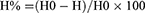Back to Journals » Infection and Drug Resistance » Volume 16
Baicalein Inhibits the Staphylococcus aureus Biofilm and the LuxS/AI-2 System in vitro
Authors Mao Y, Liu P, Chen H, Wang Y, Li C, Wang Q
Received 28 January 2023
Accepted for publication 29 April 2023
Published 9 May 2023 Volume 2023:16 Pages 2861—2882
DOI https://doi.org/10.2147/IDR.S406243
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 4
Editor who approved publication: Professor Suresh Antony
Yanni Mao, Panpan Liu, Haorong Chen, Yuxia Wang, Caixia Li, Quiqin Wang
Veterinary Pharmacology Lab, School of Animal Science and Technology, Ningxia University, Yinchuan, 750021, People’s Republic of China
Correspondence: Quiqin Wang, Email [email protected]
Introduction: Staphylococcus aureus (S. aureus) is a common cause of mastitis in dairy cows, a condition that has a significant economic impact. S. aureus displays quorum sensing (QS) system-controlled virulence characteristics, like biofilm formation, that make therapy challenging. In order to effectively combat S. aureus, one potential technique is to interfere with quorum sensing.
Methods: This study evaluated the effects of different Baicalin (BAI) concentrations on the growth and the biofilm of S. aureus isolates, including the biofilm formation and mature biofilm clearance. The binding activity of BAI to LuxS was verified by molecular docking and kinetic simulations. The secondary structure of LuxS in the formulations was characterized using fluorescence quenching and Fourier transform infrared (FTIR) spectroscopy. Additionally, using fluorescence quantitative PCR, the impact of BAI on the transcript levels of the luxS and biofilm-related genes was investigated. The impact of BAI on LuxS at the level of protein expression was also confirmed by a Western blotting investigation.
Results: According to the docking experiments, they were able to engage with the amino acid residues in LuxS and BAI through hydrogen bonding. The results of molecular dynamics simulations and the binding free energy also confirmed the stability of the complex and supported the experimental results. BAI showed weak inhibitory activity against S. aureus but significantly reduced biofilm formation and disrupted mature biofilms. BAI also downregulated luxS and biofilm-associated genes’ mRNA expression. Successful binding was confirmed using fluorescence quenching and FTIR.
Discussion: We thus report that BAI inhibits the S. aureus LuxS/AI-2 system for the first time, which raises the possibility that BAI could be employed as a possible antimicrobial drug to treat S. aureus strain-caused biofilms.
Keywords: quorum sensing system, biofilm, baicalin, Staphylococcus aureus, molecular docking
Corrigendum for this paper has been published.
Introduction
One of the most significant bacteria causing infectious mastitis in dairy cattle globally is Staphylococcus aureus (S. aureus).1 Antibiotics have played an important role in preventing and treating S. aureus infections in humans and farm animals.2 However, treatment for S. aureus infections is getting more and more challenging due to rising antibiotic resistance and biofilm formation.3,4
Bacteria use quorum sensing (QS) molecules to communicate, recognize, and react to cell density.5 QS controls biofilm formation, virulence factors, secondary metabolites, and motility, among other physiological activities in bacteria.6 QS signals have the ability to cause S. aureus to form biofilms, and the adhering cells within these biofilms are encased in a self-made matrix of extracellular polymeric substances (EPS).7 Due to acquired resistance, constrained drug transport, and antibiotic inactivation, S. aureus within the exopolymeric matrix may develop significantly increased resistance to conventional antibiotics after biofilms have formed.8 As LuxS is present in both gram-positive and gram-negative bacteria, it has been hypothesized that AI-2 may serve as an all-purpose, cross-species bacterial language,9 and this system could be a potential target for local antibiotic therapy for biofilm-associated.
Silico screening can speed up and reduce the cost of finding prospective medication candidates. In this context, protein-ligand docking, a molecular modeling method to forecast ligand-protein binding conformations, can be utilized to assist in silico screening to find leads that are similar to drugs. There is an increased interest in recent years in the reduced bacterial biofilm of herbal remedies in general.10 Among these, the majority of Scutellaria baicalensis’ biological activities are attributed to the flavonoid component baicalin (BAI), which has been used for many years as a herbal remedy in China to treat a variety of inflammatory disorders.11 As a result, BAI might be a potential therapeutic approach for infections caused by S. aureus biofilms.
In this manuscript, LuxS structural features were determined by analysis, and investigated the mechanism of LuxS/AI-2-mediated S. aureus biofilm inhibition. Through in vitro biofilm experiments and in silico molecular docking, the effect of BAI on LuxS inhibition was assessed.
Materials and Methods
Bacterial Strains and Growth Conditions
The four S. aureus strains used in this study were obtained from mastitis-related milk samples from cows. wld10 & wld19 were identified as MRSA and JY21& JY45 were MSSA. At 37 °C with shaking at 220 rpm for approximately 16 hours, S. aureus strains were grown aerobically in Mueller–Hinton broth (MHB) (Bio-Tech, Qingdao, China). S. aureus strains ATCC 29213 (National Center for Medical Culture Collections) was used as quality controls for antimicrobial susceptibility testing.12 The Vibrio harveyi (V. harveyi) BB170 (ATCC BAA-1117) and V. harveyi BB152 were, respectively, donated by professor Yongjie Liu (Nanjing Agricultural University, China) and professor Xiangan Han (Shanghai Veterinary Research Institute, Chinese Academy of Agricultural Sciences, China). In autoinducer bioassay medium (AB, Bio-Tech, Qingdao, China), the luminous reporter strain V. harveyi BB170 (sensor1-, sensor 2+) was cultured for 18 hours at 30 °C with aeration.13 V. harveyi BB152 is a mutant derived from BB120 that does not produce AI-1 and served as a positive control.14 In marine broth (MB 2216; Difco BD), strains were cultured with aeration at 30 °C and 200 rpm overnight. Meanwhile, the AI-2 signal is not produced by Escherichia coli (E. coli) DH5α, which was selected as the negative control.15 The pET-28a (+) vector (Anorun Biotechnology Co., China) was used for prokaryotic expression of the LuxS protein. For gene cloning and protein expression, DynaScience Biological Co., China’s E. coli DH5α and BL21(DE3) were employed.
Molecular Characterization of LuxS Protein
The National Center for Biotechnology (NCBI) was consulted for LuxS’s sequence, and the homologous structures were found using the NCBI BLAST program. Utilizing ClustalW and ESPript3, the multiple sequence alignment of S. aureus LuxS with other homologous proteins whose structures are known was created. The secondary structure of the LuxS was predicted using the SOPMA (https://npsa-prabi.ibcp.fr/cgi-bin/npsa_automat.pl.page=/NPSA/npsa_sopma.html). The online ConSurf server (http://consurf.tau.ac.il) computed protein surface conservation, and SWISS-MODEL (https://swissmodel.expasy.org/) was used to estimate the three-dimensional structure. With the use of MODLOOP, the disorderly loops were refined. For the modeled structure’s energy minimization, the SWISS-PDB viewer was used. The stereochemical quality of the minimized model was evaluated using many SAVES server applications, including PROCHECK, Molprobity, VERIFY3D, ERRAT ProQ, and ProSA (http://nihserver.mbi.-ucla.edu/SAVES/). The LuxS protein’s three-dimensional structure was then edited using Pymol software (version 1.7.4 Schrödinger, LLC., http://pymol.org/), which also labeled the relevant amino acid positions that had been screened, and further investigated on its amino acid conservation, protein structure, and function.
Molecular Docking
Initially, the BAI’s 2D structure was acquired from the PubChem database at https://pubchem.ncbi.nlm.nih.gov. The automated molecular docking simulations were performed using the molecular docking tool AutoDock 4.2. The receptor was saved in .pdbqt format, and Kollman charges (6.0) were added using AutoDock Tools. Gasteiger charges (−1.0002) and polar hydrogen atoms were added to BAI. Using AutoGrid4, a potential grid around the LuxS active site residues (ASP24, ARG25, LYS26, LYS27, LEU89, ARG66, ASN4, MSE3, LEU23, TYR95, THR26, ALA24, GLY25, VAL151, and LYS2) were created. Both the grid dimension and the center point coordinates are 57.75Å, with X = −21.07, Y = 18.735, and Z = 6.0. The 50 ligand poses were created using the Lamarckian genetic algorithm. To create the 9 ligand conformations in AutoDock Vina, configuration files with grid box parameters, receptor, and ligand files were used. The blind docking procedure was set up using a grid box that covered the entire protein structure. The molecular docking procedure was simulated using the docking software AutoDock Vina 1.1.2. Following these procedures, the relevant conformation scores and outcomes were chosen, and PyMOL (https://pymol.org/2/) and Discovery Studio 2020 were used to visualize them. LuxS-BAI complexes were simulated using molecular dynamics (MD) using the Sander module in Amber 12.16 On a Linux system running Ubuntu, MD simulation was performed using the Amber99sb-ildn force field in GROMACS. Using Amber99sb-ildn force field,17 the first phase of MD is the creation of topology and protein coordinates. The acypyp.py function in AmberTools generated the topology of BAI in the second step. The systems were solvated in a triclinic box with a protein-to-box edge distance of 1.2 nm using the TIP3P explicit water model. The two-step equilibrations were carried out for 100 ps at 300 K with a constant number of particles, volume, and temperature (NVT) and a constant number of particles, pressure, and temperature (NPT). Systems’ pressure and temperature were kept constant using the V-rescale temperature coupling method and the Berendsen pressure coupling approach, respectively. After all simulations were completed, we count how many protein complexes are successfully formed after all simulation trajectories are completed. Particle-mesh Ewald method18 was used to calculate the long-range electrostatic interactions. The SHAKE algorithm19 was used to fix all covalent bonds containing hydrogen atoms. Finally, the binding-free energy was calculated using the Molecular Mechanics Poisson-Boltzmann surface area (MM/PBSA) method,20 From the last 20 ns (80 to 100 ns) of molecular dynamics, the trajectory was obtained. To perform MMPBSA calculations of protein-ligand complexes, a total of 2000 snapshots taken at intervals of 10 ps across an interval of 80 to 100 ns were used. Structural conformation of the constructed models was displayed using PyMOL21.
Screening and Verification of the Highly Productive Signaling Molecule AI-2 in S. aureus
AI-2 Activity Bioassay
The AI-2 bioassays were carried out according to the procedure mentioned previously.22 All samples, along with positive and negative controls, were cultured overnight; centrifugation at 12,000 xg for 10 min was used to separate the cell-free supernatant (CFS) from each culture, which was then filtered using a Millipore filter with a 0.22-µm pore size (Millipore, Bedford, MA, USA). V. harveyi BB170 overnight culture was diluted (1:5000) on fresh AB medium. Freshly diluted V. harveyi BB170 was then mixed in various volume ratios with filtered culture supernatants, and fresh AB medium as blank control. Continued shaking during incubation, samples of the culture were taken at various time intervals for analysis.
RT-qPCR Analysis of the luxS Gene Expression Levels
The bacterial overnight cultures were subjected to centrifugation at 10,000 rpm for 10 min. Total RNA was extracted using the standard method with TRIzol reagent. RNA was treated with ABScript III RT Master Mix with gDNA remover (ABclonal, China) to remove contaminating DNA and to reverse transcribe the RNA into cDNA. In order to execute relative quantitative PCR (qPCR), 2 × University SYBR green Fast qPCR Mix (ABclonal, China) was used. The comparative Ct technique was used to calculate fold changes for the target genes using gyrB expression as a reference (primers are shown in Supplementary Table 1).
Effect of BAI on the Expression of the luxS Gene
RT-qPCR was utilized to identify transcription level variations of the gene (luxS) encoding a QS signal (Al-2) production enzyme (LuxS) in S. aureus to verify the effect of BAI on the QS system. Different concentrations of BAI (0, 128, 256, 512, and 1024 µg/mL) with S. aureus wld10 were incubated statically for 40 hours at 37 °C to allow for biofilm formation. In the final least, total RNA was isolated, and RT-qPCR was carried out as explained in RT-qPCR Analysis of the luxS Gene Expression Levels.
Construction of Plasmid for the Expression of Recombinant LuxS Protein and Purification of LuxS Protein Were Conducted
The luxS gene of S. aureus wld10 was amplified and introduced into prokaryotic expression vector pET-28a (+) in order to produce recombinant LuxS protein in a prokaryotic expression system. Sequencing was used to identify the recombinant plasmids pET-28a-luxS, and the successful ones were then transformed into E. coli Rosetta (DE3) cells and induced with 0.1 mM isopropyl-d-1-thiogalactopyranoside (IPTG) for 5 hours in SOC media to produce the soluble form of the fusion protein LuxS. SDS-PAGE was used to evaluate and identify the expressed recombinant protein, which was then stained with Coomassie blue. With the aid of a His-tagged protein purification kit from Sangon Biotech in China, the produced recombinant protein fused protein was cleaned. The rabbit vaccine was made with purified proteins. To verify that the LuxS protein had been integrated into the recombinant protein, Western blotting was used to examine the fusion protein with antibodies directed against the His tag.
Effect of BAI on the LuxS Protein
The effect of BAI on the secondary structure conformation of the LuxS protein was characterized by fluorescence spectrum and FTIR spectroscopy. The state of tryptophan and tyrosine residues in the LuxS with BAI was studied via fluorescence spectroscopy. Different concentrations of BAI (0, 128, 256, 512, and 1024 µg/mL) with Purified LuxS protein were co-cultured for 40 h at 37 °C. The fluorescence intensity of BAI binding to LuxS was measured with a fluorescence spectrophotometer (FL-970; Shanghai Prism, Shanghai, China).
FTIR was used to elucidate the effect of BAI on LuxS protein secondary structure. The secondary structure of the protein was determined by comparing amide I peaks (1700–1600 cm−1)23,24 in the absence and presence of BAI (128, 256, 512, and 1024 µg/mL). The experimental data were processed using OMNIC 8.2 and PeakFit 4.12.
In vitro Test
Susceptibility Testing and Growth Dynamics Assay
Minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC) were evaluated using the standard broth dilution method. The 100 μL of BAI was serially diluted two-fold in Mueller Hinton broth (MHB) in 96-well microtiter plates to obtain final concentrations ranging from 1 to 8192 μg/mL. The bacteria were then diluted to 106 CFU/mL for S. aureus and were seeded at 100 μL/well in a 96-well plate. MIC was defined as the activity of BAI at the lowest concentration completely inhibiting bacterial growth with no discernible turbidity.25 By transferring 20 µL from each clear MIC well onto Mueller–Hinton agar (MHA) plates, MBCs were identified. The MBC was established as the lowest concentration that, following a 24-hour incubation period at 37 °C, killed 99.9% of the final inocula. Each assay was carried out three times.26 All assays were done in triplicate. Reference strains of S. aureus ATCC29213 were used for quality control for antimicrobial susceptibility testing. The previously described approach was used to operate growth kinetics, with some minor improvements.27 S. aureus wld10 was cultured in 96-well plates at 5 × 104 CFU/100 μL with different BAI concentrations (0, 128 (1/8 MIC), 256 (1/4 MIC), and 512 (1/2 MIC) µg/mL) at 37 °C. Samples were taken every hour, serially diluted and plated onto MHA.
Transmission Electron Microscope (TEM) Was Used to Analyze S. aureus Treated with BAI
TEM was used to investigate the ultrastructural changes in S. aureus cells after BAI exposure. S. aureus suspension was incubated with a range of BAI concentrations (0, 128, and 512 µg/mL) for 40 h at 37 °C. After being separated by centrifugation, the bacteria were resuspended after being washed three times with PBS. After being submerged in the cleaned bacterial suspension for three minutes, the copper mesh was dried. The bacteria-filled copper mesh was then dyed for 15 minutes in a 3% (w/w) phosphotungstic acid solution before being dried. The product was then examined using a TEM (JSM-7001F, Japan).
Measurement of Biofilm Formation
Based on the previously published approach, the microtiter plate test was used to assess biofilm formation under static conditions.28 For the anti-biofilm assay, S. aureus wld10 was cultured in TSB medium for 12 hours at 37 °C, diluted with new TSB that had been added with 3% NaCl and 0.5% glucose (TSB-g), and then the bacterial concentration was adjusted to an OD 600 of 0.1. To generate drug concentrations ranging from 1 to 1024 µg/mL, BAI was dissolved in dimethyl sulfoxide (DMSO) and diluted with sterile TSB medium. One milliliter of the diluted BAI was then applied to each well of sterile 24-well polystyrene microtiter plates. The colonies were gently rinsed with PBS twice after the plates had been incubated at 37 °C for 40 hours. After that, they were fixed for 5 minutes in an ice-cold ethanol/acetone (1:1, v/v) solution and then stained for 30 minutes in a 0.1% crystal violet (Sigma-Aldrich GmbH, Munich, Germany) solution in water solution. After rinsing the excess dye with water, 95% ethanol was used to help the dye escape the biofilm. At a wavelength of 595 nm, OD values were recorded by a microplate reader. The lowest dose of BAI that manifested evident biofilm inhibition was designated as the minimal biofilm inhibitory concentration (MBIC).29
Confocal laser scanning microscopy (CLSM) and scanning electron microscopy (SEM) were used to study the production of biofilms. A 24-well plate with coverslips (MeVid, China) and serial dilutions of BAI (0, 128, and 512 µg/mL) was first filled with 100 µL of S. aureus (1 × 106 CFU/mL). To get rid of the planktonic and loosely bound cells, the biofilms were rinsed three times with sterile PBS after growing for 40 hours at 37 °C. Then, using a CLSM (Leica SP5, Germany), biofilms were stained for 30 min in the dark with SYTO 9 and PI. The zinc piece was soaked in 2.5% glutaraldehyde (v/v) at 4 °C for 5 hours, then dehydrated in graded ethanol (15 min for each grade) for visualization study by SEM. The SEM sample was then collected after being dried with sterile air.
Effect of BAI on the Composition of the Extracellular Matrix in S. aureus
The polysaccharide intercellular adhesin (PIA) and eDNA make up the majority of the extracellular matrix in the S. aureus biofilm. Fresh TSB with BAI (0, 128, and 512 µg/mL) and S. aureus wld10 were added to a 96-well plate, co-cultured for 16 hours to form a mature biofilm, then extracted using the Rice technique.30 Briefly, the supernatant was absorbed for centrifugation (4 °C, 18,000 rpm, 5 min) after the mature biofilm was re-suspended in TEN buffer. The supernatant was extracted after being combined with TE buffer and an organic reagent solution (phenol, chloroform, and isoamyl alcohol). The supernatant from the extract was re-extracted using chloroform and isoamyl alcohol after it had been centrifuged (4 °C, 12,000 rpm, 10 min). Thermo Fisher Scientific Inc., Waltham, Massachusetts, United States, NanoDrop One ultra-micro spectrophotometer was used to identify the release of eDNA.
Using the Congo red agar (CRA) method as described by Formosa-Dague et al,31 biofilm formation was phenotypically evaluated. The CRA medium was streaked with each co-culture, which was then incubated for 48 hours at room temperature after 24 hours at 37 °C. A four-color reference scale ranging from red to black was used to determine the color of the colonies. Black colonies denoted a favorable outcome, while pink or purple colonies were thought to be unfavorable. Almost-black colonies indicated an undecided result.
Effect of BAI on the Surface Hydrophobicity of S. aureus
Minor adjustments were made to the method used in a previous research32 to assess the hydrophobicity of the cell surface. Briefly, S. aureus wld10 was incubated with different concentrations of BAI (0, 128, 256, 512, and 1024 µg/mL), rinsed with PBS, and the bacterial suspensions were adjusted to an OD 600 of 1.0 (H0). Hexadecane (0.4 mL) and bacterial suspension (2 mL) were combined before being incubated at room temperature for one hour. The aqueous phase’s OD 600 was then determined to be H. The following formula was used to calculate the hydrophobicity of bacterial surfaces (H%):
The Effects of BAI on the Expression Levels of S. aureus Biofilm-Related Genes
To allow for the development of biofilms, S. aureus wld10 was incubated statically for 40 hours at 37 °C with various doses of BAI (0, 128, 256, 512, and 1024 µg/mL). The total RNA was then isolated in order to perform qRT-PCR analysis on the expression of genes involved in biofilms. The reaction system for the PCRs and cycling conditions were the same as described in RT-qPCR Analysis of the luxS Gene Expression Levels and Supplementary Table 1 shows the PCR primers.
Effects of BAI on Mature Biofilms of S. aureus
On accordance with RT-qPCR Analysis of the luxS Gene Expression Levels, sterile glass coverslips were positioned on 24-well tissue culture plates, where biofilms were produced. The medium was aspirated after the formation of biofilms, and non-adherent cells were then eliminated by washing the biofilms in PBS. Samples were added BAI (0, 128, 256, 512, and 1024 µg/mL), then incubated at 37 °C. Non-adherent bacteria were eliminated after 12 hours of incubation, and wells were then cleaned with PBS, fixed with 70% methanol for 15 minutes, and stained for 10 minutes with 0.1% crystal violet. PBS-washed, then air-dried. Two hundred microliters of glacial acetic acid at 30% were added and thoroughly mixed. A microplate reader was used to measure the absorbance at 595 nm.
Effect of BAI on the Activity of AI-2 of S. aureus
The effects of BAI on the AI-2 activity in the S. aureus biofilm were next assessed using a bioassay for AI-2 activity. To allow for biofilm formation, S. aureus wld10 was incubated statically for 40 hours at 37 °C with various doses of BAI (0, 128, and 512 µg/mL). Following a 10-minute, 10,000 rpm centrifugation of the samples, the supernatant was gathered and filtered to produce CFS. The CFS sample was introduced to the diluted BB170 culture at a ratio of 50:1 (v/v) after the reporter strain V. harveyi BB170 had been diluted 1:5000 with fresh AB medium. In order to identify AI-2 activity, the mixture was incubated at 30 °C for 5 hours with 180 rpm of agitation. Two hundred-microliter aliquots were then put to empty 96-well plates.
Statistical Analysis
Every experiment was performed three times. The mean and standard deviation (SD) for each piece of data were displayed. One-way analysis of variance (ANOVA) was used to examine the data, and a post hoc test was performed using the SPSS 20.0 program (IBM Corp., Armonk, NY, USA). It was deemed significant at P < 0.05.
Results and Discussion
LuxS Protein Molecular Characterisation
Studies have shown that antagonizing agr signaling by preventing AIP ligation of the AgrC receptor,33 blocking the response regulator AgrA,34 or interfering target of RNAIII-activating peptide35 has a significant preventative effect on biofilm-associated infections by S. aureus. Additionally, methicillin-resistant S. aureus (MRSA) pathogenicity is modulated by the global regulator sarA through control of key virulence components (such as adhesins and toxins) and biofilm development,36 and to search for effective inhibitors has become an active area of research. However, it is thought that the LuxS/AI-2 QS system facilitates interspecies communication in polymicrobial communities, and this regimen can be considered as a promising option for S. aureus infections. The protease LuxS involved in the synthesis of the signal molecule AI-2 is the central component of the LuxS/AI-2 system. In order to investigate the therapeutic possibility of treating S. aureus infections by disrupting the LuxS/AI-2 pathway, we conducted exploratory research on the LuxS protein.
From the NCBI database, a LuxS amino acid sequence from S. aureus was obtained. LuxS from S. aureus underwent multiple sequence alignment (MSA) with response regulators from several species. ClustalW was used to do MSA, and the illustration in Supplementary Figure 1 was produced using ESPript 3. The red color represents the conserved residues. The ADQ01866.1 belongs to autoinducer-2 production protein LuxS from Bifidobacterium longum subsp. longum BBMN68; AFC91867.1 from Streptococcus pneumoniae; 1J6X from Helicobacter pylori, and it revealed that the highest sequence similarity between LuxS and LuxS of Helicobacter pylori is 67.55%.
As shown in Supplementary Figure 2-A, the Ramachandran plot created by PROCHECK reveals that 85.8% of amino acids are in the preferred region, 14.2% are in the allowed region, and 0.0% and 0.0% are, respectively, in the generously permitted and forbidden sectors. MolProbity’s anticipated Ramachandran plot showed 91.1% of the residues were in the preferred region, 8.3% were in the permitted region, and 0.6% were in the forbidden region. Levitt-Gerstein (LG) score of 5.184 projected by ProQ for LuxS model demonstrates that the model is very good and dependable. ProSA predicted a Z-score of −6.37 for the improved LuxS model, as shown in Supplementary Figure 2-B. According to the ERRAT plot, the model’s quality factor was 93.4307%, as shown in Supplementary Figure 2-C. According to Supplementary Figure 2-D, VERIFY3D revealed that the average 3D-1D score for 80% of amino acid residues is more than or equal to 0.1. As Figure 1A showed that LuxS are dimeric proteins, which can associate to form dimeric assemblies. For the monomeric molecules (Figure 1B), alpha-helix (α-helix), beta-sheet (β-sheet), beta-turn (β-turn) and random coil content were, respectively, 41.03%, 19.87%, 1.28%, and 37.82%. Through conservativeness analysis of the LuxS protein and projection of conserved assignments onto tertiary structures according to color, the relationship between conserved amino acids and structural stability and functional performance was examined (Figure 1C). The investigation showed that the regions ASN5-PHE9, LEU11-THR15, ALA18-ARG22, HIS56-ALA63, SER77-VAL87, VAL115-ASN119, GLN122-ALA126, and SER128-LEU131 had a high degree of conservation. In terms of amino acid accessibility in solvent, 61.36% of amino acids in all examined proteins were projected to be exposed in solvent, while the percentage of buried amino acids in solvent ranged from 38.64%. Inside are several residues VAL6, PHE9, VAL16, ALA18, HIS56, SER57, ALA63, ASP75, PRO78, MET79, CYS81, GLY84, PHE85, TYR86, CYS123, LEU131, and ALA134 that stabilize the extremely conserved character of the particular protein shape. The surface-located, highly conserved amino acids are essential for biological processes, mainly MET1, GLU7, SER8, ASP12, HIS13, PRO19, ARG22, ASP33, LYS37, ASP39, ARG41, PRO45, ASN46, MET52, GLU59, HIS60, ARG67, GLY80, GLN82, THR83, ASN91, PRO116, ALA117, GLY124, TRP125, HIS129, and SER130. The investigation of the secondary and tertiary structures revealed that LuxS is structurally stable and has a highly conserved amino acid composition. These highly conserved amino acids also form a binding cavity that is ideal for the binding of other molecules.
 |
Figure 1 Structural characteristics of LuxS protein. (A) The dimer structure of LuxS. (B) The monomer structure of LuxS. (C) The analysis of LuxS protein conservativeness analysis. |
Prediction of the Binding Model Between LuxS and BAI
Numerous organic substances exhibit strong antibacterial action against S. aureus. Few natural substances have the capacity to kill drug-resistant S. aureus bacteria. In recent years, there has been a significant growth in the use of phytomedicines to treat bacterial illnesses that are resistant to conventional treatments. The plant’s phytochemical components have demonstrated strong antibacterial action against MRSA. For instance, tannins deactivate envelope proteins and enzymes to lyse the bacteria, while flavonoids in plants form a compound with the bacterial cell wall, soluble protein, and extracellular substrates. In particular, sensible selectivity is necessary when combining antivirulence drugs with antibiotics because they may reduce the effectiveness of the latter.37 For instance, resveratrol lessens the fatal effects of oxacillin by reducing the amount of ROS that S. aureus produces.37 Computer-aided design serves as a screening platform for novel inhibitors, and statistics helps in the discovery of innovative drug development uses. Aloe vera, Guava, Neem, Pomegranate, and Tea herbal extracts can be utilized as treatments against MRSA infections, according to in-silico and in vitro research.38,39 Baicalin (BAI) is a flavonoid compound extracted from Scutellaria roots, which has been shown to have anti-biofilm,40 antibacterial,41 antiviral,42 and immune-stimulating activities.43
By focusing on its QS systems, BAI was discovered to successfully prevent P. aeruginosa from forming biofilms and improve the growth inhibitory actions of antibiotics.40 Additionally, by blocking the MsrA efflux pump in ARSS,44 BAI can prevent the production of biofilms and prolong the survival rate of infected mice.45 Through literature reviews, research and brainstorming, this study proposes BAI as a potential candidate ligand for LuxS protein.
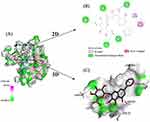
We carried out a virtual docking experiment to better comprehend how BAI and LuxS interact. The three-dimensional structure, as seen in Figure 2, demonstrates that BAI interacts with the LuxS active site and creates protein–ligand interactions with the critical LuxS amino acid residues. The two-dimensional interaction map specifically demonstrates that BAI forms hydrogen bonds with GLU7, ASN10, ANG22, LEU23, and ALA24, respectively. In addition, the amino acid residues VAL151 contributed to the interaction through Pi-Alkyl bonding; PHE152 formed Pi-Pi T-shaped bonds with BAI, respectively, facilitating the stabilization of the BAI molecule in the binding cavity by a more balanced force. In the optimal binding conformation, the binding free energy of BAI to protein LuxS was −6.9 kcal/mol. BAI is likely, therefore, to have a strong effect on protein LuxS function.
The docked complex was then subjected to a molecular-dynamics simulation of 100 ns. The RMSD, which measured the stability of the LuxS-BAI complex, is depicted in Figure 3A; after 30 ns, the complex reached equilibrium, and the systems remained stable for the entire 100 ns of the molecular simulation. These findings primarily validate the stability and reliability of the LuxS-BAI complex. RMSF was further calculated to evaluate the flexibility of the residues. The RMSF values for LuxS-BAI complex showed less RMS fluctuation, with respect to the RMSF values of LuxS (Figure 3B), and the bulk of the protein residues in the complex had RMSF values that were lower than 3Å. This is also in coherence with analysis obtained by the radius of gyration (Rg) in Figure 3C. The compactness of the protein structure is shown by the radius of gyration. Figure 3C shows that the complex structure had approximately identical Rg values from 0 to 100 ns, indicating that the LuxS-BAI complex structure was nearly stable. Binding with more hydrogen bonds was considered more stable, the number of hydrogen bonds that were generated during the simulation (Figure 3D) stayed between 2 and 5, with a maximum of 8 hydrogen bonds.
 |
Figure 3 Molecular dynamics simulation of the complex of LuxS-BAI. (A) RMSD, (B) RMS fluctuation, (C) Radius of gyration, (D) Hydrogen bonds. |
The MMPBSA module determined the LuxS-BAI complex’s binding free energy. The van der Waals energy, electrostatic energy, polar solvation energy, and SASA energy make up the binding free energy. According to Table 1, the binding free energy was −31.716 ± 15.050 kcal/mol of complex identified, with van der Waals energy and electrostatic energy making up the majority of the binding free energy overall. The participation of each LuxS amino acid residue in the calculated binding free energy was then calculated by dividing the overall binding energy of the LuxS-BAI complex into share energies for each amino acid residue. The successful results show the essential residues that positively influence the binding of the LuxS-BAI complex. LEU23 (−9.5336 kJ/moL), TYR95 (−7.5913 kJ/moL), THR26 (1.4608 kJ/mol), ALA24 (1.3526 kJ/mol), GLY25 (0.923 kJ/mol), and VAL151 (−0.8991 kJ/moL) were among the substances with the highest binding values (Figure 4).
 |
Table 1 Total Binding Free Energies Based on MM-PBSA and Their Component Energies for the Chosen Bioactive Molecules |
Screening the Strains with AI-2 Activity
AI-2 is produced by many bacteria and is used for communication both within and between species, and studies on AI-2 signal molecules are required to provide insights into LuxS/AI-2-related lifestyle of S. aureus. Therefore, the bioassays of AI-2 activity in S. aureus were optimized and performed. From Figure 5A, throughout the 8 h fermentation, all fluorescence locations showed a drop in fluorescence intensity from their original values. However, the fluorescence intensities increased during the 5 h fermentation in the samples of negative control, positive control, and 1:50 sample addition group. Since there was only diluted BB170 in the negative control, when the incubation time reached 5 h, the concentration of AI-2 produced by the indicator bacteria reached the time threshold for inducing its luminescence, and the fluorescence intensity began to rise. The sample with a sample ratio of 1:100 did not turn to rise until 7 h later, which may be due to the fact that the supernatant to be measured was added less, and the time to reach the excitation intensity of reported strain luminescence was prolonged. For the 1:10 system to be tested, the light intensity continued to decrease. It may be that more supernatant was added, and the metabolites inhibited the growth of the reporter strain. The concentration of the reporter strain was too low, and the signal receptor protein could not activate the expression of the luminescent gene. Therefore, the sample addition ratio was chosen to be 1:50, and the fluorescence intensity value at 5 h was used as a reference to calculate the relative fluorescence intensity, indicating the activity of AI-2.
According to the known reports, the production of AI-2 by most microbial strains peaked at the end of the logarithmic period, and then decreased after entering the stable period.46 As shown in Figure 5B, the strain entered the exponential phase from 3 h, reached the end of the exponential phase at 16 h, and then entered the plateau phase, and the OD600nm reached the maximum at the end of the exponential phase. OD600nm tended to be stable at 16 h, so the experiment selected 16 h culture for AI-2 activity detection. The results showed that within 16 hours, the cell density of S. aureus gradually increased with the relative fluorescence intensity of AI-2, and there was a significant positive correlation. After 16 h, the cell density remained relatively stable, while the relative fluorescence intensity began to decrease slowly. It may be that a large amount of AI-2 is reabsorbed by the bacteria or attached to the surface of the cell membrane to regulate some important physiological functions in the stationary phase.
In Figure 5C, the relative fluorescence intensity of the 4 strains of S. aureus was higher than that of the negative control, and as compared to other strains, wld10 was much greater. It has been demonstrated that the AI-2 biosynthesis pathway is largely conserved among bacterial species. S-ribosylcysteinase, also known as LuxS protein, is a key enzyme involved in the biosynthesis of AI-2, and genomic analysis revealed that more than 55 bacterial species contained the homoconserved sequence of the AI-2 synthetase LuxS.47 Biological methods were used to test the key luxS gene and the fluorescence intensity of AI-2. RT-PCR detection was performed on the 4 strains, and the results are shown in Figure 5D. The relative expression of luxS gene in strain wld10 was significantly higher than that of the other three strains, and it was positively correlated with the signal intensity of AI-2. It also shows that the combination of fluorescence detection and RT-PCR detection is a more accurate method to detect QS signal molecule AI-2. Therefore, the strain wld10 was selected for subsequent experiments.
Effect of BAI on the Conformation of LuxS Protein
Tryptophan (Trp), tyrosine (Tyr), and phenylalanine (Phe) residues are generally what cause the intrinsic fluorescence that is present in the majority of proteins. The hydrophobic Trp residues and hydrophilic Tyr residues were the main sources of endogenous fluorescence when the excitation wavelength was 280 nm.48 Fluorescence emission spectrum of LuxS is shown in Figure 6A, with a strong emission peak at 286 nm. With the increase of BAI concentration, the endogenous fluorescence of LuxS gradually weakened, and it shows that BAI interacts with LuxS and quenches the endogenous fluorescence of LuxS. The binding of BAI changed the structural conformation of LuxS, as well as the microenvironment of Trp and Tyr.
 |
Figure 6 Effect of BAI on LuxS protein conformation as determined by (A) fluorescence spectroscopy and (B) Fourier analysis. |
Infrared spectroscopy was used to further detect the structural changes of LuxS protein and the interaction between LuxS-BAI. By curve-fitting the amide I band using the peak fitting module of the Origin program (OriginLab Corporation, Northampton, MA, USA), the secondary structure of LuxS was quantitatively analyzed. The intensity of the amide I band of the LuxS protein decreased in the presence of BAI (Figure 6B), indicating that BAI changed the secondary structure of LuxS protein. It may be that the oxygen atoms and hydroxyl groups on the BAI form complexes with the C=O and C-N groups on the LuxS protein through hydrophobic interactions, resulting in rearrangement of the peptide chains of the LuxS protein and ultimately changing the secondary structure of the LuxS protein.49 We calculated the LuxS protein’s secondary structure content percentage (Table 2). As the addition of BAI was increased, β-turn, β-pleated sheet, and random coiling were increased, while α-helical was decreased. However, the structure still dominates. It may be that BAI binds to the total hydrophobic amino acid region of the α-helix structure of LuxS protein, which causes the protein molecule to unfold and change its spatial structure.
 |
Table 2 The Amide I Band Fitted the Secondary Structure of LuxS Protein Treated with Different BAI Concentrations |
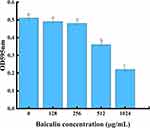
Bacteriostatic Activity of BAI and Its Effect on the Growth of S. aureus
As shown in Table 3, against S. aureus, the MIC and MBC for BAI were 1024 and 4096 µg/mL, respectively. Then, we investigated the effect of subinhibitory concentrations of BAI on the growth of S. aureus, it was found that BAI had slight effect on the growth of S. aureus at the BAI concentrations were 128 and 512 µg/mL (Figure 7). The result was consistent with the findings in Wang et al50 report and demonstrated that BAI exhibited slight anti-staphylococcal activity (MIC > 1024 µg/mL).
 |
Table 3 Antibacterial Activity Assays Against Staphylococcus aureus |
 |
Figure 7 Staphylococcus aureus growth curve with different concentrations of baicalin. |
The Effect of BAI on Bacterial Cell Morphology Was Observed with TEM
The morphology of S. aureus cells following exposure to BAI was compared using TEM. The untreated S. aureus cells maintained their plump, globose, and integrity, and the wall and membrane were intact, appearing as clearly defined cell membranes, as illustrated in Figure 8A and B. Significant alterations were seen in the bacterial cell wall following treatment with BAI at 128 µg/mL; the surface area of the cell wall became rough, and the boundary between the cell wall and cell membrane became hazy (Figure 8C). When treated with 512 µg/mL BAI, cell walls have continued to thicken and the surface exhibits obvious roughness (Figure 8D). The combined growth curves and TEM results demonstrate that these BAI-treated cells are still able to form division septa, S. aureus wld10 was not subjected to growth pressure from BAI subMIC.
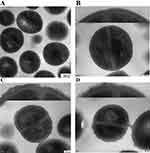 |
Figure 8 TEM images of S. aureus treated with BAI. (A and B) The TEM images of S. aureus without BAI (control group). (C) The BAI concentration was 128 µg/mL. (D) The BAI concentration was 512 µg/mL. |
The Influence on the Biofilm Formation Ability of BAI
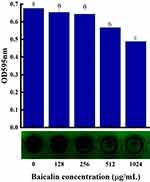
Planktonic cells interact with surfaces to create a temporary initial adhesion, which is the precursor to biofilm formation. At this stage, the presence of appendages and related proteins on the surface of the cells controls the capacity of bacteria to adhere to a surface. The irreversible attachment starts once the initial electrostatic repulsion between the cell and surface is overcome. The secretion of polysaccharides and the creation of adhesins serve as the intermediaries for this attachment.51 Therefore, reducing adhesion to surfaces may be an effective way to mitigate biofilm formation. S. aureus biofilm formation was significantly inhibited by BAI in a concentration-dependent manner, with more than 50% inhibition at 1024 µg/mL (Figure 9), and the MBIC was identified as 1024 µg/mL. The results of the crystalline violet staining demonstrated that BAI had a significant impact on the formation of S. aureus' biofilm.
The CLSM demonstrated that the BAI was successful in preventing the growth of S. aureus biofilms (Figure 10A). The test biofilm (Figure 10A1) was stained red to indicate dead S. aureus cells, while the control biofilm (Figure 10A1) was stained green to indicate live S. aureus associated cells. CLSM images revealed that BAI at concentrations of 128 and 512 µg/mL effectively inhibited the biofilm of S. aureus.
SEM was used to investigate how BAI affected the composition of the S. aureus biofilm (Figure 10B). In the control culture, S. aureus displayed a large-scale, three-dimensional structure resembling a coral with a high degree of stacking, according to SEM images (Figure 10B1). The quantity of bacteria adhering to the surface steadily dropped as the concentration of BAI rose, and the biofilm itself became thinner, looser, and less uniform (Figures 10B2 and 10B3). SEM studies therefore indicated that BAI breaks down the stereostructure of S. aureus biofilms.
Effect of BAI on the Extracellular Matrix Composition of Biofilm Communities
The biofilm is a membrane-like structure made up of extracellular macromolecules encasing the bacteria, as well as the hydrated matrix released during the growth of the bacteria that cling to the surface of living or nonliving things. The structural integrity of biofilms depends on the extracellular matrix, which is made up of PIA, eDNA, and a number of surface proteins. For bacteria to create biofilms with a sophisticated three-dimensional structure, PIA is unquestionably a necessary skeleton.31 The interaction of bacterial cells and the adherence of S. aureus to medical devices are stabilized in part by PIA. The findings shown in Figure 11A demonstrate that the PIA synthesis was gradually suppressed as the BAI concentration rose (seen as decreased color intensity of the black colonies).
In bacterial biofilms, eDNA promotes adhesion and aids in preserving the biofilm’s structural integrity. The eDNA content in the S. aureus biofilm dramatically decreased after BAI treatment, as shown in Figure 11B, and these effects were dose-dependent. Higher BAI concentrations resulted in a greater decrease of eDNA.
Hydrophobicity is also an important factor affecting the adsorption capacity. Particularly, it is known that crucial bacterial adhesion factors occur more frequently on hydrophobic surfaces, which encourages bacterial colonization.52 Table 4 shows that the addition of BAI caused lower surface hydrophobicity and the surface hydrophobicity reduced in a dose-dependent manner. It was deduced that BAI can alter cell surface hydrophobicity in ways that can inhibit bacterial adhesion.
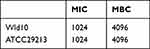 |
Table 4 Effect of BAI on Cell Surface Hydrophobicity of S. aureus |
Effect of BAI on S. aureus Biofilm-Related Genes
Bacterial surface adhesion and co-aggregation are essential for the development of biofilm communities. Therefore, this paper explores the effect of baicalin on the expression of several important adhesion-related genes in S. aureus. The results of RT-qPCR are shown in Figure 12. The expression levels of clfA, fnbA, icaD, icaA and clfB genes of S. aureus decreased after BAI treatment. Agglutination factor is one of the most important adhesion factors for S. aureus to adhere to host cells, and it is divided into clfA and clfB, and adhesion factors mainly adhere to fibrinogen and fibrin, and the mammary gland, subcutaneous, vascular, endocardial infection.53 The primary regulating mechanism for the development of MRSA biofilms is PIA (also known as polyacetyl glucosamine) dependency. The ica operon has four genes (icaADBC) and a divergently transcribed repressor (icaR) that work together to produce PIA. One of the key adhesion factors for S. aureus to stick to host cells is Fnbp. It can facilitate the attachment of S. aureus to Fn on the cell surface, causing bacteria to adhere to host cell surfaces and encouraging bacterial penetration into host tissues.54 Studies by Aart Lammers et al55 have shown that S. aureus strains lacking expression of fibronectin-binding protein cannot agglutinate cells and have significantly lower capacity to cling to and penetrate cells of the bovine mammary gland. Through the double PCR detection of the FnbA and FnbB genes of S. aureus in dairy cows, it was found that more than 95% of S. aureus had the adhesin gene FnbA, while only 10% of the FnbB gene,56 and it was confirmed that FnbA played an important role in the process of S. aureus invading the mammary gland of dairy cows.
 |
Figure 12 Transcription analysis of biofilm formation related genes. Notes: *Means difference P < 0.05, **Means difference P < 0.01, ***Means P < 0.001, ****Means P < 0.0001. |
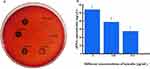
Effect of BAI on the Biofilm Clearance of S. aureus
Figure 13 shows the clearance effect of BAI on S. aureus biofilm. There were a large number of bacteria in the control group and significantly reduced the bacteria after adding the BAI. Studies have shown that many antimicrobials, including the antifungal fluconazole, can inhibit biofilm production. However, it is well established that it is not active against preformed biofilms.57 Compared to free-living bacterial cells, biofilms are more resistant to unfavorable circumstances. BAI has an inhibitory effect on biofilm development and a disruptive potential on pre-formed biofilms. Thus, it has some advantages over most anti-biofilm drugs.
Effects of BAI on the luxS Gene and AI-2 Activities in Biofilm
The main enzyme in the synthesis of Al-2 is the LuxS protein. The very conservative luxS coding genes are found in both Gram-positive and -negative bacteria. Al-2 is a byproduct of the methyl cycle in its entirety. The LuxS protein is a crucial enzyme for the synthesis of Al-2 and is important for the methyl cycle’s metabolism. However, in order to assess the impact of BAI on the expression of the luxS gene, we conducted exploratory study on the LuxS protein. The outcomes demonstrated that BAI could reduce the luxS gene expression (Figure 14A). After successfully producing recombinant LuxS protein (Supplementary Figure 3), we quantified LuxS at the protein level using a Western blot, just as it had been done earlier to validate the LuxS protein regulation with BAI. In contrast to the control circumstances, BAI caused a decrease in LuxS expression (Supplementary Figure 4), and it was discovered that BAI had an inhibitory effect on AI-2 activity in S. aureus biofilms (Figure 14B). According to earlier studies, luxS stimulates the creation of the AI-2, which may help bacteria build biofilms more effectively.58 In this work, the presence of BAI reduced AI-2 activity and luxS expression. It suggested that BAI might act as the S. aureus LuxS/AI-2 system’s QS inhibitor.
Conclusion
Not only does BAI inhibit biofilm formation, it also has properties to reduce the preformed mature biofilms of S. aureus; BAI has a strong interaction with the key active site residues of LuxS through hydrogen bonds and hydrophobic interactions. These interactions may help to constrain the conformational changes of the LuxS monomer and prevent its dimerization. The LuxS activity might be constrained by the structural requirements of the protein–protein interactions needed for dimerization. To help in the development of more effective antimicrobials, further research must be done to better understand the antimicrobial mechanism of these polymers.
Credit Author Statement
Conceptualization and supervision, Guiqin Wang; Software and visualization, Yanni Mao and Panpan Liu; Investigation, Haorong Chen and Yuxia Wang; Project administration, Caixia li; Writing—original draft preparation, Yanni Mao; Writing—review and editing, Guiqin Wang. All authors contributed significantly to the work that was published, whether it be in the ideation, study design, implementation, data collection, analysis, and interpretation, or in all of these areas. They also all participated in writing, revising, or critically evaluating the article, gave their final approval for the version that would be published, agreed on the journal to which the article would be submitted, and agreed to be responsible for all aspects of the work.
Data Sharing Statement
The corresponding author can provide the data that back up the study’s findings upon reasonable request.
Acknowledgments
We acknowledge the assistance of all the instructors and students in conducting our experiment, as well as the contributions of all the writers of this paper. We appreciate the gifts of V. harveyi BB152 and V. harveyi BB170 from Professors Yongjie Liu of Nanjing Agricultural University and Prof. Xiangan Han of Shanghai Veterinary Research Institute, Chinese Academy of Agricultural Sciences. Additionally, I’d like to thank MJEditor (www.mjeditor.com) for providing English editing services during the production of this manuscript as well as all the reviewers who took part in the review.
Funding
This work was funded by the Natural Science Foundation of China (Grant Nos.32160852).
Disclosure
The study’s authors affirm that there were no financial or commercial ties that might be viewed as having a potential conflict of interest. The authors report that they have no competing interests.
References
1. Ruegg PL. A 100-Year Review: mastitis detection, management, and prevention. J Dairy Sci. 2017;100:10381–10397. doi:10.3168/jds.2017-13023
2. Gogoi-Tiwari J, Williams V, Waryah CB, et al. Intramammary immunization of pregnant mice with staphylococcal protein a reduces the post-challenge mammary gland bacterial load but not pathology. PLoS One. 2016;11:e148383. doi:10.1371/journal.pone.0148383
3. Di Domenico EG, Cavallo I, Capitanio B, et al. Staphylococcus aureus and the cutaneous microbiota biofilms in the pathogenesis of atopic dermatitis. Microorganisms. 2019;7:301. doi:10.3390/microorganisms7090301
4. Dotto C, Lombarte SA, Ledesma M, et al. Salicylic acid stabilizes Staphylococcus aureus biofilm by impairing the agr quorum-sensing system. Sci Rep. 2021;11:2953. doi:10.1038/s41598-021-82308-y
5. Fan Q, Zuo J, Wang H, Grenier D, Yi L, Wang Y. Contribution of quorum sensing to virulence and antibiotic resistance in zoonotic bacteria. Biotechnol Adv. 2022;59:107965. doi:10.1016/j.biotechadv.2022.107965
6. Ma R, Qiu S, Jiang Q, et al. AI-2 quorum sensing negatively regulates rbf expression and biofilm formation in Staphylococcus aureus. Int J Med Microbiol. 2017;307:257–267. doi:10.1016/j.ijmm.2017.03.003
7. Rajasree K, Fasim A, Gopal B. Conformational features of the Staphylococcus aureus Agra-promoter interactions rationalize quorum-sensing triggered gene expression. Biochem Biophys Rep. 2016;6:124–134. doi:10.1016/j.bbrep.2016.03.012
8. Chen J, Zhou H, Huang J, Zhang R, Rao X. Virulence alterations in staphylococcus aureus upon treatment with the sub-inhibitory concentrations of antibiotics. J Adv Res. 2021;31:165–175. doi:10.1016/j.jare.2021.01.008
9. Wang Y, Wang Y, Sun L, Grenier D, Yi L. The LuxS/AI-2 system of Streptococcus suis. Appl Microbiol Biotechnol. 2018;102:7231–7238. doi:10.1007/s00253-018-9170-7
10. Pruteanu M, Hernandez LJ, Stach T, Hengge R. Common plant flavonoids prevent the assembly of amyloid curli fibres and can interfere with bacterial biofilm formation. Environ Microbiol. 2020;22:5280–5299. doi:10.1111/1462-2920.15216
11. Cui X, Zhang J, Sun Y, et al. Synergistic antibacterial activity of baicalin and EDTA in combination with colistin against colistin-resistant Salmonella. Poultry Sci. 2022;10:2346.
12. Abouelhassan Y, Garrison AT, Burch GM, Wong W, Norwood VT, Huigens RR. Discovery of quinoline small molecules with potent dispersal activity against methicillin-resistant Staphylococcus aureus and Staphylococcus epidermidis biofilms using a scaffold hopping strategy. Bioorg Med Chem Lett. 2014;24:5076–5080. doi:10.1016/j.bmcl.2014.09.009
13. DeKeersmaecker S, Vanderleyden J. Constraints on detection of autoinducer-2 (AI-2) signalling molecules using Vibrio harveyi as a reporter. Microbiology. 2003;149:1953–1956. doi:10.1099/mic.0.C0117-0
14. Bassler BL, Greenberg EP, Stevens AM. Cross-species induction of luminescence in the quorum-sensing bacterium Vibrio harveyi. J Bacteriol. 1997;179:4043–4045. doi:10.1128/jb.179.12.4043-4045.1997
15. Surette MG, Miller MB, Bassler BL. Quorum sensing in Escherichia coli, Salmonella typhimurium, and Vibrio harveyi: a new family of genes responsible for autoinducer production. Proc Natl Acad Sci U S A. 1999;96:1639–1644. doi:10.1073/pnas.96.4.1639
16. Case DA, Cheatham TR, Darden T, et al. The Amber biomolecular simulation programs. J Comput Chem. 2005;26:1668–1688. doi:10.1002/jcc.20290
17. Curry HB. The method of steepest descent for nonlinear minimization problems. Quart Appl Math. 1994;2:258–261. doi:10.1090/qam/10667
18. Anandakrishnan R, Onufriev AV. An N log N approximation based on the natural organization of biomolecules for speeding up the computation of long range interactions. J Comput Chem. 2010;31:691–706. doi:10.1002/jcc.21357
19. Wang T, Wang CF. On the derivation of equations of motion. J Soc Industr Appl Mathe. 1965;13:487–492. doi:10.1137/0113030
20. Sun H, Li Y, Tian S, Xu L, Hou T. Assessing the performance of MM/PBSA and MM/GBSA methods. Phys Chem. 2014;16:16719–16729. doi:10.1039/C4CP01388C
21. Makarewicz T, Kazmierkiewicz R. Improvements in GROMACS plugin for PyMOL including implicit solvent simulations and displaying results of PCA analysis. J Mol Model. 2016;22:109. doi:10.1007/s00894-016-2982-4
22. Lee KW, Jie H, Kim S, Baek MG, Yi H, Kim KS. Effect of luxS encoding a synthase of quorum-sensing signal molecule AI-2 of Vibrio vulnificus on mouse gut microbiome. Appl Microbiol Biotechnol. 2022;106:3721–3734. doi:10.1007/s00253-022-11935-w
23. Liu XH, Xi PX, Chen FJ, Xu ZH, Zeng ZZ. Spectroscopic studies on binding of 1-phenyl-3-(coumarin-6-yl)sulfonylurea to bovine serum albumin. J Photochem Photobiol B. 2008;92:98–102. doi:10.1016/j.jphotobiol.2008.04.008
24. Naik PN, Chimatadar SA, Nandibewoor ST. Interaction between a potent corticosteroid drug - dexamethasone with bovine serum albumin and human serum albumin: a fluorescence quenching and Fourier transformation infrared spectroscopy study. J Photochem Photobiol B. 2010;100:147–159. doi:10.1016/j.jphotobiol.2010.05.014
25. Belanger CR, Hancock R. Testing physiologically relevant conditions in minimal inhibitory concentration assays. Nat Protoc. 2021;16:3761–3774. doi:10.1038/s41596-021-00572-8
26. Jarkhi A, Lee AHC, Sun Z, et al. Antimicrobial effects of L-Chg10-teixobactin against Enterococcus faecalis in vitro. Microorganisms. 2022;10:1099. doi:10.3390/microorganisms10061099
27. Close Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing. M100.
28. Champion M, Portier E, Karine V. Anti-biofilm activity of a hyaluronan-like Exopolysaccharide from the marine vibrio MO245 against pathogenic bacteria. Mar Drugs. 2022;20:728. doi:10.3390/md20110728
29. Ortiz-Gomez V, Rodriguez-Ramos VD, Maldonado-Hernandez R, Gonzalez-Feliciano JA, Nicolau E. Antimicrobial polymer-peptide conjugates based on maximin H5 and PEG to prevent biofouling of E. coli and P. aeruginosa. ACS Appl Mater Interfaces. 2020;12:46991–47001. doi:10.1021/acsami.0c13492
30. Rice KC, Mann EE, Endres JL, et al. The cidA murein hydrolase regulator contributes to DNA release and biofilm development in Staphylococcus aureus. Proc Natl Acad Sci U S A. 2007;104:8113–8118. doi:10.1073/pnas.0610226104
31. Formosa-Dague C, Feuillie C, Beaussart A, et al. Sticky matrix: adhesion mechanism of the staphylococcal polysaccharide intercellular adhesin. ACS Nano. 2016;10:3443–3452. doi:10.1021/acsnano.5b07515
32. Salaheen S, Jaiswal E, Joo J, et al. Bioactive extracts from berry byproducts on the pathogenicity of Salmonella Typhimurium. Int J Food Microbiol. 2016;237:128–135. doi:10.1016/j.ijfoodmicro.2016.08.027
33. Tal-Gan Y, Ivancic M, Cornilescu G, Yang T, Blackwell HE. Highly stable, amide-bridged autoinducing peptide analogues that strongly inhibit the AgrC quorum sensing receptor in Staphylococcus aureus. Angew Chem Int Ed Engl. 2016;55:8913–8917. doi:10.1002/anie.201602974
34. Fleitas MO, Rigueiras PO, Pires A, et al. Interference with quorum-sensing signal biosynthesis as a promising therapeutic strategy against multidrug-resistant pathogens. Front Cell Infect Microbiol. 2018;8:444. doi:10.3389/fcimb.2018.00444
35. Parlet CP, Kavanaugh JS, Crosby HA, et al. Apicidin attenuates MRSA virulence through quorum-sensing inhibition and enhanced host defense. Cell Rep. 2019;27:187–198. doi:10.1016/j.celrep.2019.03.018
36. Balamurugan P, Praveen KV, Bharath D, Lavanya R, Vairaprakash P, Adline PS. Staphylococcus aureus quorum regulator SarA targeted compound, 2-[(Methylamino)methyl]phenol inhibits biofilm and down-regulates virulence genes. Front Microbiol. 2017;8:1290. doi:10.3389/fmicb.2017.01290
37. Liu Y, Zhou J, Qu Y, et al. Resveratrol antagonizes antimicrobial lethality and stimulates recovery of bacterial mutants. PLoS One. 2016;11:e153023.
38. Skariyachan S, Krishnan RS, Siddapa SB, Salian C, Bora P, Sebastian D. Computer aided screening and evaluation of herbal therapeutics against MRSA infections. Bioinformation. 2011;7:222–233. doi:10.6026/97320630007222
39. Zihadi M, Rahman M, Talukder S, Hasan M, Nahar S, Sikder M. Antibacterial efficacy of ethanolic extract of Camellia sinensis and Azadirachta indica leaves on methicillin-resistant Staphylococcus aureus and shiga-toxigenic Escherichia coli. J Adv Vet Anim Res. 2019;6:247–252. doi:10.5455/javar.2019.f340
40. Luo J, Dong B, Wang K, et al. Baicalin inhibits biofilm formation, attenuates the quorum sensing-controlled virulence and enhances Pseudomonas aeruginosa clearance in a mouse peritoneal implant infection model. PLoS One. 2017;12:e176883. doi:10.1371/journal.pone.0176883
41. Sass A, Slachmuylders L, Van Acker H, et al. Various evolutionary trajectories lead to loss of the tobramycin-potentiating activity of the quorum-sensing inhibitor baicalin hydrate in Burkholderia cenocepacia biofilms. Antimicrob Agents Chemother. 2019;63:e2018–e2092. doi:10.1128/AAC.02092-18
42. Chen Y, Yuan W, Yang Y, Yao F, Ming K, Liu J. Inhibition mechanisms of baicalin and its phospholipid complex against DHAV-1 replication. Poult Sci. 2018;97:3816–3825. doi:10.3382/ps/pey255
43. Yang X, Zhang Q, Gao Z, Yu C, Zhang L. Baicalin alleviates IL-1beta-induced inflammatory injury via down-regulating miR-126 in chondrocytes. Biomed Pharmacother. 2018;99:184–190. doi:10.1016/j.biopha.2018.01.041
44. Wang J, Jiao H, Meng J, et al. Baicalin inhibits biofilm formation and the quorum-sensing system by regulating the MsrA drug efflux pump in Staphylococcus saprophyticus. Front Microbiol. 2019;10:2800. doi:10.3389/fmicb.2019.02800
45. Peng LY, Yuan M, Wu ZM, et al. Anti-bacterial activity of baicalin against APEC through inhibition of quorum sensing and inflammatory responses. Sci Rep. 2019;9:4063. doi:10.1038/s41598-019-40684-6
46. Moslehi-Jenabian S, Gori K, Jespersen L. AI-2 signalling is induced by acidic shock in probiotic strains of Lactobacillus spp. Int J Food Microbiol. 2009;135:295–302. doi:10.1016/j.ijfoodmicro.2009.08.011
47. Bedree JK, Bor B, Cen L, et al. Quorum sensing modulates the epibiotic-parasitic relationship between Actinomyces odontolyticus and Its Saccharibacteria epibiont, a Nanosynbacter lyticus strain, TM7x. Front Microbiol. 2018;9. doi:10.3389/fmicb.2018.02049
48. Yuan E, Zhou M, Nie S, Ren J. Interaction mechanism between ZnO nanoparticles-whey protein and its effect on toxicity in GES-1 cells. J Food Sci. 2022;87:2417–2426. doi:10.1111/1750-3841.16193
49. Liu Y, Xie MX, Kang J, Zheng D. Studies on the interaction of total saponins of panax notoginseng and human serum albumin by Fourier transform infrared spectroscopy. Spectrochim Acta A Mol Biomol Spectrosc. 2003;59:2747–2758. doi:10.1016/S1386-1425(03)00055-6
50. Wang G, Gao Y, Wang H, Niu X, Wang J. Baicalin weakens Staphylococcus aureus pathogenicity by targeting sortase B. Front Cell Infect Microbiol. 2018;8:418. doi:10.3389/fcimb.2018.00418
51. Koo H, Falsetta ML, Klein MI. The exopolysaccharide matrix: a virulence determinant of cariogenic biofilm. J Dent Res. 2013;92:1065–1073. doi:10.1177/0022034513504218
52. Schiffer C, Hilgarth M, Ehrmann M, Vogel RF. Bap and cell surface hydrophobicity are important factors in Staphylococcus xylosus biofilm formation. Front Microbiol. 2019;10:1387. doi:10.3389/fmicb.2019.01387
53. Gerke C, Kraft A, Sussmuth R, Schweitzer O, Gotz F. Characterization of the N-acetylglucosaminyltransferase activity involved in the biosynthesis of the Staphylococcus epidermidis polysaccharide intercellular adhesin. J Biol Chem. 1998;273:18586–18593. doi:10.1074/jbc.273.29.18586
54. Prystopiuk V, Feuillie C, Herman-Bausier P, et al. Mechanical forces guiding Staphylococcus aureus cellular invasion. ACS Nano. 2018;12:3609–3622. doi:10.1021/acsnano.8b00716
55. Lammers A, Nuijten PJ, Smith HE. The fibronectin binding proteins of Staphylococcus aureus are required for adhesion to and invasion of bovine mammary gland cells. FEMS Microbiol Lett. 1999;180:103–109. doi:10.1111/j.1574-6968.1999.tb08783.x
56. Heimes A, Brodhagen J, Weikard R, et al. Hepatic transcriptome analysis identifies divergent pathogen-specific targeting-strategies to modulate the innate immune system in response to intramammary infection. Front Immunol. 2020;11. doi:10.3389/fimmu.2020.00715
57. Kim D, Liu Y, Benhamou RI, et al. Bacterial-derived exopolysaccharides enhance antifungal drug tolerance in a cross-kingdom oral biofilm. ISME J. 2018;12:1427–1442. doi:10.1038/s41396-018-0113-1
58. Escobar-Mucino E, Arenas-Hernandez M, Luna-Guevara ML. Mechanisms of inhibition of quorum sensing as an alternative for the Control of E. coli and Salmonella. Microorganisms. 2022;10:884. doi:10.3390/microorganisms10050884
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.