scheibel T. Engineering of recombinant spider silk proteins allows defined uptake and release of substances. J Pharm Sci. 2015;104(3):988–994. | ||
Jastrzebska K, Florczak A, Kucharczyk K, et al. Delivery of chemotherapeutics using spheres made of bioengineered spider silks derived from MaSp1 and MaSp2 proteins. Nanomedicine (Lond). 2018;13(4):439–454. | ||
Szela S, Avtges P, Valluzzi R, et al. Reduction-oxidation control of beta-sheet assembly in genetically engineered silk. Biomacromolecules. 2000;1(4):534–542. | ||
Dams-Kozlowska H, Majer A, Tomasiewicz P, Lozinska J, Kaplan DL, Mackiewicz A. Purification and cytotoxicity of tag-free bioengineered spider silk proteins. J Biomed Mater Res A. 2013;101(2):456–464. | ||
Hu X, Kaplan D, Cebe P. Determining beta-sheet crystallinity in fibrous proteins by thermal analysis and infrared spectroscopy. Macromolecules. 2006;39(18):6161–6170. | ||
Slotta UK, Rammensee S, Gorb S, Scheibel T. An engineered spider silk protein forms microspheres. Angew Chem Int Ed Engl. 2008;47(24):4592–4594. | ||
Lammel A, Schwab M, Slotta U, Winter G, Scheibel T. Processing conditions for the formation of spider silk microspheres. Chem Sus Chem. 2008;1(5):413–416. | ||
Rammensee S, Slotta U, Scheibel T, Bausch AR. Assembly mechanism of recombinant spider silk proteins. Proc Natl Acad Sci U S A. 2008;105(18):6590–6595. | ||
Tokareva O, Jacobsen M, Buehler M, Wong J, Kaplan DL. Structure-function-property-design interplay in biopolymers: spider silk. Acta Biomater. 2014;10(4):1612–1626. | ||
Seib FP, Kaplan DL. DOX-loaded silk films: drug-silk interactions and in vivo performance in human orthotopic breast cancer. Biomaterials. 2012;33(33):8442–8450. | ||
Seib FP, Pritchard EM, Kaplan DL. Self-assembling DOX silk hydrogels for the focal treatment of primary breast cancer. Adv Funct Mater. 2013;23(1):58–65. | ||
Khalid A, Mitropoulos AN, Marelli B, Tomljenovic-Hanic S, Omenetto FG. DOX loaded nanodiamond-silk spheres for fluorescence tracking and controlled drug release. Biomed Opt Express. 2016;7(1):132–147. | ||
Schierling MB, Doblhofer E, Scheibel T. Cellular uptake of drug loaded spider silk particles. Biomater Sci. 2016;4(10):1515–1523. | ||
Sanson C, Schatz C, Le Meins JF, et al. A simple method to achieve high DOX loading in biodegradable polymersomes. J Control Release. 2010;147(3):428–435. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.