Back to Journals » OncoTargets and Therapy » Volume 12
Berbamine Enhances the Efficacy of Gefitinib by Suppressing STAT3 Signaling in Pancreatic Cancer Cells
Authors Hu B, Cai H, Yang S, Tu J, Huang X, Chen G
Received 15 July 2019
Accepted for publication 26 November 2019
Published 24 December 2019 Volume 2019:12 Pages 11437—11451
DOI https://doi.org/10.2147/OTT.S223242
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Sanjeev K. Srivastava
Bingren Hu,1,2 Huajie Cai,1,2 Shouzhang Yang,1,2 Jinfu Tu,1,2 Xiaming Huang,1,2 Gang Chen1,2
1Department of Hepatobiliary Surgery, First Affiliated Hospital of Wenzhou Medical University, Wenzhou, Zhejiang 325000, People’s Republic of China; 2Key Laboratory of Diagnosis and Treatment of Severe Hepato-Pancreatic Diseases of Zhejiang Province, Wenzhou, Zhejiang 325000, People’s Republic of China
Correspondence: Gang Chen
Department of Hepatobiliary Surgery, First Affiliated Hospital of Wenzhou Medical University, Shangcai Village, Nanbaixiang Town, Ouhai District, Wenzhou 325000, People’s Republic of China
Tel +86-0577-55579451
Email [email protected]
Background: Small molecular inhibitors such as gefitinib (Gefi), which target EGF receptor (EGFR), are considered to be a viable pathway for the selective inhibition of pancreatic cancer (PC) development. However, the large difference in Gefi response between PC patient individuals and PC cell lines severely limits the clinical efficacy of Gefi. Berbamine (BBM) is a well-known natural-derived antitumor agent. However, no study yet exists on whether BBM can enhance the sensitivity of PC cells to Gefi or its underlying mechanisms.
Methods: MTS assay and clonogenic assay were used to determine whether BBM could enhance the anti-PC activity of Gefi by. Flow cytometric analysis was performed to study the cell cycle progression and rate of apoptosis after combined treatment with BBM and Gefi. Surface plasmon resonance (SPR) and Western blot experiments were carried out to detect the STAT3 binding affinity and the STAT3 inhibitory effect of BBM. Molecular docking and Molecular dynamic simulation were used to predicting the dominant interaction between BBM and STAT3.
Results: This study found that BBM synergizes with Gefi to inhibit cell growth and induce cell cycle arrest and PC cell apoptosis. Mechanistically, our results showed that BBM and Gefi have synergistic inhibitory effects on STAT3 phosphorylation, but have little effect on other EGFR downstream pathways, suggesting that BBM may exert sensitization through the inhibition of STAT3. Besides, BBM has a high affinity for STAT3 and a good inhibitory effect on STAT3 activation, further indicating that BBM was a potent direct STAT3 inhibitor. Molecular modeling between STAT3 and BBM suggested that BBM formed several key hydrophilic interactions with STAT3.
Conclusion: Our findings suggest that the combination of BBM and Gefi could be further developed as a potential PC therapy.
Keywords: berbamine, Gefi, pancreatic cancer, STAT3
Introduction
As one of the most serious malignancies, pancreatic cancer (PC) has a 5-year survival rate of less than 6%.1 The greatest obstacle to PC treatment is that the vast majority of patients (over 80%) display no symptoms until the disease reaches its terminal stage.2 Although gemcitabine-based therapies have long been established as the standard treatment for advanced PC since 1997, the toxicity and acquired resistance of these therapies require further investigation so that more targeted therapies could be developed.3 One promising area of research in the past several decades has centered around the identification of certain molecular targets, such as EGFR, K-Ras, B-Raf, PI3K/Akt and TGF-β, which are expected to yield clinical benefits in the treatment of PC.4,5
EGFR, also known as ErbB-1, is one such target that has been attracting much scholarly attention. As a member of receptor tyrosine kinases, EGFR is widely recognized as a key oncoprotein in multiple solid tumors, including lung cancer, colorectal cancer, and pancreatic cancer.6 Existing research has documented that in 30% to 50% of all PC patients, EGFR is shown to overexpress.7 Correlations have also been obtained between overexpression of EGFR and other clinical observations, such as rapid progression of disease, resistance to chemotherapy, and poor prognosis. Besides, it has been reported that EGFR pathway is frequently constitutively activated in multiple PC cell lines, and that gefitinib and erlotinib, two EGFR inhibitors, can effectively stem their proliferation.8,9 A growing body of evidence seems to suggest that EGFR-based therapy holds great promise as an effective treatment of PC.7,10 However, dismal results have been obtained in clinical trials of such therapies versus traditional chemotherapy, contrary to prior expectations.11–13 To understand the mechanisms underlying their clinical performance, researchers have continued along this line of research, in the hope of finding a feasible way to effectively apply EGFR-targeted therapies to clinical practice. For example, Thomas et al found that the combination of EGFR inhibition and Rb dephosphorylation led to a synergistic growth inhibition of PC cell lines.14 Moreover, Jiang et al reported Ginsenoside Rg3 sensitized PC cells to EGFR inhibitor erlotinib via suppressing EGFR/PI3K/Akt pathway signaling. Many of these studies have focused on finding possible ways to address erlotinib resistance in PC cells.15 However, little work has been done on how to enhance Gefi efficacy in PC cells.
One promising agent that has been shown to have anti-proliferative and pro-apoptotic effects is Berbamine (BBM). BBM is a natural bisbenzyl isoquinoline alkaloid extracted from the Chinese medicinal plant Berberis amurensis Rupr. It has been widely used in Chinese medicine for the treatment of various diseases, including autoimmune disease, inflammation, and cancer.16–20 Many studies have shown that BBM have anti-proliferative and pro-apoptotic effects for different types of cancer, including chronic myelogenous leukemia (CML), lung, liver, and breast cancer.21–24 The anti-tumor mechanisms underlying BBM have been reported in Ca2+/calmodulin-dependent protein kinase II γ (CaMKII γ), bcr/abl and NF-κB signaling pathway activation.21,25,26 Recently, a number of researchers have found that BBM also displayed unexpected activity in reverse multi-drug resistance (MDR), and identified some of the underlying mechanisms. For example, Wang et al documented that BBM exerted synergistic effects with anticancer agents celecoxib and trichostatin A by prompting the apoptosis of MDA-MB-231 cells.24 Moreover, BBM improved the effects of gemcitabine on inhibiting cell viability and inducing cell apoptosis via regulation of TGF-β/Smad signaling pathway.27 Thus, there is compelling evidence that BBM can effectively reverse chemotherapy resistance in multiple ways. However, no study yet exists that examines the potential and mechanisms of BBM in sensitizing PC cells to targeted therapy like EGFR TKI Gefi.
The present study was designed to explore the synergistic effect of BBM and Gefi on two Gefi-resistant PC cell lines Panc-1 and Miapaca-2. The results indicated that BBM remarkably increased the Gefi sensitivity of Panc-1 and Miapaca-2 cells. In addition, the Western blot and SPR assays results demonstrated that BBM might sensitize PC cell lines to Gefi by directly interacting with STAT3 to synergistically suppress the STAT3 signal. Computer simulations further revealed that BBM formed several key hydrophobic interactions with STAT3 SH2 domain, which probably dominated the STAT3 inhibitory activity of BBM.
Materials and Methods
Cell Lines and Reagents
The human pancreatic cancer cell lines Panc-1, Miapaca-2 were obtained from the American Type Culture Collection (Manassas, USA). The cells were cultured in DMEM medium (Gibco, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS, Gibco) and 100 units/mL penicillin/streptomycin. Given these treatments, the cells were kept in a room with 5% CO2 and constant temperature at 37°C. Cell Signaling Technology (Danvers, MA) provided some of the principal antibodies, such as Bcl-2, Cdk4, Cdk6, Cleaved-caspase-3, Cleaved-PARP (poly ADP-ribose polymerase), cyclin D1, Akt, Erk, p-Akt, p-Erk, p-STAT3, STAT3, and β-actin. Three other molecules include the Goat anti-rabbit IgG-HRP secondary antibody, the full length recombinant STAT3 protein, and Gefitinib and berbamine, which were procured from Santa Cruz Biotechnology (Santa Cruz, CA), Abcam (Cambridge, UK), and Selleck (Shanghai, China), respectively. SDS PAGE measured the purity of the full length recombinant STAT3 protein at over 85%, while the purity of both Gefitinib and berbamine were over 99%.
Cell Viability Assay
Cell Titer 96 Aqueous Non-Radioactive Cell Proliferation Assay Kit (Promega) was utilized to gauge the variability between different cells. First, 5×103 cells were seeded in 96-well plates for a full night. Thereafter, the culture medium was removed, and the cells were treated with different concentrations of chemicals for 2 days. At 490 nm, a microplate reader was employed to record the absorbance value of each cell. Each experimental procedure was duplicated for three times.
Clonogenic Assay
Panc-1 or Miapaca-2 cells (800 cells/well) were embedded in six-well plates for a full night. Then, the culture medium was supplemented with the addition of BBM (5 µM), DMSO, Gefi (20 µM), and Gefi (20 µM) plus BBM (5 µM). The resulting medium was kept for 24 hrs, until it was replaced by fresh culture medium, which lasted for 7–10 days on end. After this interval, PBS was used to wash the fresh medium for 1/4 h, with the number of surviving colonies then counted through crystal violet staining assay and photographed.
Cell Apoptosis Analysis
Flow cytometer analysis and Western blotting were two methods utilized to detect cell apoptosis. For the first method, the cells were first treated for 2 days. Then they were harvested by trypsin and washed with PBS twice. After that, they were re-suspended in 100 μL 1× binding buffer. The cell suspension was then supplemented with the addition of 5 μL FITC Annexin V and PI (556547, BD Biosciences, USA), and then incubated for 1/4 h at room temperature. After the cells were diluted using μL binding buffer, ACS Calibur flow cytometer (BD) was utilized to perform analysis on the samples. For the second method, Cleaved-PARP, Bcl-2, and Bax were analyzed.
Cell Cycle Analysis
Cells (2.5 × 105 cells/well) were seeded in six-well plates and cultured overnight. After treatment with different compounds for 48 hrs, they were harvested using trypsinization and pelleted by centrifugation. Then, PBS was used twice to wash the pellets, which were immersed in 75% ice-cold ethanol at 4°C overnight. After centrifugation, cold PBS was used to wash the pellets, which were then resuspended in 500 μL PBS containing 50 μg/mL propidium iodide (PI) and incubated at room temperature for 1/2 h in darkness. Finally, a FACS Calibur instrument (Becton Dickinson FACSCalibor, BD Biosciences, NJ, USA) was utilized to perform analysis on the samples.
Western Blotting
Subsequent to 24 hrs of treatment of the cells with the indicated compounds, PBS and radioimmunoprecipitation assay (RIPA) buffer were used to wash and lyse the cells, respectively. BCA protein assay kit (Beyotime Biotechnology, Shanghai, China) was also used for quantification of the cell lysates. SDS-PAGE divided the proteins into equal proportions and then electro-transferred to a 0.22-μm polyvinylidene fluoride (PVDF) membrane. The membranes were first blocked using 5% non-fat milk in TBST for 90 min. They were then incubated with specific primary antibody for a full night at 4°C. Horseradish peroxidase (HRP) (1:5000, Santa Cruz, CA) was used to conjugate the secondary antibody for 2 hrs at normal room temperature. Finally, the function of enhanced chemiluminescence (EMD Millipore, Billerica, MA, USA) in Amersham Imager 600 system (GE Healthcare Life Sciences, Shanghai, China) was used to visualize the immunoreactive bands.
Surface Plasmon Resonance (SPR) Analysis
SPR experiments were carried out on the system of Proteon XPR36 protein interaction array (Bio-Rad Laboratories, Hercules, CA, USA). In short, the STAT3 solution of 1 mg/mL PBST (5 mM, 7.4 pH) was diluted to 30 μg/mL using sodium acetate buffer (pH 4.5). The chip was activated by EDC/NHS (10 μL/min, 600 s). Then, STAT3 (5 μL/min for 400 s) was loaded and covalently fixed. About 8000 RU STAT3 is fixed on the chip. Any excessive unbounded proteins were removed by flowing PBS solution (5 mM, pH 7.4, containing 5%, w/v, DMSO). The PBS solution (5 mM, pH 7.4, containing 5%, w/v, DMSO) of Sch A was prepared and injected (10 μL/min, lasting 100 s). The associated phases with five concentrations of 120 s were injected simultaneously at a flow rate of 30/min, and then the dissociated phases were injected at 25°C for 120 s. The final chart is obtained by subtracting the blank sensing map from the duplex or quadruple sensor graph. ProteOn manager software was used for the data analysis.
Docking Protocol
The AutoDock 4.2 software was utilized to perform docking of the STAT3 and BBM. Protein Data Bank (PDB) (PDB code: 1BG1) contributed the starting protein structure of STAT3.28 Previous research has shown that the SH2 domain of STAT3 could be regarded as the binding pocket.29,30 Then, certain missing elements in the obtained structure were added, including the formal bond orders, charges, hydrogen atoms, and some residues. Thereafter, the active binding surface of STAT3 was covered using the grid box (30 Å × 30 Å × 30 Å). Lamarckian genetic algorithm (LGA) was also utilized to conduct conformational sampling with trials of 100 dockings. A predicted conformation with the best docking score was selected for further molecular dynamics (MD) simulation analysis.
Molecular Dynamics Simulation Protocol
Amber 18 software was utilized to perform MD simulation of STAT3/BBM complex.
Specifically, the HF/6-31G* was implemented by Gaussian 09 program in order to measure the electrostatic potentials of BBM.31 Then these potentials were fitted for atomic partial charges of BBM using the RESP fitting method.32 To optimize the molecular mechanics (MM), force fields of ff14SB and gaff2 were used for STAT3 and BBM to create MD parameters, respectively.33,34 Thereafter, the composite was immersed in a Tip3P tank with a distance between the protein surface and the tank boundary of 15 mm. And the counter ions were added to neutralize the system. Constrained and unconstrained composites were minimized using the steepest descent method and the conjugate gradient method. After minimization, the system was heated from 0 to 300 K for 200 ps, and the weak constraint on the solute is 5 kcal mol−1 Å−2. Then, an isothermal isostatic ensemble (NPT) MD simulation was performed on the 200 ps to adjust the solvent density. An additional 200 ps unconstrained MD simulation was performed at 300 K to unwind the system unrestricted under the NPT. Finally, a 200 ns free MD simulation was performed on the system. The Langevin temperature scalings were utilized to regulate the temperature.35 To constrain the bonds involving hydrogen atoms, the SHAKE algorithm was adopted.36 The Particle Mesh Ewald (PME) algorithm was used to adjust periodic boundary conditions and electrostatic interactions, with a cut-off value of 10 Å, for range-limited non-bonding interactions.37 The molecular mechanics/generalized born surface area (MM/GBSA) approach in Amber18 was used to evaluate the binding free energy decomposition, and altogether 500 snapshots were extracted from the equilibrated trajectory between 160 and 200 ns. Detailed descriptions of the equations were reported previously.38
Statistical Analysis
The results are presented as the means ± standard errors of the mean (S.E.M.). One-way analysis of variance (ANOVA) was performed in GraphPad Pro (GraphPad, San Diego, CA, USA) to test significant difference between groups. “ns” means no significance. A P value below 0.01 (P<0.01) indicates that the difference is highly statistically significant. All the experiments were replicated for at least three times.
Results
BBM Increased the Anti-Growth Effect of Gefi Against Panc-1 and Miapaca-2 Cells
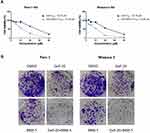
In previous studies, the anti-proliferative activity of Gefi has been closely examined in a panel of human PC cell lines, with the results showing that Panc-1 and Miapaca-2 are naturally Gefi-resistant.39 These results provide the rationale for our selection of these two cell lines in the present study. Firstly, MTS colorimetric assay was performed to identify the anti-proliferative activity of BBM on Panc-1 and Miapaca-2 cells. As Figure 1A and B suggest, the IC50 value of BBM against Panc-1 and Miapaca-2 was 8.137 and 11.16 μM, respectively. Based on the IC50 value of BBM, the concentration of BBM used in the following combination treatment was set as 5 μM, which should have little cytotoxicity on Panc-1 and Miapaca-2 cells. For Gefi, the IC50 value on Panc-1 and Miapaca-2 cells was 25.57 and 22.79 μM, respectively. Next, the sensitization effects of BBM and Gefi were further investigated. As shown in Figure 2A, the treatment combining Gefi and BBM significantly decreased the viability of Panc-1 and Miapaca-2 cells, demonstrating that such a treatment is more effective than either treatment alone. Regarding the sensitivity of Panc-1 and Miapaca-2 cells to Gefi, the IC50 values indicated that compared to the group with Gefi treatment alone, the sensitivity in the group with combined treatment of Gefi and BBM experienced an approximate 2.3-fold and 2.46-fold increase in Panc-1 and Miapaca-2 cells, respectively.
To further test whether the treatment combining Gefi and BBM leads to more efficient anti-growth effects in the long run, colony-forming assays were conducted. After the cell colonies were differentially treated for 7–10 days, they were stained with crystal violet and photographed. The results suggested that the group receiving combined treatment experienced a synergistic reduction of colony formation (Figure 2B) compared to the group treated with only one compound (20 μM Gefi or 5 μM BBM). This result was consistent with the results obtained using MTS assay.
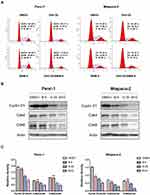
Combined Gefi and BBM Treatment Synergistically Induces Cell Cycle Arrest
To probe whether cell cycle arrest was involved in the sensitization of BBM, flow cytometry was utilized to analyze the percentage of cells at each stage of cell division. The results showed that the combination of Gefi and BBM could effectively inhibit the two PC cell lines at the G0/G1 phase (Figure 3A). In addition, Western blots were performed to verify the molecular events responsible for G1 phase blockade. The results showed that the synergistic effect of BBM enhanced the inhibitory effect of Gefi on the expression of cell cycle-related proteins (Cyclin D1, Cdk4, and Cdk6) (Figure 3B and C). These results suggested that BBM synergized with Gefi to inhibit the cells at the G0/G1 phase by reducing the expression of cell cycle-associated proteins.
Combined Gefi and BBM Treatment Synergistically Induces PC Cells Apoptosis
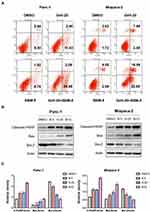
In order to study the synergistic effect of Gefi and BBM on apoptosis, single-agent treatment, or the combination therapy was used to treat Panc-1 and Miapaca-2 cells, and double staining with Annexin V-FITC and PI was performed to determine their apoptosis rate using flow cytometry. As shown in Figure 4A, the apoptosis rate of PC cells was significantly increased after the treatment combining Gefi and BBM.
To elucidate the molecular mechanisms underlying the enhancement of BBM-induced apoptosis, Western blot was performed to analyze the expression levels of the apoptosis-related proteins Cleaved-PARP, Bax, and Bcl-2. The results showed that the expression of pro-apoptotic molecules (Cleaved-PARP and Bax) was significantly upregulated, while the anti-apoptotic protein Bcl-2 was downregulated in the group receiving the combined treatment (Figure 4B and C).
BBM Binds Directly to STAT3 and Suppresses the STAT3 Activation in PC Cells
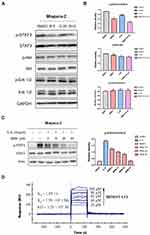
The transcription factor signal transducer and activator of transcription 3 (STAT3) is a downstream molecule involved in the signal transduction of many trans-membrane growth factor receptors (e.g. EGFR), cytosolic kinases (e.g. Src), and the Janus kinases (JAK). A heavy body of research has been dedicated to elucidating the function of STAT3 in a variety of cancer types. These studies generally show that STAT3 plays a pivotal role in pathogenesis of various hematologic and solid tumor malignancies (e.g. hepatocellular carcinoma, non-small cell lung cancer, pancreatic cancer, etc.). Additionally, several recent studies have indicated that re-activation of EGFR downstream pathway including STAT3, PI3K/Akt, and ERK is closely linked to the development of resistance to EGFR inhibitors. Therefore, it was hypothesized that the sensitizing effects of BBM might be mediated by at least one of this signaling pathways. Western blot was performed to determine the phosphorylation and total protein expression levels of STAT3, Akt, and ERK1/2 in Miapaca-2 cells, which were treated with BBM, Gefi, or BBM plus Gefi. As shown in Figure 5A and B, the results demonstrated that Gefi treatment had minimal effect on the level of p-STAT3, p-Akt and p-ERK1/2 in Miapaca-2 cells. This finding is contrary to many previous reports on NSCLC cells, but is congruent with the results of Maria et al. In addition, combining BBM and Gefi in the treatment effectively downregulated the phosphorylation of STAT3 Tyr-705, but showed no inhibitory effect on p-Akt Ser473 and p-Erk1/2 Thr202/Tyr204. These results thus provide some evidence that the mechanism underlying the synergistic effects of BBM may be mediated by the inhibition of STAT3 signal activation.
Next, to confirm the inhibitory effects of BBM on STAT3 signals, the level of p-STAT3 was determined by Western blot. As interleukin-6 (IL-6) is a common stimulator that induces STAT3 phosphorylation on tyrosine Tyr705 in many cancers, we examined the impact of BBM on IL-6-induced STAT3 phosphorylation in Miapaca-2. The results (Figure 5C) demonstrated that BBM inhibited p-STAT3 induced by IL-6 in a dose-dependent manner, even at a concentration as low as 5 μM.
Furthermore, to investigate whether BBM could directly bind to STAT3, SPR experiments were conducted. As shown in Figure 5D, the binding affinity of STAT3 to BBM increased with the level of concentration, and a low equilibrium dissociation constant (KD) of 12.1 μM was calculated. Taken together, the above results suggested that STAT3 may be a direct target of BBM and that the Gefi-sensitization effects of BBM were mainly mediated by its inhibition of STAT3.
Analysis of the BBM Binding Site by Molecular Modeling
To predict possible binding patterns between STAT3 and BBM, molecular docking experiments were first performed. Then, 200 ns MD simulations were carried out to obtain a series of equilibrated and stable structures. To confirm that the simulated complex reached equilibrium and to investigate the dynamic stability of the STAT3/BBM complex, the root-mean-square deviations (RMSDs) of all the backbone atoms (Cα) of STAT3 and the heavy atoms of BBM were monitored, as plotted in Figure 6A. The RMSDs of the backbone atoms of STAT3 showed dynamic fluctuations (<1 Å) after 50 ns simulation. The RMSDs of BBM Cα were relatively stable during the whole MD simulation (Figure 6A). Principal Component Analysis (PCA) was a method commonly used to reduce the dimensionality of data and to identify the conformational space occupied by proteins/molecules during MD simulations. In this study, the input data are Cartesian coordinates from the MD simulation trajectory. That is to say, the principal component (PC) represents the variance of the coordinate space. When PCs are plotted against each other, similar structures cluster together. As shown in Figure 6B, the PCA results showed that the conformational space of STAT3 is dynamically distributed, and that most of the structures show a high degree of similarity. The scatter plot also suggests that the plots are closely plotted against each other. These observations indicated that a relatively stable STAT3/BBM complex binding conformation was determined by our 200 ns MD simulation. Subsequently, the residue-free energy decomposition was performed using the last 40 ns trajectory of the MD simulation. As illustrated in Figure 6C, the most five contributed residues were Glu-594, Glu-638, Ser-636, Gln-635, Ile-634. The predominant residues were hydrophilic amino acids, indicating that polar interactions may contribute to the binding of BBM to STAT3. The BBM embedded in the SH2 field of STAT3 seems to match very well to implement the binding pocket as a lock and key (Figure 6D).
In addition, the potential energy surface of the electrostatic molecule clearly presents information on the negative charge of the bound band, which was consistent with the results of per-residue free energy decomposition of BBM binding to STAT3 (Figure 6D). The structural analysis showed that the majority of interactions were hydrogen bond to stabilize the conformation between BBM and STAT3 (Figure 6E and F).
Discussion
During the past decade, the rapid development of immunotherapy, coupled with the increasing number of FDA-approved targeted therapies, has led to enormous progress in the overall clinical prognosis of breast cancer, NSCLC, melanoma, and other tumors. In contrast, PC treatment seems to be lagging behind. For decades, researchers have been trying to find a practical and sustainable target, but only the EGFR inhibitor erlotinib has been approved by the FDA for use in combination with gemcitabine. However, the results of the third phase of the program turned out to be unsatisfactory. Compared with the treatment using gemcitabine alone, the combination therapy showed little improvement in DFS and OS. Gefitinib (Gefi) is another first representative of EGFR inhibitors and has been widely used in patients with EGFR-sensitive mutations. In some preclinical studies, Gefi has also been shown to have a good inhibitory effect on PC cells. However, the sensitivity of Gefi varies greatly among different PC cell lines, which may be one of the reasons for the poor clinical efficacy of Gefi. For example, Maria et al reported that CfPac1 and BxPC-3 cells are highly sensitive to Gefi, while Panc-1 and Miapaca-2 cells are highly insensitive to Gefi.39 Therefore, finding an effective sensitizer that overcomes the natural resistance of Panc-1 and Miapaca-2 cells to Gefi may be a promising method to improve the clinical efficacy of Gefi.
There is increasing evidence that many natural products not only inhibit the growth of cancer cells but also effectively reverse tumor resistance. For example, it has been reported that betulinic acid, a pentacyclic lupane-type triterpene extracted from plants, can reverse the resistance of cancer cells to gemcitabine in PC cells and enhance the sensitivity of non-small cell lung cancer cells to sorafenib.40,41 Therefore, in this study, we focused specifically on another safe and effective anti-cancer natural product, berbamine (BBM), and investigated whether it might enhance the sensitivity of Gefi to drug-resistant cells. Although there have been some studies on the chemical sensitization of BBM (Carmofur, gemcitabine), the effects of BBM on Gefi resistance or other targeted therapies have not been studied. In this study, we found that combined treatment with BBM significantly enhanced the anti-proliferative, cell cycle arrest and pro-apoptotic effects of Gefi in Panc-1 and Miapaca-2 cells. In addition, although some studies have shown that BBM has significant anti-proliferative activity against human lung cancer, liver cancer and pancreatic cancer cells in vitro, its anti-tumor mechanism remains elusive. Our results of Western blot and SPR analysis provide evidence that BBM is a potential STAT3 inhibitor. By measuring the effects of STAT3 signaling after single-agent or combination therapy, it was further confirmed that the anti-PC activity and synergistic effect of BBM was primarily mediated by the inhibition of STAT3 signals (Figure 7).
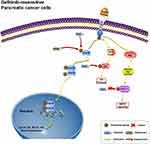 |
Figure 7 Hypothetical schema of the mechanism of the Gefi-sensitization effects of BBM. |
Since 2003, the FDA has approved gefitinib for patients with metastatic or advanced non-small cell lung cancer. Epidermal growth factor receptor-targeted therapy has been applied to clinical practice for nearly 16 years. In the applications, a persistent problem has been its drug resistance. Thanks to the work of many researchers, a number of resistance mechanisms have been discovered. In general, EGFR second-site mutations (T790M, C797S, etc.), bypass signal pathway compensation (such as MET amplification, HER2 upregulation or constitutive NF-κB activities), and EGFR downstream signals reactivation (such as ERK and Akt reactivation) are considered as the most important factors in the development of drug resistance.42 The important role of STAT3 in this process, in particular, has been attracting much attention in recent years. As a key transcription factor for cell survival and proliferation, STAT3 can be phosphorylated in Tyr-705 and subsequently activated by various receptor tyrosine kinases (EGFR, FGFR, and IGF-1R, etc.), receptor-associated kinases such as JAK, and non-receptor kinases such as Src and Abl.43 This means that cunning cancer cells can be steered by upstream signal blockade and choose an alternative way to activate STAT3. Besides, a lot of studies have demonstrated that targeting STAT3 could be a promising way to treat PC. However, there is no STAT3 inhibitor approved for clinical use yet. Currently, a large number of STAT3 have been identified. However, improving the drug-making ability of these molecules remains a big conundrum. In China, BBM has been applied to the clinical treatment of patients with imatinib-induced neutropenia and inflammation. The safety and anti-tumor effects of BBM have also been validated in clinical practice, which will provide great convenience for rapid development of combination therapy of gefitinib and BBM.
The well-known STAT3 inhibitors, such as OPB-31121, TTI-101, and S31-201, have entered early clinical research or preclinical research.44 But their specific mode of action is still obscure. The structural characteristics of BBM differ from those of these inhibitors in that it exhibits high rigidity and planarity, making the binding conformation of BBM and STAT3 more unique. Therefore, this study conducted molecular docking and MD simulation to explore potential interaction patterns and key binding sites between BBM and STAT3. Our molecular docking and simulation analyses showed that BBM depended mainly on polar interactions with several hydrophilic residues near STAT3 Ser-636. In order to overcome the low water solubility of BBM, which is one of the important issues in BBM research, it is extremely important to improve its drug-forming properties by changing its structure. According to our molecular modeling results, introducing some hydrophilic functional group such as piperazinyl and N, N-dimethylaminoethyl into the hydroxyl of BBM may effectively raise its solubility, and provide more effective inhibitory activity. In addition, the preparation of BBM in the form of liposomes or nanoparticles can also increase the bioavailability and tumor targeting of BBM.
Conclusions
To sum up, the present study indicated that BBM could enhance the effects of Gefi against Panc-1 and Miapaca-2 cells on inhibiting proliferation, inhibiting cell cycle, and promoting apoptosis. Regarding the underlying mechanisms, the results in this study demonstrated that BBM directly binds to STAT3, and facilitates Gefi in reducing the STAT3 signals in PC cells. Finally, molecular docking and molecular simulations of the BBM/STAT3 complex revealed that stable binding between these two molecules depended on hydrophilic interactions.
Acknowledgments
This research was supported by the Wenzhou Science and Technology Bureau of China (Y20170041).
Disclosure
The authors declare no conflict of interest.
References
1. Kamisawa T, Wood LD, Itoi T, Takaori K. Pancreatic cancer. Lancet. 2016;388(10039):73–85. doi:10.1016/S0140-6736(16)00141-0
2. Michl P, Gress TM. Current concepts and novel targets in advanced pancreatic cancer. Gut. 2013;62(2):317–326. doi:10.1136/gutjnl-2012-303588
3. Olson JL, Bold RJ. Currently available first-line drug therapies for treating pancreatic cancer. Expert Opin Pharmacother. 2018;19(17):1927–1940. doi:10.1080/14656566.2018.1509954
4. Matsuoka T, Yashiro M. Molecular targets for the treatment of pancreatic cancer: clinical and experimental studies. World J Gastroenterol. 2016;22(2):776–789. doi:10.3748/wjg.v22.i2.776
5. Strimpakos AS, Syrigos KN, Saif MW. The molecular targets for the diagnosis and treatment of pancreatic cancer. Gut Liver. 2010;4(4):433–449. doi:10.5009/gnl.2010.4.4.433
6. Tebbutt N, Pedersen MW, Johns TG. Targeting the ERBB family in cancer: couples therapy. Nat Rev Cancer. 2013;13(9):663–673. doi:10.1038/nrc3559
7. Nedaeinia R, Avan A, Manian M, Salehi R, Ghayour-Mobarhan M. EGFR as a potential target for the treatment of pancreatic cancer: dilemma and controversies. Curr Drug Targets. 2014;15(14):1293–1301. doi:10.2174/1389450115666141125123003
8. Zhou X, Zheng M, Chen F, et al. Gefitinib inhibits the proliferation of pancreatic cancer cells via cell cycle arrest. Anat Rec (Hoboken). 2009;292(8):1122–1127. doi:10.1002/ar.20938
9. Kelley RK, Ko AH. Erlotinib in the treatment of advanced pancreatic cancer. Biologics. 2008;2(1):83–95. doi:10.2147/btt.s1832
10. Sidaway P. Pancreatic Cancer: EGFR inhibition is effective against KRAS-wild-type disease. Nat Rev Clin Oncol. 2017;14(9):524–525.
11. Hammel P, Huguet F, van Laethem JL, et al. Effect of chemoradiotherapy vs chemotherapy on survival in patients with locally advanced pancreatic cancer controlled after 4 months of gemcitabine with or without erlotinib: the LAP07 randomized clinical trial. JAMA. 2016;315(17):1844–1853. doi:10.1001/jama.2016.4324
12. Brell JM, Matin K, Evans T, et al. Phase II study of docetaxel and gefitinib as second-line therapy in gemcitabine pretreated patients with advanced pancreatic cancer. Oncology. 2009;76(4):270–274. doi:10.1159/000206141
13. Ignatiadis M, Polyzos A, Stathopoulos GP, et al. A multicenter phase II study of docetaxel in combination with gefitinib in gemcitabine-pretreated patients with advanced/metastatic pancreatic cancer. Oncology. 2006;71(3–4):159–163. doi:10.1159/000106064
14. Thomas NA, Abraham RG, Dedi B, Krucher NA. Targeting retinoblastoma protein phosphorylation in combination with EGFR inhibition in pancreatic cancer cells. Int J Oncol. 2019;54(2):527–536. doi:10.3892/ijo.2018.4658
15. Jiang J, Yuan Z, Sun Y, Bu Y, Li W, Fei Z. Ginsenoside Rg3 enhances the anti-proliferative activity of erlotinib in pancreatic cancer cell lines by downregulation of EGFR/PI3K/Akt signaling pathway. Biomed Pharmacother. 2017;96:619–625. doi:10.1016/j.biopha.2017.10.043
16. Ren Y, Lu L, Guo TB, et al. Novel immunomodulatory properties of berbamine through selective down-regulation of STAT4 and action of IFN-gamma in experimental autoimmune encephalomyelitis. J Immunol. 2008;181(2):1491–1498. doi:10.4049/jimmunol.181.2.1491
17. Li SY, Ling LH, Teh BS, Seow WK, Thong YH. Anti-inflammatory and immunosuppressive properties of the bis-benzylisoquinolines: in vitro comparisons of tetrandrine and berbamine. Int J Immunopharmacol. 1989;11(4):395–401. doi:10.1016/0192-0561(89)90086-6
18. Parhi P, Suklabaidya S, Kumar Sahoo S. Enhanced anti-metastatic and anti-tumorigenic efficacy of Berbamine loaded lipid nanoparticles in vivo. Sci Rep. 2017;7(1):5806. doi:10.1038/s41598-017-05296-y
19. Zhang H, Jiao Y, Shi C, et al. Berbamine suppresses cell proliferation and promotes apoptosis in ovarian cancer partially via the inhibition of Wnt/beta-catenin signaling. Acta Biochim Biophys Sin (Shanghai). 2018;50(6):532–539. doi:10.1093/abbs/gmy036
20. Zhao Y, Lv JJ, Chen J, et al. Berbamine inhibited the growth of prostate cancer cells in vivo and in vitro via triggering intrinsic pathway of apoptosis. Prostate Cancer Prostatic Dis. 2016;19(4):358–366. doi:10.1038/pcan.2016.29
21. Xu R, Dong Q, Yu Y, et al. Berbamine: a novel inhibitor of bcr/abl fusion gene with potent anti-leukemia activity. Leuk Res. 2006;30(1):17–23. doi:10.1016/j.leukres.2005.05.023
22. Hou ZB, Lu KJ, Wu XL, Chen C, Huang XE, Yin HT. In vitro and in vivo antitumor evaluation of berbamine for lung cancer treatment. Asian Pac J Cancer Prev. 2014;15(4):1767–1769. doi:10.7314/APJCP.2014.15.4.1767
23. Wang GY, Lv QH, Dong Q, Xu RZ, Dong QH. Berbamine induces Fas-mediated apoptosis in human hepatocellular carcinoma HepG2 cells and inhibits its tumor growth in nude mice. J Asian Nat Prod Res. 2009;11(3):219–228. doi:10.1080/10286020802675076
24. Wang S, Liu Q, Zhang Y, et al. Suppression of growth, migration and invasion of highly-metastatic human breast cancer cells by berbamine and its molecular mechanisms of action. Mol Cancer. 2009;8:81. doi:10.1186/1476-4598-8-81
25. Gu Y, Chen T, Meng Z, et al. CaMKII gamma, a critical regulator of CML stem/progenitor cells, is a target of the natural product berbamine. Blood. 2012;120(24):4829–4839. doi:10.1182/blood-2012-06-434894
26. Liang Y, Xu RZ, Zhang L, Zhao XY. Berbamine, a novel nuclear factor kappaB inhibitor, inhibits growth and induces apoptosis in human myeloma cells. Acta Pharmacol Sin. 2009;30(12):1659–1665. doi:10.1038/aps.2009.167
27. Jin X, Wu Y. Berbamine enhances the antineoplastic activity of gemcitabine in pancreatic cancer cells by activating transforming growth factor-beta/Smad signaling. Anat Rec (Hoboken). 2014;297(5):802–809. doi:10.1002/ar.22897
28. Becker S, Groner B, Muller CW. Three-dimensional structure of the Stat3beta homodimer bound to DNA. Nature. 1998;394(6689):145–151. doi:10.1038/28101
29. Li H, Liu A, Zhao Z, et al. Fragment-based drug design and drug repositioning using multiple ligand simultaneous docking (MLSD): identifying celecoxib and template compounds as novel inhibitors of signal transducer and activator of transcription 3 (STAT3). J Med Chem. 2011;54(15):5592–5596. doi:10.1021/jm101330h
30. Xiao H, Bid HK, Jou D, et al. A novel small molecular STAT3 inhibitor, LY5, inhibits cell viability, cell migration, and angiogenesis in medulloblastoma cells. J Biol Chem. 2015;290(6):3418–3429. doi:10.1074/jbc.M114.616748
31. Frisch M, Trucks G, Schlegel HB, et al. Gaussian 09. Wallingford (CT): Gaussian, Inc; 2009.
32. Wang J, Wang W, Kollman PA, Case DA. Antechamber: an accessory software package for molecular mechanical calculations. J Am Chem Soc. 2001;222:U403.
33. Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA. Development and testing of a general amber force field. J Comput Chem. 2004;25(9):1157–1174. doi:10.1002/(ISSN)1096-987X
34. Lindorff‐Larsen K, Piana S, Palmo K, et al. Improved side‐chain torsion potentials for the Amber ff99SB protein force field. Proteins. 2010;78(8):1950–1958.
35. Izaguirre JA, Catarello DP, Wozniak JM, Skeel RD. Langevin stabilization of molecular dynamics. J Chem Phys. 2001;114(5):2090–2098. doi:10.1063/1.1332996
36. Krautler V, Van Gunsteren WF, Hunenberger PH. A fast SHAKE: algorithm to solve distance constraint equations for small molecules in molecular dynamics simulations. J Comput Chem. 2001;22(5):501–508. doi:10.1002/1096-987X(20010415)22:5<501::AID-JCC1021>3.0.CO;2-V
37. Shan Y, Klepeis JL, Eastwood MP, Dror RO, Shaw DE. Gaussian split Ewald: a fast Ewald mesh method for molecular simulation. J Chem Phys. 2005;122(5):54101. doi:10.1063/1.1839571
38. Xie X, Tu J, You H, Hu B. Design, synthesis, and biological evaluation of novel EF24 and EF31 analogs as potential IkappaB kinase beta inhibitors for the treatment of pancreatic cancer. Drug Des Devel Ther. 2017;11:1439–1451. doi:10.2147/DDDT.S133172
39. Pino MS, Shrader M, Baker CH, et al. Transforming growth factor alpha expression drives constitutive epidermal growth factor receptor pathway activation and sensitivity to gefitinib (Iressa) in human pancreatic cancer cell lines. Cancer Res. 2006;66(7):3802–3812. doi:10.1158/0008-5472.CAN-05-3753
40. Sun L, Cao J, Chen K, et al. Betulinic acid inhibits stemness and EMT of pancreatic cancer cells via activation of AMPK signaling. Int J Oncol. 2019;54(1):98–110. doi:10.3892/ijo.2018.4604
41. Kutkowska J, Strzadala L, Rapak A. Synergistic activity of sorafenib and betulinic acid against clonogenic activity of non-small cell lung cancer cells. Cancer Sci. 2017;108(11):2265–2272. doi:10.1111/cas.2017.108.issue-11
42. Chong CR, Janne PA. The quest to overcome resistance to EGFR-targeted therapies in cancer. Nat Med. 2013;19(11):1389–1400. doi:10.1038/nm.3388
43. Siveen KS, Sikka S, Surana R, et al. Targeting the STAT3 signaling pathway in cancer: role of synthetic and natural inhibitors. Biochim Biophys Acta. 2014;1845(2):136–154. doi:10.1016/j.bbcan.2013.12.005
44. Johnson DE, O’Keefe RA, Grandis JR. Targeting the IL-6/JAK/STAT3 signalling axis in cancer. Nat Rev Clin Oncol. 2018;15(4):234–248. doi:10.1038/nrclinonc.2018.8
 © 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2019 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.