Back to Journals » Infection and Drug Resistance » Volume 14
Bacteriophage – A Promising Alternative Measure for Bacterial Biofilm Control
Authors Tian F, Li J, Nazir A, Tong Y
Received 3 November 2020
Accepted for publication 23 December 2020
Published 20 January 2021 Volume 2021:14 Pages 205—217
DOI https://doi.org/10.2147/IDR.S290093
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Suresh Antony
Fengjuan Tian,* Jing Li,* Amina Nazir, Yigang Tong
Beijing Advanced Innovation Center for Soft Matter Science and Engineering, College of Life Science and Technology, Beijing University of Chemical Technology, Beijing, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Yigang Tong Email [email protected]
Abstract: Bacterial biofilms can enhance bacteria’s viability by providing resistance against antibiotics and conventional disinfectants. The existence of biofilm is a serious threat to human health, causing incalculable loss. Therefore, new strategies to deal with bacterial biofilms are needed. Bacteriophages are unique due to their activity on bacteria and do not pose a threat to humans. Consequently, they are considered safe alternatives to drugs for the treatment of bacterial diseases. They can effectively obliterate bacterial biofilms and have great potential in medical treatment, the food industry, and pollution control. There are intricate mechanisms of interaction between phages and biofilms. Biofilms may prevent the invasion of phages, and phages can kill bacteria for biofilm control purposes or influence the formation of biofilms. At present, there are various measures for the prevention and control of biofilms through phages, including the combined use of drugs and the application of phage cocktails. This article mainly reviews the function and formation process of bacterial biofilms, summarizes the different mechanisms between phages and biofilms, briefly explains the phage usage for the control of bacterial biofilms, and promotes phage application maintenance human health and the protection of the natural environment.
Keywords: biofilm, phage, drug-resistant bacteria, antibiotic substitute, bacterial biofilm control
Introduction
Pathogenic bacteria pose severe threats to food, the medical system, and other industries.1 Consequently, biofilm production by bacterial strains increases their survival ability and virulence, making them more potent pathogenic factors.1–3 Broad-spectrum antibiotics have gained momentum for preventing and controlling bacteria, resulting in the constant emergence of drug-resistant bacteria.4–6 Before the foreseeable problem of super bacterial infection, antibiotic substitutes that can be effectively used quickly become an important research field to deal with pathogens potential hazards.
Phage is a green alternative to antibiotics isolated from the natural environment. Phages present varied characteristics, such as safe and environment friendly. Thousands of different bacteriophages are already present in the human body and environment.7,8 Future development of phages can focus on optimizing their efficacy and safety.9 It is crucial to understand the formation process of bacterial biofilm and the infection mechanism of bacteriophages for the better use bacteriophages’ unique functions, which have a great significance for removing biofilm and the protection of biological safety in all walks of life.
Therefore, it is needed to expand our understanding of phage disrupting biofilm and develop new safe and effective biofilm treatment methods in deteriorating bacterial hazards.
The Bacterial Biofilm
Composition and Function of Bacterial Biofilm
The bacterial biofilm was first discovered on tooth surfaces in 1676, observed by Van Leeuwenhoek with a simple microscope, describing microorganisms’ growth attached to bare surfaces.10 However, it was until 1978 that the theory of biofilms was proposed by Costerton et al.11 Bacterial biofilms are aggregates of microorganisms and play a significant role in the persistence of bacterial infections.12 Bacteria multiply to form communities and adhere to each other or on the surface. They secrete polysaccharide matrix, fibrin, lipoprotein, and other substances to form extracellular polymers and embed themselves in the self-produced extracellular polymer matrix to form a biofilm.13–15 Extracellular polymers are critical to the structure and stability of biofilms.16 In most bacterial biofilms, the proportion of microorganisms in dry matter is less than 10%. In comparison, the matrix composed of different extracellular polymers can account for more than 90%, forming a three-dimensional structure of the biofilm responsible for supporting the surface adhesion of the biofilm and cohesion.17 The extracellular polymer matrix acts as a diffusion barrier or directly binds antibacterial agents to prevent it from entering bacterial cells.18 Simultaneously, bacteria can adapt to the surrounding environment with biofilm, obtain nutrients, and discharge waste through the gaps between bacterial communities.
The Formation of Biofilm
The biofilm formation is a complex process and can be divided into three stages in general (Figure 1). Bacterial cells attach to a surface first and then grow into a fixed biofilm community. After this, the bacterial cells disperse from the biofilm community to the new surroundings and start a new cycle.19–21 In the first stage of biofilm formation, bacterial adhesion on an object’s surface is divided into reversible and irreversible attachment. Reversible adhesion is mediated by non-specific Vander Waals force, Lewis acid-based electronic force, an electrostatic force. In contrast, irreversible adhesion relies on bacterial specific adhesion pili flagella to the attachment surface.22–24 When free bacterial cells attach to the surface irreversibly, they reproduce and are accompanied by an extracellular polymer matrix synthesis. The extracellular polymer matrix conduces to more bacterial cells, mutual adsorption and attaching to the surface of the object.16 The continuous growth of bacterial cells on the attached surface gradually matures, forming a biofilm consisting of millions of closely-spaced cells. The final stage of biofilm formation is that cells are separated from the biofilm population and diffuse into the environment to form the next biofilm. This is an essential stage of biofilm formation, contributing to the biological spread, bacterial survival, and disease transmission.20,25
Biofilms can develop from the same bacteria, but also can form by different bacteria.26 For example, dental plaque can be formed by hundreds of bacteria, and all kinds of bacteria colonization on the tooth surface follows an orderly development process.27 Early bacteria adhere to the tooth enamel surface, and later bacteria adhere to that already existed to form a new biofilm.28 Biofilm diffusion can also be divided into three different stages. First, the bacterial cells are detached from the biofilm colonies. Second, bacterial cells move to a new location. At last, the bacterial cells move to a suitable location to finish attachment. Biofilm diffusion includes active and passive diffusion. The former is actively carried out by bacteria, and the latter is the transfer of biofilm cells due to external forces. Diffusion patterns include: erosion, sloughing, and seeding. The continuous release of single cells or small clusters of cells from biofilms at low levels is called erosion. The rapid massive loss of biofilm is known as sloughing. The rapid release of a mount of single cells or small cell clusters from a hollow cavity formed inside a biofilm colony are called seeding.20,29–33
Impact of Bacterial Biofilm Presence
Most bacteria can form biofilms; once the biofilm is formed, it will exacerbate the host’s immune mechanism and antibiotics resistance, improving bacteria’s survival rate. Bacterial biofilms that are difficult to remove from medical devices or wounds are more likely to cause infection. When the body’s immunity or resistance is weakened, bacteria that survive in the biofilm can be released to cause new infections.34 The presence of biofilms allows bacteria to persist in pathological environments leading to chronic infections during the healing process.35 Bacterial biofilms have a significant impact on the virulence and viability of bacteria, not only in the medical field but also in industries that require a highly sterile environment, such as food processing, where biofilms can significantly affect people’s manufacture and life.36,37
Phage Interacts with Biofilm
Common disinfectants and antibiotics are more effective in removing free cells but do not readily penetrate and destroy biofilms and are less effective against antibiotic-resistant bacteria, with a reported 100–1000-fold increase in antibiotics concentration required to eradicate bacteria containing biofilms.38–41 Due to different microorganisms, the type of mixed biofilm increases with symbiotic microorganisms, which are likely to be more resistant to antibiotics and increase the therapeutic burden.42 Simultaneously, antibiotics are being developed at a much slower rate than the rate at which bacteria develop resistance. There are many applications on clearing biofilms by physical and chemical methods, yet little is known about the interactions between biofilms and phages, and the role of phages as effective bacterial scavengers has not been developed, thus a deeper understanding of the mechanisms involved between phages and biofilms are necessary to facilitate phages application.
Mechanism of Phage Prevention and Control of Biofilm
Phages are considered to be the most abundant microorganisms on the planet. The diversity of phages comes from their dynamic adaptability in the face of selective pressure.43 Phages are viruses that only infect bacteria, and the number in nature is about 1031,at least ten times more than bacteria.44 There are two types of phages: the lysogenic phage and the other is the lytic phage. The lysogenic phage integrates itself with the host bacterial genome to achieve a coexisting state. In contrast, the lytic phage injects its genome into the host bacteria and utilizes the host ribosomes to manufactures its proteins.
Bacterial cell resources rapidly manufactured capsid proteins and viral genomes, which assemble into the phage progeny to lyse the host bacteria.45,46 Phages use receptor binding proteins (RBPs) to interact with receptors on the surface of bacteria. Following the initial reversible attachment, phages fix on the bacteria’s surface irreversibly and inject the phage genome into the host cytoplasm.47 The infection of phage into the host can be divided into several steps: (i) attachment to bacterial surface receptors (usually sugar or protein) through phage receptor binding proteins, (ii) injection of phage genome into bacterial cytoplasm, (iii) phage protein assembly, (iv) phage progeny release.48
Phages can clear biofilms in a variety of ways. (i) Lysogenic phages can integrate into the bacterial genome and affect the formation of biofilms.49 For example, the integration of phage Bxb1 inactivates the groEL1 gene of Mycobacterium smegmatis. After integration, the bacteria can float naturally but prevent the formation of mature biofilm.50 (ii) Phage clears bacterial biofilm and lyses cells by encoding lyase (Figure 2).51,52 Lu et al 2007, designed and engineered a bacteriophage capable of expressing the enzyme DspB to degrade the β-1,6-N-acetyl-D-glucosamine during infection, which is a crucial adhesion needed for biofilm. Phage-encoded DspB is released into the environment with host cell lysis, causing more degradation of the biofilm, thereby controlling the formation of the biofilm. This engineered bacteriophage attacks both on biofilm and the bacteria in the biofilm matrix, removing 99.997% of the biofilm with the dual action of DspB and phage. This phenomenon demonstrating the feasibility of using engineered phages with biofilm-degrading enzymatic activity to reduce bacterial biofilms.53 (iii) The phage expresses enzymes that degrade extracellular polymers, destroys the polysaccharide matrix and proteins in the biofilm that encapsulates the bacteria, and clears the bacterial protective barrier, and then enters the biofilm to destroy the bacteria. Pires et al 2016, summarized 160 hypothetical depolymerises, including glucanase, acetylase, etc., and revealed the diversity of understanding depolymerases.54 Hanlon et al 2001, found that phages can degrade Pseudomonas aeruginosa (P. aeruginosa) exopolysaccharides by the enzyme to reach the cell surface.55 The depolymerase produced by most phages only recognizes host-derived polysaccharides, and has certain limitations for expanding the host spectrum of phages.56 With the gradual maturity of genetic engineering technology, this limitation can be lifted using genetic engineering techniques to modify phage depolymerase and expand the future’s phage host spectrum.
Bacterial Biofilm Resists Phage Infection
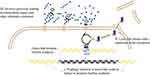
Although phages have good results in the application of biofilm removal, there are still a large number of studies suggesting the limitations of phage applications. The prolonged arms race between bacteria and phages has likewise enabled bacteria to acquire the ability to resist phages, including alterations in phage-recognition receptors in bacteria, abortive infection systems (Abi) that can lead to cell death or metabolic arrest (Figure 3). In contrast, phages infect the RM system and DISARM of bacteria following the phage genome’s entry and remove the genome by the CRISPR system.48,57–61 The complex material composition and interlocking membrane structure of the biofilm, as the first physical line of bacterial defense against phages, makes the invasion of phages more difficult. In general, bacteria utilize the over-expression of substances in the biofilm to capture viral particles to block the invasion of phages and protect them from damage.62 Colonic acid is an extracellular polysaccharide (EPS); a mutant strain of Escherichia coli (E. coli) that over expresses colonic acid, which protects Enterobacteriaceae bacteria from phage invasion by forming protective capsules.63 Vidakovic et al 2015, used E. coli biofilms and lysing T7 phages as models. They found that CsgA (curli polymer) protects bacteria by covering the cell surface and binding phage particles to prevent them from contacting the bacterial surface.64 The interaction of multiple bacteria will produce a biofilm that is more resistant to phage. The biofilm in which multiple bacteria coexist can be a mixture of multiple bacterial biofilms or dominated by a single bacterial biofilm. Biofilm mixture can effectively reduce phage invasion, but only specific strains can enhance the anti-phage effect. The competition between the bacterial strains may also reduce the resistance.65–67 Complex mixed biofilms are one of the critical barriers for phage clearance of biofilms.
 |
Figure 3 Biofilms capture phages through extracellular polymers. |
The Other Relationship Between Phage and Biofilms
Phages are regarded as natural enemies of bacteria and bacterial biofilms; interestingly, some phages have a “cooperative” relationship with the bacterial host. Secor et al 2015, reported that filamentous phage interacted with the host and microbial polymers to form a higher-order liquid crystal structure of the biofilm matrix, which enhanced the biofilm’s adhesion, the survival rate in the dry state, and antibiotic resistance.68 Filamentous phage also affects biofilms, leading to tolerance, virulence, dispersion of biofilms, and colony variation.69 Rice et al 2019, demonstrated that filamentous phages play an important role in P. aeruginosa and biofilms’ virulence formation process. P. aeruginosa that infects filamentous phages has shown more significant toxicity, while P. aeruginosa lacking filamentous phage’s gene cannot develop hollow centers or undergo cell death.70 The presence of phages may also cause changes in the structure of the bacteria themselves or the biofilm. Davies et al 2016, found that mild phages, especially transposable phages, may promote bacterial pathogens’ adaptive changes in the host. They used an artificial sputum medium to simulate sputum after a cystic fibrosis lung infection. Cultivated phage-infected and phage-uninfected P. aeruginosa, the results show that both bacteria can adapt to the biofilm environment. However, the phage population showed a higher degree of parallel evolution and faster selective elimination, which promoted the bacteria in the host’s faster adaptive evolution in the body.71 Bull et al 2018, demonstrated that spatially structured phage death provides cells with a means of protection that can boost cell densities an order of magnitude above that attained under mass action. The extracellular polysaccharides and cell fragments of the bacteria produce high-density bacterial regions in the biofilm space structure environment. Phage sinks formed around this area, which prevented it from further infecting the living bacteria, reducing the infection of the living bacteria by the phage from the physical barrier. Besides, the presence of different phages has different effects on the development of biofilms.72 Tan et al 2015, found that Vibrio anguillarum (V. anguillarum) phage ΦH20 has an excellent inhibitory effect on biofilm. In contrast, phage KVP40 stimulates V. anguillarum to produce more biofilms to prevent the invasion of phage KVP40. The long-term co-evolutionary relationship between phages and bacteria exists which produces more complex mechanisms. There are many challenges in studying the interaction mechanism between phages and bacteria.73
Application of Phage for Prevention and Control of Biofilm
Application of Phage to Destroy Biofilm of Pathogenic Bacteria
Antibacterial drugs have a screening effect and inhibit the bacterial growth but if bacterial resistance occur, then antibiotics have no effect to bacteria. Antibiotic resistance has become a high concern in global public health.52,74,75 Bacterial biofilms often involve many chronic infections that are difficult to treat and are associated with moist object surfaces. Bacterial biofilms are prone to develop tolerance to antibiotics and host immunity.76 It is estimated that more than 700,000 deaths occur each year due to resistant bacteria.77 Therefore, it is urgent need to develop new treatment methods for antibiotic-resistant bacteria and biofilms. Phage therapy is now accepted as a good way to inhibit antibiotic-resistant bacteria.78,79 Only a few countries in the world use phages for bacterial therapy, and as technology continues to advance, other regions have been experimenting with phage therapies as well.79 Phages have shown great potential in the treatment of antibiotic-resistant bacterial infections.
Phage Clears Biofilm
Phage has been successfully used to reduce contamination in medical catheters and is being developed as a new drug to control bacterial biofilm infection.52 Proteus mirabilis (P. mirabilis) forms a dense crystalline biofilm on the catheter, thereby blocking the flow of urine. In the early stage of bacterial infection, the phage can eliminate P. mirabilis. Although the level of planktonic cells has not been reduced in the late stage of infection, it can significantly reduce the formation of coating.80 Patients who underwent total knee arthroplasty experienced recurrent prosthetic infections due to the presence of Klebsiella pneumoniae biofilms. Fortunately, intravenous phage therapy has achieved a satisfactory outcome where antibiotics have not worked well.81 Biofilms formed by Enterococcus faecalis (E. faecalis) and other pathogenic bacteria in the oral cavity are compromising oral health.82,83 Khalifa et al 2015, characterized a lytic phage EFDG1 that efficiently infects and kills planktonic and biofilm cultures of E. faecalis both in vitro and in vivo experimental model of tooth root canal infection.84 Advances in phage therapy may offer potential ways for better treatment of biofilm-derived oral infections. As a common Gram-negative pathogen in hospitals, P. aeruginosa has formed a highly stable biofilm and often exhibits antibiotic resistance, which has attracted widespread attention, designated as a key priority for developing new therapies by the World Health Organization.85–88 Phage φMR299-2 (podovirus) and φNH-4 (myovirus) have been shown to eliminate P. aeruginosa in the mouse lung and cystic fibrosis lung airway cells.89
Combination of Phage and Antibacterial Drugs
Using antibiotics that reduce biofilm will usually lead to the emergence of many antibiotic-resistant bacteria. In different treatment methods, the combination of phage and antibacterial drugs can significantly increase the lethality of bacteria.90–92 Lu et al 2009, used engineered phages to inhibit the SOS network in E. coli from enhancing the lethality of quinolones in vitro, proving that engineered phages can enhance the killing effect of antibiotics on biofilms.93 Srinivasan et al 2019, found that the addition of calcium can enhance the lytic activity of Vibrio parahaemolyticus phage.94 Appropriate antibiotic treatment combined with phage can dissolve the biofilms of Staphylococcus aureus (S. aureus) and P. aeruginosa, while the effect on S. aureus is more obvious.95 The combination of phage PEV20 and ciprofloxacin has a synergistic antibacterial effect on eliminating P. aeruginosa and its biofilm, protecting the lung epithelial fibroblast cells from P. aeruginosa.96,97 The presence of antibiotics may interfere with bacterial metabolism activity, and the activity of bacterial metabolism may affect the binding of phages.
Nevertheless, the antibacterial function of phages still exists.98 When phages are used in combination with antibiotics, the ratio of the two and their administration requirements greatly impact the therapeutic effect. Therefore, it will be the focus of future research to adequately evaluate the therapeutic potential of phages in order to optimize the conditions of the combination and ensure its safety and efficacy.99,100
Application of Phage Cocktail Therapy to Prevent and Control Biofilm
Phage cocktail therapy treats bacteria with a mixture of multiple phages, and it shows great potential for treating complicated bacterial infections and other issues supported by the findings of recent successful compassionate treatments in Europe and the United States. Latz et al 2017, mixed three multi-drug-resistant P. aeruginosa phages and found that compared to a single phage, the mixed phages showed the best effect without the synergy antagonism.101 Alves et al 2016, reported that a new type of phage cocktail reduce and disperse P. aeruginosa biofilm under static and flowing conditions and can be used as an agent for treating P. aeruginosa infection.86 Forti et al 2018, designed a cocktail of six phages that worked well with P. aeruginosa isolates from clinical samples, demonstrating the ability to clear multi-drug resistant strains of bacteria. The release of bacteriophages was capable of almost complete clearance to form a biofilm for 48 hours. Meanwhile, in an attempt to treat respiratory tract infections in mice with phages, it was found that the use of phage cocktails with different MOIs can reduce the P. aeruginosa infections in mice and that appropriately timed phage cocktails were effective in rescuing P. aeruginosa-infected wax moth (G. mellonella) through treating bacteremia.102 Alves et al 2014, found that phage DRA88 and phage K combination could lyse a variety of S. aureus.103 S. aureus multiple cocktails show 100% lysis efficiency against clinically resistant S. aureus.2 The use of phage cocktails allows more bacteria to be controlled, so more types of phage can be used as treatment options, therefore modification of phage will also become one of the promising fields for phage research. Technically, to screen multiple effective phages quickly may earn time for clinical treatment.
Application of Lysin to Remove Biofilm
Lysin is a phage-derived enzyme that can degrade bacterial peptidoglycan. Lysin has a vital role in controlling biofilms. Schuch et al 2017, evaluated the ability of phage lysin CF-301 to clear biofilms in a variety of situations, and CF-301 was able to completely clear S. aureus biofilms that were resistant to high concentrations of antibiotics (daptomycin and ciprofloxacin). In experiments to treat S. aureus infection in vivo, low concentrations of CF-301 were effective in destroying all biofilms and killing free bacteria, making it a good potential antimicrobial agent.104 Lysin P128 is a chimeric protein with potent anti-staphylococcal activity.105 Lysin P128 has a therapeutic effect on various Staphylococci and its biofilm, it is very effective in treating rats infected with S.aureus.106,107 Díez-Martínez et al 2015, exchanged the structural components of two lysins and successfully constructed a new chimeric enzyme with high bactericidal activity and different substrate ranges.108 The combination of lysin and antibiotics has a synergistic effect in treating Streptococcus suis biofilms and is expected to become a new anti-biofilm class drugs.109,110 Lood et al 2015, reported that phage lysin kill multi-resistant gram-negative bacterium Acinetobacter baumannii (A. baumannii) in a mouse bacteremia model.111 Thandar et al 2016, found that the thirty amino acids in the C-terminal portion of the phage lysin PlyF307 were sufficient to kill A. baumannii, and they subsequently modified this portion of the short peptide with amino acids and found that the bactericidal efficiency of the short peptide could be increased tenfold. The target of this lysin-derived short peptide seems to be the cell membrane, and it can significantly reduce A. baumannii activity in mouse skin.112 Yang et al 2016, reported that an engineered lysin ClyR, which has activity on Streptococcus mutans biofilm in vitro and in vivo, is expected to be a preventive or therapeutic agent prevention of dental caries.113 Vázquez et al 2019, reported that the two lysins Cpl-711 and PL3 have synergistic effects in promoting purified cell wall degradation, inhibiting bacterial cell growth in vitro, and killing plankton and cells in the biofilm.114 Extensive research on lysin has proved its potential ability to treat various pathogenic bacteria, and it is expected to be developed as an alternative drug for future biofilms control.
Application of Phage for Prevention of Biofilm in Water Environment
Biofilms formed by microorganisms during waste water treatment are the main problems encountered during membrane filtration and are particularly serious in circulating aquaculture systems.115,116 Phages can solve aggravating water infrastructure corrosion and biological fouling due to the generation of bacterial biofilms. They can also selectively suppress the proliferation and foaming of bacteria that can prevent sludge clarification and reduce the antibiotics-resistant strains in biological wastewater treatment systems diffusion.41
P. aeruginosa biofilms often clog filters in drinking water treatment plants and increase cleaning costs. Zhang et al 2013, achieved a removal rate of 89% using phage isolated from sewage to clean P. aeruginosa biofilm, and reached a 96% removal rate when used in combination with chlorine.117 Goldman et al 2009, found that phage can reduce the microbial attachment rate on ultra filtration membrane modules by 40% to 60% and proved that the simultaneous use of multiple phages could prevent multifactorial formation biofilms.118 The use of phages to control water pollution and potential biological threats caused by biofilms is a cost-effective and environmentally friendly new technology. Sewage contains a large number of microorganisms, including most pathogenic bacteria, and the large-scale use and discharge of antibiotics have also caused the continuous emergence of antibiotic-resistant, thus posing a great threat to species and environment.115,119,120 Increasing numbers of antibiotic-resistant virulent Vibrio species are being isolated from the dam, boreholes, and tap water, and the screening of phages that lyse antibiotic-resistant Vibrio species by Maje et al 2020, is essential for safeguarding the biosecurity of water supplies.121 Aquaculture farmers usually rely on much preventive use of antibiotics in farmed fish to reduce pathogenic vibrio and its biofilm, which has gradually caused the emergence of vibrio resistance and increased the aquaculture industry’s burden. Nandita et al 2019, found a characteristic lysin in V. parahaemolyticus phage, with lytic activity against various vibrio species, thus expected to become a promising bio-bactericide process for the treatment of vibrio resistance to solve the problem of antibiotics overuse in aquatic industry.122
Application of Phage for Prevention of Biofilm in Food Industry
Listeria monocytogenes (L. monocytogenes) can cause food-borne diseases. It has a higher risk of treatment in the elderly, immuno-compromised, pregnant women and children, and the mortality rate is exceptionally high. The biofilm can colonize in the food production environment and help food-borne pathogens survive under harsh living conditions.123–126 L. monocytogenes is challenging to eradicate. It’s ability to form a biofilm protects it from the effects of conventional cleaning, which is the critical factor in the survival of L. monocytogenes in the food industry. The strong colonization ability of this bacteria could be easily detected in production environment.123,124,127 Iacumin et al 2016, found that L. monocytogenes phage P100 was active against five strains or serotypes, and the bactericidal effect of phages at different concentrations on bacteria was counted. When the phage level was 8 log PFU/cm2, it had a bactericidal activity effect, and it can be used to reduce the concentration of L. monocytogenes.128 Soni et al 2010, determined the activity of P100 against 21 L. monocytogenes and determined that P100 could also reduce the cell population of L. monocytogenes in the presence of biofilm.129 The mixed-use of multiple phages to kill bacteria that are difficult to remove in the food industry shows good therapeutic potential. Phages and phage-lysing proteins that can eradicate biofilms are promising to become future disinfectants.130,131
Side Effects of Phage Therapy
The success of phage therapy in humans is still being checked, and certain major hurdles remain to be studied. During phage therapy,132 phage insensitive mutants (BIM) can occur, which will contribute to several complexities in phage therapy. Recent researchers have shown, though that the advent of BIM can minimize bacterial virulence, making it easier to cleanse the immune system.133 Phages are complex species that can transmit toxin genes among bacteria, particularly lysogenic phages.134 Hence, lytic phages must be chosen for phage therapy after screening and toxicity tests. Additionally, when infected with unfamiliar species or several infections, the clinical significance of bacteriophages is absurd. Furthermore, phage therapy requires a broad range of phages and high purity. Currently, commonly used approaches are very difficult and ineffective for extracting and purifying bacteriophages. In addition, the processing of phage preparations is complex and the procedure is complicated. For acute or special circumstances, phage therapy is also not appropriate.
Conclusion
Bacteria are ubiquitous in the natural environment. More and more health events are caused by bacteria and biofilm, which deserves more attention in recent years. The biofilm structure formed by the accumulation of bacterial communities provides a barrier for bacteria, challenging to remove by conventional means. The traditional physical and chemical disinfection methods cannot altogether remove the biofilm. Even the use of antibiotics will pressure the selection of bacteria, leading to the emergence of more resistant bacteria. The presence of bacteria causes severe problems in most industries that require aseptic environments, such as the food industry, medical, and pollution treatment. Bacteriophages are the virus against bacteria, and almost all bacteria have their corresponding phages. Therefore, the phage can be used as a promising substitute for antibiotics or other antibacterial drugs. Phages and bacteria in nature have experienced a prolonged arms race. Phages have been developing methods to infect bacteria, and bacteria have also developed mechanisms to resist phages.135 With the advancements in identifying interaction mechanisms, such as the immune and evasive immune relationship between bacterial CRISPR and phage anti-CRISPR, being discovered. Phosphorothioation-sensing bacterial protection system,136 have been characterized in recent years, and the diversity of the arms race between the two is increasingly more clearly illustrated. This gives one a deeper understanding of the relationship between the two and offers a stronger foundation for the use of phages as removal/control biofilm formation.137
Bacteria can prevent the adsorption of phage by forming a complex biofilm. Therefore, making phages break through the defense of biofilm becomes a research hotspot of prevention and treatment of phage biofilm. Some hospitals have begun to use aerosol phage environmental cleaners to remove drug-resistant bacteria. The research on phage cocktails has also shown good momentum. Some bacteriophage agents have been successfully developed to eliminate specific bacteria and control biofilm.138
However, the standard methods of biofilm removal are still physical and chemical. At present, there is no established phage-based method to eliminate biofilms, so we should continue to study the mechanism of phages and bacteria to explore potential and environment-friendly methods for biofilm control. Current phage measures used to prevent and control biofilm still have some limitations. For example, the host spectrum of bacteriophages is usually narrow. The biofilm is usually composed of various bacteria, limiting the application of phages to a certain extent. At the same time, host bacteria can use their defense system to prevent phage reproduction, making it more challenging to find suitable phages that can break through the bacterial restrictions.
Future research should aim to expand the scope of application of phages by promoting phage mixtures and engineered phage transformation. Breaking through the restrictions of phages on biofilm control is essential. But the genome and function of most phages remain unknown. Many unknown bacteriophage enzymes are expected to be potential drugs for eliminating biofilms, and relevant functions should be emphasized to make them green alternatives of antibiotic drugs for prevention and control of biofilms. The evaluation of phage application should also be carried out. The mature phage strategy could be a powerful weapon against potential bacterial threats and receive government support and vigorous development.
Author Contributions
All authors contributed to data analysis, drafting or revising the article, have agreed on the journal to which the article will be submitted, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Funding
This research was supported by a grant from The National Key Research and Development Program of China (2018YFA0903000), the National Natural Science Foundation of China (81672001), and China MoST Emergency Project on COVID-19 (2020YFC0840800).
Disclosure
The authors declare that this research has been conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1. Del Pozo JL. Biofilm-related disease. Expert Rev Anti Infect Ther. 2018;16(1):51–65. doi:10.1080/14787210.2018.1417036
2. Racenis K, Kroica J, Rezevska D, et al. S. aureus colonization, biofilm production, and phage susceptibility in peritoneal dialysis patients. Antibiotics. 2020;9:9. doi:10.3390/antibiotics9090582
3. Preda VG, Săndulescu O. Communication is the key: biofilms, quorum sensing, formation and prevention. Discoveries. 2019;7(3):e100. doi:10.15190/d.2019.13
4. Di Domenico EG, Farulla I, Prignano G, et al. Biofilm is a major virulence determinant in bacterial colonization of chronic skin ulcers independently from the multidrug resistant phenotype. Int J Mol Sci. 2017;18:5. doi:10.3390/ijms18051077
5. Del Pozo JL, Patel R. Are antibiotics and surgery sufficient to treat biofilm-associated infections? Enferm Infecc Microbiol Clin. 2013;31(10):641–642.
6. Nazir A, Zhao Y, Li M, et al. Structural Genomics of repA, repB1-Carrying IncFIB Family pA1705-qnrS, P911021-tetA, and P1642-tetA, multidrug-resistant plasmids from klebsiella pneumoniae. Infection and Drug Resistance. 2020;13:1889. doi:10.2147/IDR.S228704
7. Moye ZD, Woolston J, Sulakvelidze A. Bacteriophage applications for food production and processing. Viruses. 2018;10:4. doi:10.3390/v10040205
8. Schooley RT, Biswas B, Gill JJ, et al. Development and use of personalized bacteriophage-based therapeutic cocktails to treat a patient with a disseminated resistant acinetobacter baumannii infection. Antimicrob Agents Chemother. 2017;61:10. doi:10.1128/AAC.00954-17
9. Philipson CW, Voegtly LJ, Lueder MR, et al. Characterizing phage genomes for therapeutic applications. Viruses. 2018;10:4.
10. Donlan RM. Biofilms: microbial life on surfaces. Emerg Infect Dis. 2002;8(9):881–890. doi:10.3201/eid0809.020063
11. Costerton JW, Geesey GG, Cheng KJ. How bacteria stick. Scientific American. 1978;238(1):86–95.
12. Fulaz S, Vitale S, Quinn L, Casey E. Nanoparticle-biofilm interactions: the role of the EPS matrix. Trends Microbiol. 2019;27(11):915–926. doi:10.1016/j.tim.2019.07.004
13. Sharma D, Misba L, Khan AU. Antibiotics versus biofilm: an emerging battleground in microbial communities. Antimicrob Resist Infect Control. 2019;8:76. doi:10.1186/s13756-019-0533-3
14. Costerton JW, Stewart PS, Greenberg EP. Bacterial biofilms: a common cause of persistent infections. Science. 1999;284(5418):1318–1322. doi:10.1126/science.284.5418.1318
15. Donlan RM. Preventing biofilms of clinically relevant organisms using bacteriophage. Trends Microbiol. 2009;17(2):66–72. doi:10.1016/j.tim.2008.11.002
16. Halan B, Buehler K, Schmid A. Biofilms as living catalysts in continuous chemical syntheses. Trends Biotechnol. 2012;30(9):453–465. doi:10.1016/j.tibtech.2012.05.003
17. Flemming H-C, Wingender J. The biofilm matrix. Nat Rev Microbiol. 2010;8(9):623–633. doi:10.1038/nrmicro2415
18. Mah TFC, O’Toole GA. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 2001;9(1):34–39. doi:10.1016/S0966-842X(00)01913-2
19. Rabin N, Zheng Y, Opoku-Temeng C, Du Y, Bonsu E, Sintim HO. Biofilm formation mechanisms and targets for developing antibiofilm agents. Future Med Chem. 2015;7(4):493–512. doi:10.4155/fmc.15.6
20. Kaplan JB. Biofilm dispersal: mechanisms, clinical implications, and potential therapeutic uses. J Dent Res. 2010;89(3):205–218. doi:10.1177/0022034509359403
21. Muhammad MH, Idris AL, Fan X, et al. Beyond risk: bacterial biofilms and their regulating approaches. Front Microbiol. 2020;11:928.
22. Renner LD, Weibel DB. Physicochemical regulation of biofilm formation. MRS Bull. 2011;36(5):347–355. doi:10.1557/mrs.2011.65
23. Palmer J, Flint S, Brooks J. Bacterial cell attachment, the beginning of a biofilm. J Ind Microbiol Biotechnol. 2007;34(9):577–588. doi:10.1007/s10295-007-0234-4
24. Moormeier DE, Bayles KW. Staphylococcus aureus biofilm: a complex developmental organism. Mol Microbiol. 2017;104(3):365–376. doi:10.1111/mmi.13634
25. Toyofuku M, Inaba T, Kiyokawa T, Obana N, Yawata Y, Nomura N. Environmental factors that shape biofilm formation. Biosci Biotechnol Biochem. 2016;80(1):7–12. doi:10.1080/09168451.2015.1058701
26. Tolker-Nielsen T. Biofilm development. Microbiol Spectr. 2015;3(2):
27. Graves DT, Corrêa JD, Silva TA. The oral microbiota is modified by systemic diseases. J Dent Res. 2019;98(2):148–156. doi:10.1177/0022034518805739
28. Marsh PD. Dental plaque as a microbial biofilm. Caries Research. 2004;38(3):204–211. doi:10.1159/000077756
29. Lappin-Scott HM, Bass C. Biofilm formation: attachment, growth, and detachment of microbes from surfaces. Am J Infect Control. 2001;29(4):250–251. doi:10.1067/mic.2001.115674
30. Wilson S, Hamilton MA, Hamilton GC, Schumann MR, Stoodley P. Statistical quantification of detachment rates and size distributions of cell clumps from wild-type (PAO1) and cell signaling mutant (JP1) Pseudomonas aeruginosa biofilms. Appl Environ Microbiol. 2004;70(10):5847–5852. doi:10.1128/AEM.70.10.5847-5852.2004
31. Purevdorj-Gage B, Costerton WJ, Stoodley P. Phenotypic differentiation and seeding dispersal in non-mucoid and mucoid Pseudomonas aeruginosa biofilms. Microbiology. 2005;151((Pt5)):1569–1576. doi:10.1099/mic.0.27536-0
32. McDougald D, Rice SA, Barraud N, Steinberg PD, Kjelleberg S. Should we stay or should we go: mechanisms and ecological consequences for biofilm dispersal. Nat Rev Microbiol. 2011;10(1):39–50. doi:10.1038/nrmicro2695
33. Characklis WG, Marshall KCNYW. Biofilm processes. In Biofilms. 1990;pp195–231.
34. Lebeaux D, Ghigo JM, Beloin C. Biofilm-related infections: bridging the gap between clinical management and fundamental aspects of recalcitrance toward antibiotics. Microbiol Mol Biol Rev. 2014;78(3):510–543.
35. Hughes G, Webber MA. Novel approaches to the treatment of bacterial biofilm infections. Br J Pharmacol. 2017;174(14):2237–2246. doi:10.1111/bph.13706
36. Al-Wrafy F, Brzozowska E, Gorska S, Gamian A. Pathogenic factors of Pseudomonas aeruginosa - the role of biofilm in pathogenicity and as a target for phage therapy. Postepy Hig Med Dosw. 2017;71:78–91. doi:10.5604/01.3001.0010.3792
37. Yan Z, Huang M, Melander C, Kjellerup BV. Dispersal and inhibition of biofilms associated with infections. J Appl Microbiol. 2019;128:1279–1288. doi:10.1111/jam.14491
38. Ceri H, Olson ME, Stremick C, Read RR, Morck D, Buret A. The calgary biofilm device: new technology for rapid determination of antibiotic susceptibilities of bacterial biofilms. J Clin Microbiol. 1999;37(6):1771–1776. doi:10.1128/JCM.37.6.1771-1776.1999
39. Fauvart M, De Grootet VN, Michiels J. Role of persister cells in chronic infections: clinical relevance and perspectives on anti-persister therapies. J Med Microbiol. 2011;60(6):699–709. doi:10.1099/jmm.0.030932-0
40. Roy R, Tiwari M, Donelli G, Tiwari V. Strategies for combating bacterial biofilms: A focus on anti-biofilm agents and their mechanisms of action. Virulence. 2018;9(1):522–554. doi:10.1080/21505594.2017.1313372
41. Mathieu J, Yu P, Zuo P, Da Silva MLB, Alvarez PJJ. Going viral: emerging opportunities for phage-based bacterial control in water treatment and reuse. Acc Chem Res. 2019;52(4):849–857. doi:10.1021/acs.accounts.8b00576
42. Sanchez-Vizuete P, Le Coq D, Bridier A, Herry J-M, Aymerich S, Briandet R. Identification of ypqP as a new bacillus subtilis biofilm determinant that mediates the protection of staphylococcus aureus against antimicrobial agents in mixed-species communities. Appl Environ Microbiol. 2015;81(1):109–118. doi:10.1128/AEM.02473-14
43. Labrie SJ, Samson JE, Moineau S. Bacteriophage resistance mechanisms. Nat Rev Microbiol. 2010;8(5):317–327. doi:10.1038/nrmicro2315
44. Clokie MR, Millard AD, Letarov AV, Heaphy S. Phages in nature. Bacteriophage. 2011;1(1):31–45. doi:10.4161/bact.1.1.14942
45. Cisek AA, Dabrowska I, Gregorczyk KP, Wyzewski Z. Phage therapy in bacterial infections treatment: one hundred years after the discovery of bacteriophages. Current Microbiology. 2017;74(2):277–283. doi:10.1007/s00284-016-1166-x
46. Lin DM, Koskella B, Lin HC. Phage therapy: an alternative to antibiotics in the age of multi-drug resistance. World J Gastrointestinal Pharmacol Therapeutics. 2017;8(3):162–173. doi:10.4292/wjgpt.v8.i3.162
47. Nobrega FL, Vlot M, de Jonge PA, et al. Targeting mechanisms of tailed bacteriophages. Nat Rev Microbiol. 2018;16(12):760–773. doi:10.1038/s41579-018-0070-8
48. Azam AH, Tanji Y. Bacteriophage-host arm race: an update on the mechanism of phage resistance in bacteria and revenge of the phage with the perspective for phage therapy. Appl Microbiol Biotechnol. 2019;103(5):2121–2131.
49. Fan X, Li W, Zheng F, Xie J. Bacteriophage inspired antibiotics discovery against infection involved biofilm. Critical Reviews Eukaryotic Gene Expression. 2013;23(4):317–326. doi:10.1615/CritRevEukaryotGeneExpr.2013007717
50. Ojha A, Anand M, Bhatt A, Kremer L, Jacobs WR, Hatfull GF. GroEL1: A dedicated chaperone involved in mycolic acid biosynthesis during biofilm formation in mycobacteria. Cell. 2005;123(5):861–873. doi:10.1016/j.cell.2005.09.012
51. Karatan E, Watnick P. Signals, regulatory networks, and materials that build and break bacterial biofilms. Microbiol Mol Biol Rev. 2009;73(2):
52. Bruessow H. Bacteriophage-host interaction: from splendid isolation into a messy reality. Curr Opin Microbiol. 2013;16(4):500–506. doi:10.1016/j.mib.2013.04.007
53. Lu TK, Collins JJ. Dispersing biofilms with engineered enzymatic bacteriophage. Proc Nat Acad Sci United States America. 2007;104(27):11197–11202. doi:10.1073/pnas.0704624104
54. Pires DP, Oliveira H, Melo LDR, Sillankorva S, Azeredo J. Bacteriophage-encoded depolymerases: their diversity and biotechnological applications. Appl Microbiol Biotechnol. 2016;100(5):2141–2151. doi:10.1007/s00253-015-7247-0
55. Hanlon GW, Denyer SP, Olliff CJ, Ibrahim LJ. Reduction in exopolysaccharide viscosity as an aid to bacteriophage penetration through Pseudomonas aeruginosa biofilms. Appl Environ Microbiol. 2001;67(6):2746–2753. doi:10.1128/AEM.67.6.2746-2753.2001
56. Pei R, Lamas-Samanamud GR. Inhibition of biofilm formation by T7 bacteriophages producing quorum-quenching enzymes. Appl Environ Microbiol. 2014;80(17):5340–5348. doi:10.1128/AEM.01434-14
57. Commons J, Morley D, Westra ER, van Houte S. CRISPR-Cas immunity leads to a coevolutionary arms race between Streptococcus thermophilus and lytic phage. Philosophical Transactions Royal Society B-Biol Sci. 2019;374:1772.
58. Cohen D, Melamed S, Millman A, et al. Cyclic GMP-AMP signalling protects bacteria against viral infection. Nature. 2019;574(7780):
59. Ofir G, Melamed S, Sberro H, et al. DISARM is a widespread bacterial defence system with broad anti-phage activities. Nat Microbiol. 2018;3(1):90–98. doi:10.1038/s41564-017-0051-0
60. Molineux IJ. Host-parasite interactions: recent developments in the genetics of abortive phage infections. New Biol. 1991;3(3):230–236.
61. Chopin MC, Chopin A, Bidnenko E. Phage abortive infection in lactococci: variations on a theme. Curr Opin Microbiol. 2005;8(4):473–479. doi:10.1016/j.mib.2005.06.006
62. Briandet R, Lacroix-Gueu P, Renault M, et al. Fluorescence correlation spectroscopy to study diffusion and reaction of bacteriophages inside biofilms. Appl Environ Microbiol. 2008;74(7):2135–2143. doi:10.1128/AEM.02304-07
63. Vidakovic L, Singh PK, Hartmann R, Nadell CD, Drescher K. Dynamic biofilm architecture confers individual and collective mechanisms of viral protection. Nature Microbiology. 2018;3:1. doi:10.1038/s41564-017-0050-1
64. Kim MS, Kim YD, Hong SS, Park K, Ko KS, Myung H. Phage-encoded colanic acid-degrading enzyme permits lytic phage infection of a capsule-forming resistant mutant escherichia coli strain. Appl Environ Microbiol. 2015;81(3):900–909.
65. Milho C, Silva MD, Alves D, et al. Escherichia coli and Salmonella Enteritidis dual-species biofilms: interspecies interactions and antibiofilm efficacy of phages. Sci Rep. 2019;9:9. doi:10.1038/s41598-019-54847-y
66. Tan D, Zhang Y, Cheng M, et al. Characterization of Klebsiella pneumoniae ST11 isolates and their interactions with lytic phages. Viruses-Basel. 2019;11:11.
67. Testa S, Berger S, Piccardi P, Oechslin F, Resch G, Mitri S. Spatial structure affects phage efficacy in infecting dual-strain biofilms of Pseudomonas aeruginosa. Communications Biol. 2019;2. doi:10.1038/s42003-019-0633-x
68. Secor PR, Sweere JM, Michaels LA, et al. Filamentous bacteriophage promote biofilm assembly and function. Cell Host Microbe. 2015;18(5):549–559. doi:10.1016/j.chom.2015.10.013
69. Lee Y, Song S, Sheng L, Zhu L, Kim J-S, Wood TK. Substrate binding protein DppA1 of ABC transporter DppBCDF increases biofilm formation in pseudomonas aeruginosa by inhibiting pf5 prophage lysis. Front Microbiol. 2018;24:9.
70. Rice SA, Tan CH, Mikkelsen PJ, et al. The biofilm life cycle and virulence of Pseudomonas aeruginosa are dependent on a filamentous prophage. Isme Journal. 2009;3(3):271–282. doi:10.1038/ismej.2008.109
71. Davies EV, James CE, Williams D, et al. Temperate phages both mediate and drive adaptive evolution in pathogen biofilms. Proc Nat Acad Sci United States America. 2016;113(29):8266–8271. doi:10.1073/pnas.1520056113
72. Bull JJ, Christensen KA, Scott C, Jack BR, Crandall CJ, Krone SM. Phage-bacterial dynamics with spatial structure: self organization around phage sinks can promote increased cell densities. Antibiotics-Basel. 2018;7:1.
73. Tan D, Dahl A, Middelboe M. Vibriophages differentially influence biofilm formation by vibrio anguillarum strains. Appl Environ Microbiol. 2015;81(13):4489–4497. doi:10.1128/AEM.00518-15
74. Dancer SJ. The effect of antibiotics on methicillin-resistant Staphylococcus aureus. J Antimicrobial Chemother. 2008;61(2):246–253. doi:10.1093/jac/dkm465
75. Cepas V, López Y, Muñoz E, et al. Relationship between biofilm formation and antimicrobial resistance in gram-negative bacteria. Microb Drug Resist. 2019;25(1):72–79. doi:10.1089/mdr.2018.0027
76. Pires DP, Melo LDR, Boas DV, Sillankorva S, Azeredo J. Phage therapy as an alternative or complementary strategy to prevent and control biofilm-related infections. Curr Opin Microbiol. 2017;39:48–56. doi:10.1016/j.mib.2017.09.004
77. Moulton-Brown CE, Friman V-P. Rapid evolution of generalized resistance mechanisms can constrain the efficacy of phage-antibiotic treatments. Evolutionary Applications. 2018;11(9):1630–1641. doi:10.1111/eva.12653
78. Ormälä A-M, Jalasvuori M. Phage therapy: should bacterial resistance to phages be a concern, even in the long run? Bacteriophage. 2013;3(1):e24219–e24219. doi:10.4161/bact.24219
79. Cesta N, Di Luca M, Corbellino M, Tavio M, Galli M, Andreoni M. Bacteriophage therapy: an overview and the position of italian society of infectious and tropical diseases. Infez Med. 2020;28(3):322–331.
80. Nzakizwanayo J, Hanin A, Alves DR, et al. Bacteriophage can prevent encrustation and blockage of urinary catheters by proteus mirabilis. Antimicrob Agents Chemother. 2016;60(3):1530–1536. doi:10.1128/AAC.02685-15
81. Cano EJ, Caflisch KM, Bollyky PL, et al. Phage therapy for limb-threatening prosthetic knee klebsiella pneumoniae infection: case report and in vitro characterization of anti-biofilm activity. Clin Infect Dis. 2020. doi:10.1093/cid/ciaa705
82. Shlezinger M, Khalifa L, Houri-Haddad Y, et al. Phage therapy: a new horizon in the antibacterial treatment of oral pathogens. Curr Top Med Chem. 2017;17(10):1199–1211. doi:10.2174/1568026616666160930145649
83. Shlezinger M, Friedman M, Houri-Haddad Y, Hazan R, Beyth N. Phages in a thermoreversible sustained-release formulation targeting E. faecalis in vitro and in vivo. PLoS One. 2019;14(7):e0219599. doi:10.1371/journal.pone.0219599
84. Khalifa L, Brosh Y, Gelman D, et al. Targeting Enterococcus faecalis biofilms with phage therapy. Appl Environ Microbiol. 2015;81(8):2696–2705. doi:10.1128/AEM.00096-15
85. Selezska K, Kazmierczak M, Muesken M, et al. Pseudomonas aeruginosa population structure revisited under environmental focus: impact of water quality and phage pressure. Environ Microbiol. 2012;14(8):1952–1967.
86. Alves DR, Perez-Esteban P, Kot W, et al. A novel bacteriophage cocktail reduces and disperses Pseudomonas aeruginosa biofilms under static and flow conditions. Microb Biotechnol. 2016;9(1):61–74.
87. Burrows LL. The therapeutic pipeline for pseudomonas aeruginosa Infections. ACS Infectious Diseases. 2018;4(7):1041–1047.
88. Fong SA, Drilling AJ, Ooi ML, et al. Safety and efficacy of a bacteriophage cocktail in an in vivo model of Pseudomonas aeruginosa sinusitis. Translational Res. 2019;206:41–56.
89. Alemayehu D, Casey PG, McAuliffe O, et al. Bacteriophages phi MR299-2 and phi NH-4 can eliminate pseudomonas aeruginosa in the murine lung and on cystic fibrosis lung airway cells. Mbio. 2012;3:2.
90. Coulter LB, McLean RJC, Rohde RE, Aron GM. Effect of bacteriophage infection in combination with tobramycin on the emergence of resistance in escherichia coli and pseudomonas aeruginosa biofilms. Viruses-Basel. 2014;6(10):3778–3786.
91. Kumaran D, Taha M, Yi Q, et al. Does treatment order matter? investigating the ability of bacteriophage to augment antibiotic activity against staphylococcus aureus biofilms. Front Microbiol. 2018;9:127.
92. Shlezinger M, Coppenhagen-Glazer S, Gelman D, Beyth N, Hazan R. Eradication of vancomycin-resistant enterococci by combining phage and vancomycin. Viruses-Basel. 2019;11:10.
93. Lu TK, Collins JJ. Engineered bacteriophage targeting gene networks as adjuvants for antibiotic therapy. Proc Nat Acad Sci United States America. 2009;106(12):4629–4634.
94. Srinivasan R, Chaitanyakumar A, Subramanian P, et al. Recombinant engineered phage-derived enzybiotic in Pichia pastoris X-33 as whole cell biocatalyst for effective biocontrol of Vibrio parahaemolyticus in aquaculture. International J Biol Macromolecules. 2019.
95. Yilmaz C, Colak M, Yilmaz BC, Ersoz G, Kutateladze M, Gozlugol M. Bacteriophage therapy in implant-related infections an experimental study. J Bone Joint Surgery-Am. 2013;95A(2):117–125.
96. Chang RYK, Das T, Manos J, Kutter E, Morales S, Chan HK. Bacteriophage PEV20 and ciprofloxacin combination treatment enhances removal of pseudomonas aeruginosa biofilm isolated from cystic fibrosis and wound patients. AAPS J. 2019;21(3):49.
97. Lin Y, Chang RYK, Britton WJ, Morales S, Kutter E, Chan HK. Synergy of nebulized phage PEV20 and ciprofloxacin combination against Pseudomonas aeruginosa. Int J Pharm. 2018;551(1–2):158–165.
98. Abedon ST. Phage-antibiotic combination treatments: antagonistic impacts of antibiotics on the pharmacodynamics of phage therapy? Antibiotics. 2019;8(4):182.
99. Casey E, van Sinderen D, Mahony J. In vitro characteristics of phages to guide ‘real life’ phage therapy suitability. Viruses. 2018;10:4.
100. Tagliaferri TL, Jansen M, Horz HP. Fighting pathogenic bacteria on two fronts: phages and antibiotics as combined strategy. Front Cell Infect Microbiol. 2019;9:22. doi:10.3389/fcimb.2019.00022
101. Latz S, Kruettgen A, Haefner H, Buhl EM, Ritter K, Horz H-P. Differential effect of newly isolated phages belonging to PB1-Like, phiKZ-Like and LUZ24-like viruses against multi-drug resistant pseudomonas aeruginosa under varying growth conditions. Viruses-Basel. 2017;9:11.
102. Forti F, Roach DR, Cafora M, et al. Design of a broad-range bacteriophage cocktail that reduces pseudomonas aeruginosa biofilms and treats acute infections in two animal models. Antimicrob Agents Chemother. 2018;62:6. doi:10.1128/AAC.02573-17
103. Alves DR, Gaudion A, Bean JE, et al. Combined use of bacteriophage k and a novel bacteriophage to reduce staphylococcus aureus biofilm formation. Appl Environ Microbiol. 2014;80(21):6694–6703. doi:10.1128/AEM.01789-14
104. Schuch R, Khan BK, Raz A, Rotolo JA, Wittekind M. Bacteriophage lysin CF-301, a potent antistaphylococcal biofilm agent. Antimicrob Agents Chemother. 2017;61:7.
105. Paul VD, Rajagopalan SS, Sundarrajan S, et al. A novel bacteriophage Tail-Associated Muralytic Enzyme (TAME) from Phage K and its development into a potent antistaphylococcal protein. Bmc Microbiology. 2011;11.
106. Poonacha N, Nair S, Desai S, et al. Efficient killing of planktonic and biofilm-embedded coagulase-negative staphylococci by bactericidal protein P128. Antimicrob Agents Chemother. 2017;61:8. doi:10.1128/AAC.00457-17
107. Channabasappa S, Durgaiah M, Chikkamadaiah R, Kumar S, Joshi A, Sriram B. Efficacy of novel antistaphylococcal ectolysin P128 in a rat model of methicillin-resistant staphylococcus aureus bacteremia. Antimicrob Agents Chemother. 2018;62:2.
108. Diez-Martinez R, De Paz HD, Garcia-Fernandez E, et al. A novel chimeric phage lysin with high in vitro and in vivo bactericidal activity against Streptococcus pneumoniae. J Antimicrobial Chemother. 2015;70(6):1763–1773.
109. Meng X, Shi Y, Ji W, et al. Application of a bacteriophage lysin to disrupt biofilms formed by the animal pathogen streptococcus suis. Appl Environ Microbiol. 2011;77(23):8272–8279. doi:10.1128/AEM.05151-11
110. Rios AC, Moutinho CG, Pinto FC, et al. Alternatives to overcoming bacterial resistances: state-of-the-art. Microbiological Research. 2016;2016(191):51–80. doi:10.1016/j.micres.2016.04.008
111. Lood R, Winer BY, Pelzek AJ, et al. Novel phage lysin capable of killing the multidrug-resistant gram-negative bacterium acinetobacter baumannii in a mouse bacteremia model. Antimicrob Agents Chemother. 2015;59(4):1983–1991. doi:10.1128/AAC.04641-14
112. Thandar M, Lood R, Winer BY, Deutsch DR, Euler CW, Fischetti VA. Novel engineered peptides of a phage lysin as effective antimicrobials against multidrug-resistant acinetobacter baumannii. Antimicrob Agents Chemother. 2016;60(5):2671–2679. doi:10.1128/AAC.02972-15
113. Yang H, Bi Y, Shang X, et al. Antibiofilm activities of a novel chimeolysin against streptococcus mutans under physiological and cariogenic conditions. Antimicrob Agents Chemother. 2016;60(12):7436–7443.
114. Vazquez R, Garcia P. Synergy between two chimeric lysins to kill streptococcus pneumoniae. Front Microbiol. 2019;10:10. doi:10.3389/fmicb.2019.01251
115. Muziasari WI, Pitkanen LK, Sorum H, Stedtfeld RD, Tiedje JM, Virta M. The resistome of farmed fish feces contributes to the enrichment of antibiotic resistance genes in sediments below baltic sea fish farms. Front Microbiol. 2017;7.
116. Wang J, Liu Q, Wu B, et al. Quorum sensing signaling distribution during the development of full-scale municipal wastewater treatment biofilms. Sci Total Environ. 2019;685:28–36. doi:10.1016/j.scitotenv.2019.05.249
117. Zhang Y, Hu Z. Combined treatment of Pseudomonas aeruginosa biofilms with bacteriophages and chlorine. Biotechnol Bioengineering. 2013;110(1):286–295. doi:10.1002/bit.24630
118. Goldman G, Starosvetsky J, Armon R. Inhibition of biofilm formation on UF membrane by use of specific bacteriophages. J Membrane Sci. 2009;342(1–2):145–152. doi:10.1016/j.memsci.2009.06.036
119. Cabello FC, Godfrey HP, Tomova A, et al. Antimicrobial use in aquaculture re-examined: its relevance to antimicrobial resistance and to animal and human health. Environ Microbiol. 2013;15(7):1917–1942.
120. Tomova A, Ivanova L, Buschmann AH, et al. Antimicrobial resistance genes in marine bacteria and human uropathogenic Escherichia coli from a region of intensive aquaculture. Environmental Microbiology Reports. 2015;7(5):803–809. doi:10.1111/1758-2229.12327
121. Maje MD, Kaptchouang Tchatchouang CD, Manganyi MC, Fri J, Ateba CN. Characterisation of vibrio species from surface and drinking water sources and assessment of biocontrol potentials of their bacteriophages. Int J Microbiol. 2020;2020:8863370. doi:10.1155/2020/8863370
122. Matamp N, Bhat SG. Phage endolysins as potential antimicrobials against multidrug resistant vibrio alginolyticus and vibrio parahaemolyticus: current status of research and challenges ahead. Microorganisms. 2019;7:3. doi:10.3390/microorganisms7030084
123. Coughlan LM, Cotter PD, Hill C, Alvarez-Ordonez A. New weapons to fight old enemies: novel strategies for the (bio) control of bacterial biofilms in the food industry. Front Microbiol. 2016;7. doi:10.3389/fmicb.2016.01641
124. Colagiorgi A, Bruini I, Di Ciccio PA, Zanardi E, Ghidini S, Ianieri A. Listeria monocytogenes biofilms in the wonderland of food industry. Pathogens. 2017;6:3. doi:10.3390/pathogens6030041
125. Galie S, Garcia-Gutierrez C, Miguelez EM, Villar CJ, Lombo F. Biofilms in the food industry: health aspects and control methods. Front Microbiol. 2018;9:9. doi:10.3389/fmicb.2018.00898
126. Van Houdt R, Michiels CW. Biofilm formation and the food industry, a focus on the bacterial outer surface. J Appl Microbiol. 2010;109(4):1117–1131. doi:10.1111/j.1365-2672.2010.04756.x
127. Gray JA, Chandry PS, Kaur M, Kocharunchitt C, Bowman JP, Fox EM. Novel biocontrol methods for listeria monocytogenes biofilms in food production facilities. Front Microbiol. 2018;9:9. doi:10.3389/fmicb.2018.00605
128. Iacumin L, Manzano M, Comi G. Phage inactivation of listeria monocytogenes on san daniele dry-cured ham and elimination of biofilms from equipment and working environments. Microorganisms. 2016;4:1. doi:10.3390/microorganisms4010004
129. Soni KA, Nannapaneni R. Removal of listeria monocytogenes biofilms with bacteriophage P100. J Food Prot. 2010;73(8):1519–1524. doi:10.4315/0362-028X-73.8.1519
130. Arachchi GJG, Cridge AG, Dias-Wanigasekera BM, et al. Effectiveness of phages in the decontamination of Listeria monocytogenes adhered to clean stainless steel, stainless steel coated with fish protein, and as a biofilm. J Ind Microbiol Biotechnol. 2013;40(10):1105–1116. doi:10.1007/s10295-013-1313-3
131. Gutierrez D, Rodriguez-Rubio L, Martinez B, Rodriguez A, Garcia P. Bacteriophages as weapons against bacterial biofilms in the food industry. Front Microbiol. 2016;7:7. doi:10.3389/fmicb.2016.00825
132. Pires DP, Dötsch A, Anderson EM, et al. A genotypic analysis of five P. aeruginosa strains after biofilm infection by phages targeting different cell surface receptors. Front Microbiol. 2017;8:1229. doi:10.3389/fmicb.2017.01229
133. Sumrall ET, Shen Y, Keller AP, et al. Phage resistance at the cost of virulence: listeria monocytogenes serovar 4b requires galactosylated teichoic acids for InlB-mediated invasion. PLoS Pathogens. 2019;15(10):e1008032. doi:10.1371/journal.ppat.1008032
134. Novick RP. Pathogenicity islands and their role in staphylococcal biology. Gram-Positive Pathogens. 2019;536–548.
135. Koskella B, Brockhurst MA. Bacteria-phage coevolution as a driver of ecological and evolutionary processes in microbial communities. FEMS Microbiol Rev. 2014;38(5):916–931. doi:10.1111/1574-6976.12072
136. Xiong X, Wu G, Wei Y, et al. SspABCD–SspE is a phosphorothioation-sensing bacterial defence system with broad anti-phage activities. Nature Microbiology. 2020;1–12.
137. Stanley SY, Maxwell KL. Phage-encoded anti-CRISPR defenses. Annu Rev Genet. 2018;52:445–464. doi:10.1146/annurev-genet-120417-031321
138. Sharma U, Vipra A, Channabasappa S. Phage-derived lysins as potential agents for eradicating biofilms and persisters. Drug Discov Today. 2018;23(4):848–856. doi:10.1016/j.drudis.2018.01.026
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.