Back to Journals » Cancer Management and Research » Volume 11
BHLHE41 promotes U87 and U251 cell proliferation via ERK/cyclinD1 signaling pathway
Authors Wang C, Zhao N, Zheng Q, Zhang D, Liu Y
Received 6 May 2019
Accepted for publication 24 July 2019
Published 14 August 2019 Volume 2019:11 Pages 7657—7672
DOI https://doi.org/10.2147/CMAR.S214697
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Rudolph Navari
Chen Wang,1–3 Na Zhao,1–3 Qin Zheng,1–3 Di Zhang,1–3 Yang Liu1–3
1Department of Pathology, College of Basic Medical Sciences, China Medical University, Shenyang 110122, People’s Republic of China; 2Department of Pathology, The First Affiliated Hospital, China Medical University, Shenyang 110001, People’s Republic of China; 3Department of Pathology, Institute of Pathology and Pathophysiology, China Medical University, Shenyang 110122, People’s Republic of China
Purpose: The biological functions of BHLHE41 in the proliferation of glioblastoma remained unexplored. We aimed to investigate the biological roles and underlying molecular mechanisms of BHLHE41 in glioblastoma.
Materials and methods: We used multiple methods, including Western blot analysis, soft agar colony-formation assay, CCK8 assay, and flow cytometry, to evaluate the changes in multiple cellular functions after BHLHE41 knockdown or overexpression in U87 and U251 cell lines. The TCGA database was then used to analyze the associations between BHLHE41 expression with clinicopathological factors and the overall survival (OS) of glioma patients.
Results: This study determined that overexpression of BHLHE41 promoted glioma cell proliferation and colony formation. Besides, BHLHE41 upregulated cyclinD1, cyclinD3, and cyclinE1 expression and drove phase transition from G1 to S and G2 phases by upregulating these cyclins. In contrast, knockdown of BHLHE41 had an opposite effect on all of these parameters. However, BHLHE41 had no effect on apoptosis. Moreover, BHLHE41 activated MAPK/ERK signaling pathway to upregulate cyclinD1 expression. After the ERK signal pathway was blocked by a specific inhibitor, SCH772984, cyclinD1 upregulation was reversed. Furthermore, the median OS of low-grade glioma (LGG) patients with low to median level of BHLHE41 was 22.6 months, longer than that of the patients with high level of BHLHE41 (21.0 months).
Conclusion: BHLHE41 has an important role in the proliferation of glioblastoma and could serve as a novel candidate for targeted therapy of glioblastoma.
Keywords: BHLHE41, cyclinD1, ERK, cell cycle, proliferation, glioblastoma
Introduction
Basic helix-loop-helix E41 (BHLHE41) is located in 12p12.1 and functions as a transcription factor. It expresses in all human tissues and has the greatest expression abundance in the central nervous system (GTEx database: https://gtexportal.org/). In addition to regulating cell proliferation, apoptosis and hypoxia response, recent studies show BHLHE41 is also involved in regulating invasion and metastasis.1–5 It is not yet clear whether BHLHE41 is an oncogene or a tumor suppressor. BHLHE41 regulation of either proliferation or apoptosis is not yet consistently understood and is still being debated. In esophageal, osteosarcomas, prostate, breast, and endometrial cancer cells, BHLHE41 functions as an anti-apoptotic factor and promotes cell proliferation and invasiveness.6–11 It suppresses cell proliferation and metastasis, and also facilitates pancreatic as well as gastric cancer cell apoptosis.12,13 These might be attributed to the different BHLHE41 regulatory pathways in different cancers.
The MAPK/ERK pathway plays a critical part in integrating external signals from binding between a signaling molecule and a receptor in the cell surface into signaling events promoting cell growth and proliferation in many mammalian cell types.14–17 ERK phosphorylation results in a cascade activation and leads to numerous downstream target phosphorylations involved in cell proliferation regulation.15 In some tumors, sustained ERK activity may activate downstream genes that induce cell cycle entry, suppress negative cell cycle regulators, and lead to excessive tumor cell proliferation.18,19 Targets include cyclin D complexes with Cdk4 and Cdk6 (Cdk4/6), which ERK phosphorylates.20–22 This suggests that BHLHE41 might regulate cyclinD1 and, possibly, cell cycling via the ERK pathway.
Most studies of BHLHE41 functions focus on neurodevelopment and the circadian rhythm. Little is known about its role in central nervous system tumors. This study analyzed BHLHE41 biological functions in U251 and U87 cells. Using bi-directional regulation of BHLHE41, it was found that BHLHE41 increased cell proliferation and colony formation. BHLHE41 also upregulated cyclinD1, cyclinD3, and cyclinE1 expression and drove a phase transition from G1 to S and G2 phases. Adding SCH772984 reversed BHLHE41-induced cyclinD1 upregulation as well as ERK phosphorylation. The results indicate that BHLHE41 promoted U87 and U251 cell proliferation via the ERK/cyclinD1 axis.
Materials and methods
Cell culture
The U87 and U251 cell lines were obtained from the Shanghai Genechem Co., Ltd. (Shanghai, China) and were cultured in Dulbecco’s modified Eagle medium (DMEM) (GibCo BRL, Grand Island, NY, USA) with 10% fetal bovine serum, and 100 U/mL penicillinstreptomycin mixture (GibCo BRL) at 37°C, and 5 % CO2. The medium was changed every 2–3 days, and cultures were split using 0.25% trypsin. Since the data from Human Protein Atlas showed that BHLHE41 had the highest expression in U138, U251 and U87 cells, we select U251 and U87 cell lines for the experiments (Figure S1).
Plasmid and siRNA
Plasmid pcDNA3.1, pcDNA3.1-BHLHE41 was a generous gift from Prof. Kijima and the efficiency was demonstrated in the previous research. BHLHE41-siRNA and non-coding (NC)-siRNA were constructed by Taihe Biotechnology Co., Beijing, China. The sequences for the sense and antisense BHLHE41 siRNA#1 were 5ʹ-r(CGACAGGCAUAAACAAGAA) d (TT)-3ʹ and 5ʹ-r(UUCUUGUUUAUGCCUGUCG) d (TG) −3ʹ. The sequences for the sense and antisense BHLHE41 siRNA#2 were 5ʹ-r(CGUUGCAACCUAUUCUGAA) d (TT)-3ʹ and 5ʹ-r((UUCAGAAUAGGUUGCAACG) d (TG)-3ʹ. The sequences for the sense and antisense BHLHE41 siRNA#3 were 5ʹ-r(GCCUAACCAUUAGUACUUA) d (TT)-3ʹ and 5ʹ-r(UAAGUACUAAUGGUUAGGC) d (TG)-3ʹ. U251 and U87 cells were seeded at 1×105 cells per 35 mm well. After 24 h, BHLHE41 were transfected into the cells using Lipofectamine LTX (Invitrogen). After transfection, the cells were incubated for 24 h and subjected to various analysis. For the siRNA transfection experiments, U251 and U87 cells were seeded at 5×104 cells per 35 mm well. Twenty-four hours later, the siRNA was transfected into the cells using Lipofectamine 3000 reagent (Invitrogen) according to the manufacturer’s instructions. Transfected cells were incubated for another 48 h and subjected to various analyses. Total amounts of siRNA were adjusted with NC siRNA.
Western blot (WB)
Total protein was extracted using lysis buffer (Pierce, Rockford, IL) and quantified with the Bradford method.23 A total of 50 µg total protein per sample were separated via 15%, 10%, or 8% SDS-PAGE, and transferred onto polyvinylidene fluoride membranes (PVDF; Millipore, Billerica, MA). Membranes were incubated overnight at 4°C with the following primary antibodies: BHLHE41(1:500, Abcam, Cambridge, UK), GAPDH (1:2000, Abcam, Cambridge, UK), His-tag (1:1000, CWBio, Shanghai, China), Myc-tag (1:1000, ImmunoWay Biotechnology Company, Plano, TX), cyclinA2, cyclinB1, cyclinD1, cyclinD2, cyclinD3, cyclinE1, cyclinE2, cyclinH (1:1000, #9869, CST). ERK (1:1000, #4695, CST), phospho (p)-ERK (1:1000, #4370, CST). PI3K (1:1000, #4249, CST), phospho (p)-PI3K (1:1000, #4228 CST). AKT (1:1000, #4691, CST), phospho (p)-AKT (1:1000, #13038, CST). I-κB (1:1000, #4812, CST), NF-κB (1:1000, #8242S CST), Bax (1:1000, 50599-2-lg Proteintech), Bcl-2(1:1000, 12789-1-AP Proteintech). Membranes were washed and incubated with peroxidase-conjugated anti-mouse or anti-rabbit IgG (Santa Cruz Biotechnology, Santa Cruz, CA) at 37°C for 2 h. Bound proteins were visualized using electrochemiluminescence (Pierce, Rockford, IL) and detected with a bio-imaging system (DNR Bio-Imaging Systems, Jerusalem, Israel).
CCK8 assay
Cell proliferative rates of U87 and U251 cell lines were evaluated with the CCK8 assay (MedChemExpress, China) according to the instruction of the manufacturer. The above glioma cells were transfected with pcDNA or BHLHE41, or glioma cells transfected with NC-siRNA or BHLHE41-siRNA, and plated into 96-well plates (2×103 cells per well) containing media. Then, the glioma cells were cultured for 24 h, 48 h, 72 h and 96 h and incubated with CCK-8 regent at a final concentration of 10 μL/mL for 4 h at 37°C. Before reading the plate, it is important to mix gently on an orbital shaker for 1 min to ensure homogeneous distribution of color. Then, the absorbance was measured at 450 nm with a Thermomax microplate reader (Molecular Devices, Sunnyvale, CA, USA). Each experiment was repeated three times.
Flow cytometry analysis of cell cycle
Flow cytometry was used to analyze the cell cycle according to standard methods on a FACS Calibur cytometer (Becton-Dickinson Biosciences, San Jose, CA, USA). For DNA content analysis, U87 and U251 cells transfected with NC-siRNA or BHLHE41- siRNA, or U87 and U251 cells transfected with either pcDNA or Myc-His-BHLHE41. Then we seeded U87 and U251 cells in 6-cm dishes in growth medium with 10% Fetal bovine serum (FBS) overnight. The cultures were rinsed by phosphate buffered saline (PBS) and changed to serum-free medium. After serum starvation for 24 h, the cells were released into cell cycle by addition of 10% FBS. For flow cytometrical analysis of cell cycle distribution, cells were collected and fixed in 70% ethanol overnight at 4°C, washed with PBS, and then incubated with 1 mg/mL RNase A for 30 min at 37°C. DNA staining was performed by treating the cells with RNase A (10 mg/ml) for 30 mins and with propidium iodide (10 μg/ml; Sigma) for 5 mins.
Flow cytometry analysis of cell apoptosis
The Annexin V-fluorescein isothiocyanate (FITC)-binding assay was used to quantify the percentage of early and late apoptosis. We seeded U87 and U251 cells in 6-cm dishes and then transfected with pcDNA or Myc-His-BHLHE41. Following 24 h of transfection, the cells were cultured with DMSO or 50 µg/ml, 100 µg/ml, 150 µg/ml cisplatin for another 20 h. For flow cytometrical analysis of cell apoptosis distribution, cells were washed with PBS. And then, add 5 μl of Annexin V and 5 μl PI to each tube. After 15-min incubation at room temperature in the dark, add 400 μl of 1× Binding Buffer to each tube. Both adherent and nonadherent cells were harvested prior to staining with Annexin V-FITC and PI according to the manufacturer’s instructions.
Plate colony formation assay
U87 and U251 cells transfected with NC-siRNA, BHLHE41-siRNA, pcDNA, and Myc-His-BHLHE41 (800 cells/plate) were cultured in 5 mL of DMEM supplemented with 10% FBS in a 6-cm plate. After 14 days, colonies were washed with PBS, fixed with methanol for 30 min, and stained with crystal violet for 30 min. Clearly visible colonies (>50 mm in diameter) were counted as positive for growth.
The cancer genome atlas (TCGA) analysis
A total of 662 patients were enrolled in this study. BHLHE41 expression level and clinicopathological characteristics in 509 LGG patients (281 men and 228 women, TCGA-LGG) was obtained from TCGA-LGG (https://portal.gdc.cancer.gov/projects/TCGA-LGG). BHLHE41 expression level and clinicopathological characteristics in 153 GBM (glioblastoma) patients (99 men and 54 women, TCGA-GBM) were obtained from TCGA-GBM (https://portal.gdc.cancer.gov/projects/TCGA-GBM). GBM Patients were between 21 and 89 years of age, and most of them received chemotherapy and radiotherapy. LGG Patients were between 14 and 87 years old. Correlation between BHLHE41 expression and clinicopathological features in GBM and LGG patients was plotted using GraphPad Prism 6 and SPSS software.
Statistical analysis
SPSS version 22.0 for Windows (SPSS, Chicago, IL, USA) and GraphPad Prism 7.0 (USA, GraphPad Software) for Windows was used for all analyses. Ratio paired t-test was employed to analyze differences in the protein expression of cyclins and BHLHE41. Mann-Whitney U test was employed to analyze differences in CCK8 and plate colony formation assay. P < 0.05 was considered statistically significant. Prognosis about LGG and GBM patients were analyzed by UALCAN database(http://ualcan.path.uab.edu/index.html).24
Results
BHLHE41 overexpression enhances U87 and U251 cell proliferation and colony formation
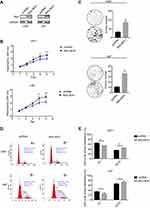
U87 and U251 cell lines were transfected with Myc-His-BHLHE41, which used the vector (pcDNA) as a control to verify BHLHE41 functions in glioblastoma cell proliferation. BHLHE41 plasmid validity was confirmed using Western blot (WB) (Figure 1A). CCK8 assays and Soft agar colony-formation assays were performed to analyze the BHLHE41 role in cell proliferation and colony formation (Figure 1B and C). Compared with the control group, BHLHE41 significantly enhanced U87 and U251 cell proliferation and colony formation along with foci number (U87: pcDNA vs BHLHE41, 10.7±1.5 vs 36.3±4.5, p<0.05; U251: pcDNA vs BHLHE41, 31.3±3.5 vs 86.3±9.1, p<0.01).
Flow cytometry was performed after transfection with Myc-His-BHLHE41 and pcDNA to measure BHLHE41 effects on cell cycles. Cell percentages in the G1/G0 phase decreased from 64.89±1.31% to 54.74±0.30% (U251) and from 33.99±2.98% to 25.70±4.21% (U87). S and G2 phase cell percentages increased from 35.11±1.31% to 45.26±0.30% (U251), and 66.01±2.98% to 74.30±4.21% (U87) in the BHLHE41 transfected group compared to control group (Figure 1D and E). This suggests that BHLHE41 might drive phase transition from G1 to S and G2 phases or promote S-phase entry in U87 and U251 cells.
BHLHE41 silence lessens U87 and U251 cell proliferation and colony formation
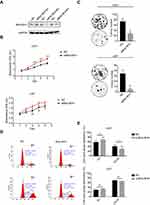
Three kinds of siRNA were prepared for use with BHLHE41. Their efficiencies were confirmed by WB analysis (Figure 2A). BHLHE41 siRNA#2 was selected as it had the greatest inhibitory effect. CCK8 assays showed that BHLHE41 knockdown inhibits U87 and U251 cell growth. Colony-formation assays showed that BHLHE41 silencing attenuates U87 and U251 cell proliferation, colony formation, and foci number (U87: pcDNA vs BHLHE41, 34.3±7.0 vs 4.5±2.5, p<0.01; U251: pcDNA vs BHLHE41, 73.7±11.2 vs 28.3±7.0, p<0.05) (Figure 2B and C).
Flow cytometry revealed that siRNA-induced BHLHE41 silence had higher cell proportions in the G1 phase (U251: 59.06±7.90% vs 69.94±11.60%; U87: 29.74±1.17% vs 34.48±1.83%) and a lower proportion in the S and G2 phase (U251: 40.94±7.90% vs 30.06±11.60%; U87: 70.26±1.17% vs 65.52±1.83%) compared with the control group (Figure 2D and E).
BHLHE41 regulates cyclin expression
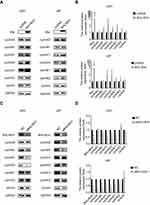
BHLHE41 was noted as possibly causing cell decrease in G1/G0-phase cells and facilitating cell entry in the S phase compared with the controls. Differing cyclin expression levels are known to cause differing cell cycle distributions. Cyclin expression in U87 and U251 cells was detected using WB analysis. CyclinD1 and cyclinE1 were significantly upregulated after BHLHE41 transfection in U251 cells. CyclinD1 and cyclinD3 were significantly upregulated in U87 cells (Figure 3A and B). Silencing BHLHE41 obtained the opposite results. CyclinD1 and cyclinE1 were significantly downregulated in U251 cells. CyclinA2, cyclinD1, cyclinD3, and cyclinE1 were significantly downregulated in U87 cells. The other cyclins were either unchanged or only slightly changed after BHLHE41 bi-directional regulation (Figure 3C and D).
BHLHE41 promotes cell proliferation through the ERK/cyclinD1 pathway
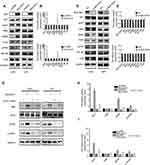
A set of core signaling pathway-related proteins was examined to determine by which molecular mechanism BHLHE41 enhanced proliferation. As shown in Figure 4A–C, overexpression of BHLHE41 significantly enhanced MAPK-ERK pathway activation in U87 and U251 cells (Figure 4A–C), while BHLHE41 knockdown decreased the phosphorylation of ERK (Figure 4D–F). Other signaling pathways involved in regulating proliferation, such as NF-κB and PI3K-Akt-mTOR, were examined. No changes in p-AKT, p-PI3K or I-κB were found. This suggests MAPK/ERK might be one of the pathways correlated with the BHLHE41-induced upregulation of cyclinD1. Pathway inhibitors were used to analyze ERK pathway effects on the BHLHE41-induced upregulation of cylcinD1.
SCH772984 significantly reduced p-ERK expression (Figure 4G–I). The BHLHE41-induced cyclinD1 upregulation was reversed after blocking ERK pathway in both U87 and U251 cells (Figure 4G–I). This suggests that BHLHE41 promoted cell proliferation through the ERK/cyclinD1 pathway in U87 and U251 cells.
BHLHE41 overexpression has no effect on cell apoptosis in U87 and U251 cells
To determine whether the overexpression of BHLHE41 affected cell apoptosis, U87 and U251 cells were transfected with pcDNA or BHLHE41 expression plasmid after treatment with DMSO or cisplatin (50, 100, or 150 μg/mL). Apoptosis was quantified at 20 h by Annexin V-FITC/PI double staining assay and WB analysis after DMSO or cisplatin treatment in U87 and U251 cells. As shown in Figure 5A and B, cisplatin administration at 50 μg/mL or DMSO failed to induce cell apoptosis in U87 and U251 cells, while treatment with a higher dose of cisplatin (100 or 150 μg/mL) significantly induced cell apoptosis in both U87 and U251 cells (Figure 5A and B). However, compared with the control group, the number of apoptosis cells in U87 and U251 cells was only slightly changed after transfected with BHLHE41. To further study the effect of BHLHE41 on cell apoptosis, we detected the cellular levels of apoptosis-related proteins such as Bax and Bcl-2 by WB analysis. As expected, we found that BHLHE41 overexpression in the absence or presence of cisplatin (50, 100 or 150 μg/mL) showed little effect on the expression of Bax and Bcl-2 (Figure 6A and B). Our results demonstrate that BHLHE41 overexpression had no effect on cell apoptosis in U87 and U251 cells.
Correlation between BHLHE41 expression and clinicopathological factors in 509 cases of LGG and 153 cases of GBM
Since we found that BHLHE41 expression enhanced cell proliferation in glioblastoma cells, we further investigated the association of BHLHE41 expression with clinical and pathological characteristics in LGG and GBM patients by TCGA analysis. The correlation between the BHLHE41 expression levels and clinicopathological characteristics in LGG is summarized in Table 1. The data demonstrated that BHLHE41 expression was significantly correlated with histological type (p<0.001), but did not significantly vary depending on age (p=0.352), gender (p=0.715), or WHO grading (p=0.109). The high level of BHLHE41 was observed more frequently in astrocytoma compared with oligodendroglioma. As shown in Figure 7A, the overall survival of LGG patients with the high level of BHLHE41 was much lower than that of group with the low to median level of BHLHE41 (Figure 7A, p=0.0019). In GBM patients’ group, as shown in Table 2, there was no correlation between BHLHE41 expression and age (p=0.722) or gender (p=0.559). As shown in Figure 7B, the overall survival of GBM patients with the high level of BHLHE41 was also lower than that of the patients with the low to median level of BHLHE41 (Figure 7B), but without a statistical difference (p=0.17). As shown in Figure 7C, the high level of BHLHE41 indicates poor prognosis of patients with WHO grades 2 and 3 (Figure 7C, p<0.0001).
 |
Table 1 Correlation of BHLHE41 expression with clinicopathological features in 509 LGG cases |
 |
Table 2 Correlation of BHLHE41 expression with clinicopathological features in 153 GBM cases |
Discussion
BHLHE41, also known as SHARP-1/DEC2/BHLHB3, is a member of the DEC subfamily within the bHLH gene family. DEC subfamily contains two highly conserved members: BHLHE40 and BHLHE41. Initial studies on them have focused on embryonic development and tissue differentiation.3 Recent studies have shown that the deregulation of their expression is associated with tumor progression.5,7,9,10,12,13 Unlike BHLHE40, which is broadly expressed in many tissues, BHLHE41 showed a more restricted pattern of mRNA expression. It has the greatest expression abundance in the central nervous system.25 Recent studies indicate that oscillatory bHLH factor expression is important in nervous system development.26,27 Sun et al demonstrated that BHLHE41 plays critical roles in regulating cell death and Neuro2a cell neurite outgrowth.28 BHLHE41 has been identified as a likely regulator of neuronal differentiation. However, its function remains largely unknown. This is particularly true for its roles in oncogenesis and central nervous system tumor progression. This study suggests the importance of BHLHE41 in regulating glioblastoma cell proliferation. In U251 and U87 cells, BHLHE41 overexpression significantly increases cell proliferation and colony formation. CyclinD1 and cyclinE1 expression also increased after BHLHE41 overexpression treatment, which resulted in cell cycle acceleration. The results indicate BHLHE41 is a positive regulator for glioblastoma cell proliferation. Silencing endogenous BHLHE41 confirmed this. In many tumors, BHLHE41 has been found to promote cell proliferation, migration, and invasion.9,10,28–30
Cell proliferation is driven by a series of proteins called cyclins, which are the key components of cell cycle machinery. They operate in the cell nucleus and are involved in binding, activating, and providing substrate specificity to cyclin-dependent kinases (CDKs).31 CyclinD1 is frequently deregulated in cancer and is a cancer phenotype and disease progression biomarker.32 Previous findings suggest that cyclinD1 expression upregulation and CDK4 activation directly lead to some cancer hallmarks by inducing proliferation.33–35 Of the cyclins, the variation among cyclinD1, cyclinD3, and cyclinE1 expression is the most apparent after BHLHE41 treatment. BHLHE41 transfection increased cell percentages in the S and G2 phases, while decreasing it in the G1 phase. This indicates that the BHLHE41 presence facilitated cell proliferation and is consistent with results of the CCK8 and colony assay. Consistent with this is the fact that silencing endogenous BHLHE41 led to significant cyclin (cyclinD1 and cyclinE1) decreases in both U251 and U87 cells. Correspondingly, the proportion of the cells in S and G2 phase decreased and in G1 phase increased. Silencing BHLHE41 suppressed proliferation and colony formation.
Cell growth is determined by the balance between proliferation and apoptosis. Since we have found the effect of BHLHE41 on cell proliferation, next we performed Annexin V and PI double staining by flow cytometry to measure whether BHLHE41 may suppress apoptosis, and therefore promoted cell growth. However, our results demonstrate that BHLHE41 has no effect on apoptosis in U251 and U87 cells. Consistent with the data of flow cytometry, Bax and Bcl-2 expression did not change much after BHLHE41 overexpression.
Several signaling pathways have been reported to be involved in regulating cell proliferation, including Ras-MAPK, JAK-STAT, and PI3K-Akt-mTOR.15,35 MEK/ERK signaling pathway activation plays a crucial role in almost all cell functions, such as apoptosis, autophagy, and senescence. It also enhances cell cycle progression and proliferation.17 Wu et al suggested that BHLHE41 was a target gene of the PI3K/AKT signaling pathway and promoted cell proliferation by upregulating c-Myc.10 In our study, the phosphorylation status of several key factors of these pathways, such as p-AKT, p-ERK, p-PI3K, and I-κB, was measured to analyze how BHLHE41 regulated cyclins. The phosphorylation status of most proteins was unchanged. The only exception to this was p-ERK. After blocking the MAPK/ERK signaling pathway with the inhibitor SCH772984, the BHLHE41-induced upregulation of cyclinD1 reversed. This indicates that BHLHE41 mediated the MAPK/ERK signaling pathway and facilitated cell proliferation by upregulating cyclinD1.
The TCGA analysis demonstrated that the high level of BHLHE41 is closely associated with poor prognosis in LGG. In GBM group, we also found the same situation, but without statistical difference. It should be noted that the patients in GBM group generally undertook preoperative radiotherapy and temozolomide treatment. We speculate these preoperative therapies might have affected BHLHE41 transcription and caused a divergence in the statistical result.
Conclusion
In conclusion, this study is believed to be the first to demonstrate that BHLHE41 enhances U251 and U87 cell proliferation and colony formation abilities. This positive regulation was obtained through the ERK signaling pathway and caused cyclinD1 upregulation, which was followed by a cell cycle acceleration. In addition, the high level of BHLHE41 was correlated with poor prognosis. These results suggest that targeting the BHLHE41/ERK/cyclinD1 axis may provide a new diagnostic and therapeutic approach for managing glioblastoma patients.
Acknowledgments
This work was supported by the Natural Science Foundation of Liaoning Province (grant no. 20170540989 to Yang Liu) and the General Project of Liaoning Provincial Education Department Foundation (grant no. LK201638 to Di Zhang).
Author contributions
All authors contributed to data analysis, drafting and revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Rossner MJ, Dorr J, Gass P, Schwab MH, Nave KA. Sharps: mammalian enhancer-of-split- and hairy-related proteins coupled to neuronal stimulation. Mol Cell Neurosci. 1997;10(3–4):460–475.
2. Honma S, Kawamoto T, Takagi Y, et al. Dec1 and Dec2 are regulators of the mammalian molecular clock. Nature. 2002;419(6909):841–844. doi:10.1038/nature01123
3. Liu Z, Li Z, Mao K, et al. Dec2 promotes Th2 cell differentiation by enhancing Il-2r signaling. J Immunol. 2009;183(10):6320–6329. doi:10.4049/jimmunol.0900975
4. Ozaki N, Noshiro M, Kawamoto T, et al. Regulation of basic helix-loop-helix transcription factors Dec1 and Dec2 by roralpha and their roles in adipogenesis. Genes Cells. 2012;17(2):109–121. doi:10.1111/j.1365-2443.2011.01574.x
5. Sato F, Bhawal UK, Yoshimura T, Muragaki Y. Dec1 and Dec2 crosstalk between circadian rhythm and tumor progression. J Cancer. 2016;7(2):153–159. doi:10.7150/jca.13748
6. Sato H, Wu Y, Kato Y, et al. Dec2 expression antagonizes cisplatininduced apoptosis in human esophageal squamous cell carcinoma. Mol Med Rep. 2017;16(1):43–48. doi:10.3892/mmr.2017.6571
7. Liu Q, Wu Y, Yoshizawa T, et al. Basic helix-loop-helix transcription factor Dec2 functions as an anti-apoptotic factor during paclitaxel-induced apoptosis in human prostate cancer cells. Int J Mol Med. 2016;38(6):1727–1733. doi:10.3892/ijmm.2016.2798
8. Asanoma K, Liu G, Yamane T, et al. Regulation of the mechanism of twist1 transcription by Bhlhe40 and Bhlhe41 in cancer cells. Mol Cell Biol. 2015;35(24):4096–4109. doi:10.1128/MCB.00678-15
9. Jiang B, Mu W, Wang J, et al. Microrna-138 functions as a tumor suppressor in osteosarcoma by targeting differentiated embryonic chondrocyte gene 2. J Exp Clin Cancer Res. 2016;35:69. doi:10.1186/s13046-016-0444-6
10. Wu Y, Sato H, Suzuki T, et al. Involvement of C-Myc in the proliferation of Mcf-7 human breast cancer cells induced by Bhlh transcription factor Dec2. Int J Mol Med. 2015;35(3):815–820. doi:10.3892/ijmm.2014.2042
11. Hu T, He N, Yang Y, Yin C, Sang N, Yang Q. Dec2 expression is positively correlated with Hif-1 activation and the invasiveness of human osteosarcomas. J Exp Clin Cancer Res. 2015;34:22. doi:10.1186/s13046-015-0135-8
12. Li P, Jia YF, Ma XL, et al. Dec2 suppresses tumor proliferation and metastasis by regulating Erk/Nf-Kappab pathway in gastric cancer. Am J Cancer Res. 2016;6(8):1741–1757.
13. Sato F, Kawamura H, Wu Y, et al. The basic helix-loop-helix transcription factor Dec2 inhibits Tgf-beta-induced tumor progression in human pancreatic cancer Bxpc-3 cells. Int J Mol Med. 2012;30(3):495–501. doi:10.3892/ijmm.2012.1037
14. Hymowitz SG, Malek S. Targeting the Mapk pathway in Ras mutant cancers. Cold Spring Harb Perspect Med. 2018;8(11):a031492. doi:10.1101/cshperspect.a031492
15. Sun Y, Liu WZ, Liu T, Feng X, Yang N, Zhou HF. Signaling pathway of Mapk/Erk in cell proliferation, differentiation, migration, senescence and apoptosis. J Recept Signal Transduct Res. 2015;35(6):600–604. doi:10.3109/10799893.2015.1030412
16. Papa S, Choy PM, Bubici C. The Erk and Jnk pathways in the regulation of metabolic reprogramming. Oncogene. 2018;38(13):2223–2240. doi:10.1038/s41388-018-0582-8
17. Liu F, Yang X, Geng M, Huang M. Targeting Erk, an Achilles’ Heel of the Mapk pathway, in cancer therapy. Acta Pharm Sin B. 2018;8(4):552–562. doi:10.1016/j.apsb.2018.01.008
18. Wakimoto R, Ono M, Takeshima M, Higuchi T, Nakano S. Differential anticancer activity of pterostilbene against three subtypes of human breast cancer cells. Anticancer Res. 2017;37(11):6153–6159. doi:10.21873/anticanres.12064
19. Bahrami A, Hesari A, Khazaei M, Hassanian SM, Ferns GA, Avan A. The therapeutic potential of targeting the Braf mutation in patients with colorectal cancer. J Cell Physiol. 2018;233(3):2162–2169. doi:10.1002/jcp.25952
20. Ebisuya M, Kondoh K, Nishida E. The duration, magnitude and compartmentalization of Erk Map kinase activity: mechanisms for Providing Signaling Specificity. J Cell Sci. 2005;118(Pt 14):2997–3002. doi:10.1242/jcs.02505
21. Marshall CJ. Specificity of receptor tyrosine kinase signaling: transient versus sustained extracellular signal-regulated kinase activation. Cell. 1995;80(2):179–185. doi:10.1016/0092-8674(95)90401-8
22. Dent P. Erk plays the baddie (again). Cancer Biol Ther. 2013;14(11):997–998. doi:10.4161/cbt.26377
23. Fujimoto K, Shen M, Noshiro M, et al. Molecular cloning and characterization of Dec2, a new member of basic helix-loop-helix proteins. Biochem Biophys Res Commun. 2001;280(1):164–171. doi:10.1006/bbrc.2000.4133
24. Chandrashekar DS, Bashel B, Balasubramanya SAH, et al. UALCAN: a portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia. 2017;19(8):649–658. doi:10.1016/j.neo.2017.05.002
25. Kageyama R, Shimojo H, Ohtsuka T. Dynamic control of neural stem cells by Bhlh factors. Neurosci Res. 2018;138:12–18. doi:10.1016/j.neures.2018.09.005
26. Lasky JL, Wu H. Notch signaling, brain development, and human disease. Pediatr Res. 2005;57(5 Pt 2):104R–09R. doi:10.1203/01.PDR.0000159632.70510.3D
27. Sun Y, Zhang H, Wang L, et al. Loss of the basic helix-loop-helix transcription factor Bhlhe41 induces cell death and impairs neurite outgrowth in neuro2a cells. Mol Cell Biochem. 2018;450(1–2):167-174. doi:10.1007/s11010-018-3383-z
28. Liang Y, Zhang P, Li S, Li H, Song S, Lu B. Microrna-873 acts as a tumor suppressor in esophageal cancer by inhibiting differentiated embryonic chondrocyte expressed gene 2. Biomed Pharmacother. 2018;105:582–589. doi:10.1016/j.biopha.2018.05.152
29. Sato F, Muragaki Y, Kawamoto T, Fujimoto K, Kato Y, Zhang Y. Rhythmic expression of Dec2 protein in vitro and in vivo. Biomed Rep. 2016;4(6):704–710. doi:10.3892/br.2016.656
30. Whittaker SR, Mallinger A, Workman P, Clarke PA. Inhibitors of cyclin-dependent kinases as cancer therapeutics. Pharmacol Ther. 2017;173:83–105. doi:10.1016/j.pharmthera.2017.02.008
31. VanArsdale T, Boshoff C, Arndt KT, Abraham RT. Molecular pathways: targeting the cyclin D-Cdk4/6 axis for cancer treatment. Clin Cancer Res. 2015;21(13):2905–2910. doi:10.1158/1078-0432.CCR-14-0816
32. Choi YJ, Li X, Hydbring P, et al. The requirement for Cyclin D function in tumor maintenance. Cancer Cell. 2012;22(4):438–451. doi:10.1016/j.ccr.2012.09.015
33. Yaguchi K, Yamamoto T, Shimada M, et al. Ploidy-dependent change in cyclin D2 expression and sensitization to Cdk4/6 inhibition in human somatic haploid cells. Biochem Biophys Res Commun. 2018;504(1):231–237. doi:10.1016/j.bbrc.2018.08.160
34. Bousoik E, Montazeri Aliabadi H. “Do we know jack” about Jak? A closer look at Jak/Stat signaling pathway. Front Oncol. 2018;8:287. doi:10.3389/fonc.2018.00287
35. Forrest MP, Waite AJ, Martin-Rendon E, Blake DJ. Knockdown of human Tcf4 affects multiple signaling pathways involved in cell survival, epithelial to mesenchymal transition and neuronal differentiation. PLoS One. 2013;8(8):e73169. doi:10.1371/journal.pone.0073169
Supplementary materials
 |
Figure S1 Expression of BHLHE41 in different organs.Notes: Reproduced from Human Protein Atlas. Available from: https://www.proteinatlas.org/ENSG00000123095-BHLHE41/tissue. Creative Commons license and disclaimer available from: http://creativecommons.org/licenses/by/4.0/legalcode.1 The data demonstrated that BHLHE41 had the highest expression in U138, U251 and U87 cell lines in brain.Abbreviation: BHLHE41, Basic helix-loop-helix family member e41. |
Reference
1. Human Protein Atlas. Available from: https://www.proteinatlas.org/ENSG00000123095-BHLHE41/tissue. Creative Commons license and disclaimer available from: http://creativecommons.org/licenses/by/4.0/legalcode. Accessed August 08, 2019.
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.