Back to Journals » Cancer Management and Research » Volume 12
B3GNT3, a Direct Target of miR-149-5p, Promotes Lung Cancer Development and Indicates Poor Prognosis of Lung Cancer
Authors Sun Y, Liu T, Xian L, Liu W, Liu J, Zhou H
Received 29 October 2019
Accepted for publication 24 February 2020
Published 31 March 2020 Volume 2020:12 Pages 2381—2391
DOI https://doi.org/10.2147/CMAR.S236565
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Yong Teng
Yu Sun,1,* Tao Liu,1,* Lei Xian,1 Wenzhou Liu,1 Jun Liu,2 Huafu Zhou2
1Department of Cardio-Thoracic Surgery, The Second Affiliated Hospital of Guangxi Medical University, Guangxi, People’s Republic of China; 2Department of Cardio-Thoracic Surgery, The First Affiliated Hospital of Guangxi Medical University, Guangxi, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Huafu Zhou
Department of Cardio-Thoracic Surgery, The First Affiliated Hospital of Guangxi Medical University, Guangxi, People’s Republic of China
Email [email protected]
Background: B3GNT3 (β 1, 3-N-acetylglucosaminyltransferase-3) belongs to the β 3GlcNAcT family and is essential to form extended core 1 oligosaccharides. Previous studies revealed that B3GNT3 expression was dysregulated in multiple cancers. Here, we aimed to understand the expression profile and function of B3GNT3 in lung cancer.
Materials and Methods: The expression of B3GNT3 was measured by immunohistochemistry and public database analysis. B3GNT3 was knocked down to evaluate the lung cancer cell proliferation, migration and invasion in in vitro and in vivo tumor formation experiments. miR-149-5p targeting B3GNT3 was identified with TargetScan analysis and confirmed with reporter assay. Overexpression of miR-149-5p was achieved using microRNA mimics and function of microRNA-149-5p/B3GNT3 axis was tested in vitro.
Results: B3GNT3 was upregulated in lung cancer, and B3GNT3 overexpression was associated with poor prognosis of lung cancer patients. High expression of B3GNT3 was associated with advanced TNM stages, larger tumor size, tumor metastasis and recurrence. Functionally, we demonstrated that knockdown of B3GNT3 suppressed lung cancer cell growth and invasion in vitro. Knockdown of B3GNT3 suppressed lung cancer development in a xenograft tumor model. Moreover, miR-149-5p was validated to negatively regulate B3GNT3 expression through directly targeting B3GNT3 3ʹ-UTR. Overexpression of miR-149-5p could antagonize the tumorigenesis effect of B3GNT3 in vitro.
Conclusion: In summary, our study demonstrated that B3GNT3 overexpression was correlated with poor prognosis of lung cancer patient, indicating that B3GNT3 could be a promising prognostic biomarker for lung cancer. miR-149-5p negatively regulated B3GNT3 expression, which might be utilized for therapeutic target in lung cancer.
Keywords: B3GNT3, lung cancer, miR-149-5p, tumor progression
Introduction
Lung cancer is one of the most common causes of cancer-related death, which has high morbidity and mortality worldwide.1 Though great advance has been achieved during the past decades regarding the new therapeutic strategies such as surgery and adjuvant therapy, lung cancer prognosis remains poor due to the high chance of lung cancer recurrence and metastasis.2,3 Thus, it is crucial to identify new biomarkers and therapeutic targets for lung cancer treatment.
B3GNT3 (β1, 3-N-acetylglucosaminyltransferase-3) belongs to the β3GlcNAcT family and is essential to form extended core 1 oligosaccharides.4,5 Multiple β3GlcNAcT family members are demonstrated to be involved in tumor development and progression.6,7 B3GNT3, which is constitutively expressed in neutrophils, lymphocytes, colon, stomach and small intestine, has been discovered playing in crucial roles in various tumors.4,8,9 Mounting evidence has shown that B3GNT3 functions as an oncogene and promotes tumor development and metastasis, such as cervical cancer, pancreatic cancer.10,11 Meanwhile, B3GNT3 impeded tumor cell migration and invasion in neuroblastoma, which indicated its tumor suppressor role.9 In non-small cell lung cancer patients, upregulated B3GNT3 expression was significantly correlated with poor overall survival.12 However, the detailed function of B3GNT3 and how B3GNT3 expression is regulated in lung cancer remains unclear.
MicroRNAs (miRNAs) are a group of small non-coding RNAs, post-transcriptionally regulating target gene expression via base-pairing to the target gene mRNAs.13 Accumulating evidence has shown that miRNAs have important roles in lung cancer development, progression and metastasis.14,15 MicroRNAs, such as miR-494, miR-153 and miR-218, inhibited lung cancer development and metastasis via suppressing cellular proliferation, inhibiting cell migration and invasion and EMT.16–18
Here, we found that B3GNT3 was overexpressed in lung cancer and upregulated expression of B3GNT3 was associated with unfavorable prognosis of lung cancer patients. Silencing B3GNT3 inhibited lung cancer cell growth in vitro and suppressed lung tumor development in vivo. We further identified miR-149-5p negatively regulated B3GNT3 via targeting its 3ʹ-UTR. In summary, our data indicate that miR-149-5p/B3GNT3 could be utilized as a potential therapeutic target in lung cancer.
Materials and Methods
Human Specimen
120 paired lung cancer tissue sections and adjacent non-cancer tissues were obtained via surgical section at the Second Affiliated Hospital of Guangxi Medical University. All patients provided written informed consent. The study was reviewed and approved by the Research Ethics Committee of Guangxi Medical University and was carried out following the principles of the Declaration of Helsinki. The clinicopathological features of 120 lung cancer patients were shown in Supplementary Table 1.
Cell Culture
Lung cancer cell line (A549, H1299), and HEK293 were from Cell Bank of Shanghai (Shanghai, China) and maintained in DMEM cell culture medium containing 10% FBS, 1% streptomycin and penicillin (all from Life Technologies, CA, USA) in cell culture incubator (5% CO2, 37°C).
qRT-PCR
QRT-PCR was conducted as previously described.19 Primers used were listed as following: B3GNT3 Forward primer: AGGCACAGACTCACGGAGACAT; Reverse primer: GTTGAGCACGAAGCTGGCGTTG; GAPDH Forward primer: GTCTCCTCTGACTTCAACAGCG; Reverse primer: ACCACCCTGTTGCTGTAGCCAA; PCR primers for detecting miR-149-5p and U6 RNA were obtained from Ribobio. Relative B3GNT3 or miR-149-5p expression was normalized to GAPDH or U6 RNA level using the 2−ΔΔCt method, respectively.
siRNA/shRNA/miRNA Mimics or Inhibitor Transfection
B3GNT3 siRNA or shRNA vectors and relative negative controls, miR-149-5p mimics or miR-149-5p inhibitor was purchased from GenePharma (Shanghai, China). Lipofectamine 3000 (Thermo fisher Scientific, CA USA) was utilized for cell transfection. The sequences of synthetic oligonucleotides about miR-149-5p mimics, inhibitor and relative negative controls in Supplementary Table 2.
Western Blot Analysis
Western blot was conducted as previously described.19 Antibodies used in this study were listed as following: anti-B3GNT3 (ab96267, Abcam, United Kingdom), anti-GAPDH (ab8245, Abcam, United Kingdom).
Cell Growth Assays
Cell growth was assessed using CCK-8 assay, EdU staining assay and colony formation assays as previously described.20
Transwell Assay
Cell invasion was assessed using transwell assay. Briefly, 2 × 104 cells were seeded into the top chambers with Matrigel-coated membrane (Corning, USA) in 100 μL serum-free medium. Bottom chamber was filled with complete medium. Cells were cultured for 36 h and then cells invaded to the lower chamber were counted under a microscope with 0.5% crystal violet staining.
Tissue Microarray and Immunohistochemistry
Tissue microarrays (TMAs) were established with lung cancer tissues and paired normal tissues. Immunohistochemical staining (IHC) was carried out on sections using antibodies against B3GNT3 (1:100, ab96267, Abcam, Cambridge, United Kingdom) and Ki-67 (D2H10, Cell signalling technology, USA) as previously described.21 Sections were semi-quantitatively scored for the B3GNT3 staining patterns as follows: the staining extent in each sample was scored as 1+ (<25% staining region), 2+ (25–50% staining region), 3+ (50% to 75% staining region), or 4+ (>75% staining region). The staining intensity was quantified as 0 (negative), 1+ (weak), 2+ (intermediate), or 3+ (strong). The final score was obtained by multiplying the intensity and extension values (range 0–12) and the samples were grouped as 1+ (score 0), 2+ (score 1–2), 3+ (score 3–4), 4+ (score 6–8), and 5+ (score 9–12) staining. Meanwhile, for statistical purposes, scores of 4+ and 5+ were defined as B3GNT3 high expression and the other final scores were considered as B3GNT3 low expression. For quantification of relative IHC staining intensity of B3GNT3 and Ki-67 in mouse xenograft tumors, we used the ImageJ software (NIH, USA) to quantify the staining intensity and set the control group (sh-NC) as relative staining intensity of 1. IHC staining intensity was scored and categorized by two independent investigators.
Luciferase Reporter Assay
B3GTN3 WT or mutated 3ʹ-UTR sequences were constructed into the luciferase reporter vector (Genecopoeia, Rockville, MD, USA). HEK293 cells were transfected with reporter vectors containing WT or mutated 3ʹ-UTR of B3GTN3 sequences, together with negative control or miR-149-5p mimics. 48 hrs later, the relative luciferase activity was analyzed as previously described.19
Bioinformatics Analysis
Lung cancer patient datasets in The Cancer Genome Atlas (TCGA) were analysed and mRNA expression profiles of B3GNT3 and patient-related clinical information were obtained and analysed as previously described.22 Overall survival (OS) and disease-free survival (DFS) were examined by Kaplan-Meier survival analysis and Log-rank tests.
Xenograft Tumor Model
BALB/c nude mice (5–6 weeks) were obtained from Shanghai Laboratory Animal Center (Shanghai, China). 5 × 106 lung cancer A549 control cells or with B3GNT3 stable knockdown were subcutaneously injected into nude mice. Tumor size was measured with a vernier caliper and the tumor volume was determined (length×width2×1/2). Tumorigenesis was also recorded by a bioluminescence imaging system (Caliper, USA). The xenograft tumor experiment was approved by the Experimental Animal Ethics Committee of Guangxi Medical University. In addition, Guide for the Care and Use of Laboratory Animals (8th edition) was strictly followed in the present study.
Statistics
GraphPad Prism 6 was used for statistical analysis. All the results were showed as mean ± standard deviation (SD). Student’s t-test or one-way analysis of variance was used for calculation of p values where necessary. Statistical significance was defined as a p value < 0.05.
Results
B3GNT3 is Overexpressed in Lung Cancer Tissues and Upregulated Expression of B3GNT3 Expression is Associated with Unfavorable Prognosis in TCGA Lung Cancer Cohort
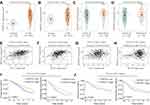
We first analyzed the B3GNT3 expression pattern in lung cancers by bioinformatics analysis of TCGA cohort. B3GNT3 expression was significantly higher in lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC) than that in control tissues in TCGA cohort (Figure 1A and B). Furthermore, lung cancers with advanced TNM stages (TNM III–IV) had markedly higher B3GNT3 expression than that in lung cancers with TNM I–II (Figure 1C and D). In addition, B3GNT3 expression was positively correlated with the expression of Ki-67 and PCNA (Figure 1E–H). We further investigated patient prognosis in lung TCGA LUAD cohort and TCGA LUSC cohort. Lung cancer patients with high B3GNT3 expression exhibited statistically lower OS and DFS rates than patients with low B3GNT3 expression (Figure 1I and J).
High Expression Level of B3GNT3 is Correlated with Advanced Lung Cancer Stages and Unfavorable Prognosis in TMA Cohort
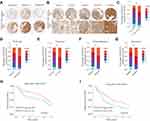
To further validate the level of B3GNT3 and the correlation of B3GNT3 levels with prognosis, we analyzed B3GNT3 expression in tissue microarray (TMA) containing lung cancer tissues and control tissues. Immunohistochemical staining (IHC) showed that B3GNT3 expression was remarkably enhanced in lung cancer tissue sections (Figure 2A). We established a scoring system for measuring the expression levels of B3GNT3 based on the IHC staining intensity of B3GNT3 (Figure 2B). Lung cancer tissues had markedly higher B3GNT3 staining intensity scores than that in normal tissues (Figure 2C). Moreover, our analysis revealed that B3GNT3 expression was remarkably enhanced in lung cancers with late TNM stages (Figure 2D), bigger tumor size, distant metastasis and recurrence (Figure 2D–G). Consistently, lung cancer patients with high B3GNT3 expression exhibited markedly lower OS and DFS rates than that in lung cancer patients with low level of B3GNT3 in TMA cohort (Figure 2H and I). Taken together, our results suggested that high B3GNT3 expression was associated with unfavorable prognosis in lung cancer patients.
Silencing B3GNT3 Inhibits Lung Cancer Cell Growth and Invasion in vitro
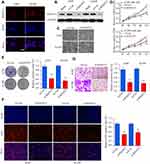
Immunofluorescence staining of B3GNT3, together with DAPI staining, was conducted to reveal the cellular localization of B3GNT3. As shown in Figure 3A, B3GNT3 was mainly distributed in the cytosol of A549 or H1299 cells. To further investigate the function of B3GNT3, we used siRNA targeting B3GNT3 to silence the expression of B3GNT3. B3GNT3 Knockdown was confirmed by Western blot in A549 or H1299 cells (Figure 3B). Functionally, CCK-8 cell proliferation assay demonstrated that knockdown of B3GNT3 markedly inhibited lung cancer cell proliferation in A549 or H1299 cells (Figure 3C and D). In addition, knockdown of B3GNT3 showed markedly decreased colony formation and DNA synthesis in lung cancer cells (Figure 3E and F). Furthermore, cell invasive ability of lung cancer cell A549 or H1299 was significantly impeded by B3GNT3 knockdown (Figure 3G). Collectively, our findings indicated that B3GNT3 knockdown suppressed lung cancer cell growth and invasion in vitro.
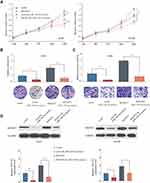
Knockdown of B3GNT3 Suppresses Lung Cancer Development in vivo
To dissect the role of B3GNT3 in vivo, A549 cells with stable B3GNT3 knockdown or control cells were injected into nude mice subcutaneously to establish xenograft lung tumor model. Knockdown of B3GNT3 significantly impeded the lung tumor growth, with much smaller size and less tumor weight in sh-B3GNT3 group compared with that in sh-NC group (Figure 4A–4D). Moreover, we analyzed the B3GNT3 and proliferation marker Ki-67 expression in tumor sections by IHC staining. While B3GNT3 expression was markedly lower in sh-B3GNT3 group, Ki-67 expression was also significantly decreased after B3GNT3 knockdown (Figure 4E and F). In summary, B3GNT3 knockdown could suppress lung cancer development in vivo.
B3GNT3 Is Negatively Regulated by miR-149-5p
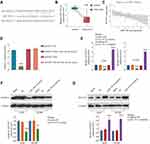
To understand how B3GNT3 expression is regulated in lung cancer, we conducted bioinformatics analysis to seek for the miRNAs targeting B3GNT3. MiR-149-5p was predicted to have the complementary binding sequences against 3ʹ-UTR of B3GNT3 (Figure 5A). MiR-149-5p was demonstrated to have dual roles in tumor, function as either tumor suppressor or oncogene.23 MiR-149-5p expression profile was analyzed in lung cancer tissue and control tissues and the result exhibited that lung cancer tissues had significantly lower levels of miR-149-4p (Figure 5B). Intriguingly, Pearson correlation analysis revealed that B3GNT3 expression was negatively associated with miR-149-5p expression (Figure 5C). We further confirmed the regulation between B3GNT3 and miR-149-5p via luciferase reporter assay. MiR-149-5p mimics specifically suppressed the luciferase activity in HEK293 with luciferase reporter vector containing WT 3ʹ-UTR of B3GNT3, but not with mutated 3ʹ-UTR of B3GNT3 (Figure 5D). Moreover, we demonstrated that miR-149-5p overexpression dampened B3GNT3 expression, while inhibition of miR-149-5p enhanced B3GNT3 expression (Figure 5E–G). These results suggested that miR-149-5p might suppress the expression of B3GNT3 via binding to 3ʹ-UTR of B3GNT3.
MiR-149-5p Antagonizes the Tumorigenesis Effect of B3GNT3 in vitro
To further evaluate miR-149-5p/B3GNT3 axis in regulating lung cancer tumor progression, A549 or H1299 cells were transfected with control vector, miR-149-5p mimics, pcDNA3.1-B3GNT3, or miR-149-5p mimics+pcDNA3.1-B3GNT3. Overexpression miR-149-5p suppressed cell growth and B3GNT3 overexpression enhanced cell proliferation in A549 or H1299 cells (Figure 6A and B). MiR-149-5p mimics abrogated the promotion of cell proliferation induced by B3GNT3 overexpression (Figure 6A and B). Further, transwell experiments showed that B3GNT3 overexpression enhanced cell invasion while miR-149-5p mimics reversed the tumorigenesis effect of B3GNT3 overexpression in A549 cells (Figure 6C). Consistently, we demonstrated miR-149-5p mimics inhibited B3GNT3 expression in A549 or H1299 cells, and overexpression B3GNT3 together with miR-149-5p mimics transfection partially rescued B3GNT3 expression (Figure 6D). Collectively, our data supported that miR-149-5p antagonized the tumorigenesis effect of B3NGT3 in vitro.
Discussion
B3GNT3 is constitutively expressed in various normal tissues including stomach, small intestine, colon and placenta.4 The function of B3GNT3 has been studied in the regulation of tumorigenesis in multiple cancers, such as neuroblastoma, pancreatic cancer, cervical cancer, and non-small cell lung cancer.7,9–12 While B3GNT3 expression is highly upregulated in most of malignant tumors and associated with tumor progression, malignant transformation, and metastasis,8,24,25 B3GNT3 was also reported to suppress the metastasis of neuroblastoma cells and predict favorable outcomes.9 In this study, we revealed the overexpression pattern of B3GNT3 in lung cancers by both bioinformatics analyzing TCGA lung cancer cohort and IHC analyzing lung cancer tissue microarray. Moreover, we verified that upregulated expression of B3GNT3 was correlated with unfavorable prognosis in lung cancer.
Our results revealed that silencing B3GNT3 inhibited lung cancer cell growth and invasion in vitro, and impeded lung tumor development in xenograft tumor model in vivo. Though the underlying mechanisms by which B3GNT3 promoted lung cancer development was not clear, previous studies indicated that B3GNT3 regulated the biosynthesis of L-selectin ligand and affected cancer cell trafficking.26 Other studies showed that B3GNT3 could catalyze PD-L1 glycosylation and enhance the immune checkpoint therapy in triple-negative breast cancer.27 These results indicated that B3GNT3 might have different roles and complicated function in tumor progression and prognosis, which needs further investigation.
We further identified miR-149-5p negatively regulated B3GNT3 expression by binding to its 3ʹ-UTR. MiR-149-5p was upregulated in prostate cancer, acute myeloid leukemia, and glioblastoma.28–30 Other studies demonstrated miR-149-5p functioned as tumor suppressor and was downregulated in gastric cancer, hepatocellular carcinoma and neuroblastoma.31–33 Ke et al showed that miR-149 suppressed lung cancer cell EMT via targeting FOXM1.34 In the current study, we identified that miR-149-5p was lower expressed in lung cancer than that in normal tissues and B3GNT3 expression was negatively correlated with the expression of miR-149-5p. Previous studies have reported that miR-149-5p regulated AKT1/mTOR signaling pathway and was sponged by circular RNA circNRIP1.35 Rui-Da Xu et al demonstrated that miR-149-5p directly regulated TNFRSF12A and inhibited human osteosarcoma development via regulating TWEAK/Fn14/PI3K/AKT pathway.36 Due to the complicated regulatory mechanisms of miRNA, miR-149-5p could have multiple targets involved in the regulation of lung cancer development and progression.
There are several limitations in our study that should be addressed in the future study. First, the underlying mechanism and signaling pathway by which B3GNT3 regulates lung cancer cell proliferation and invasion was not elucidated. Second, whether B3GNT3 is controlled by other miRNAs or whether miR-149-5p functions solely through negatively regulating B3GNT3 needs further clarification. In addition, it is worthwhile to further investigate the potential value of B3GNT3 as a prognosis biomarker.
In summary, for the first time, we have demonstrated that B3GNT3 is significantly upregulated in lung cancer both at mRNA and protein levels, and miR-149-5p negatively regulates B3GNT3 expression and suppresses lung cancer development. Thus, miR-149-5p/B3GNT3 axis might be further explored as a promising prognosis biomarker and therapeutic strategy in lung cancer.
Author Contributions
All authors contributed to data analysis, drafting or revising the article, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
The authors report no conflicts of interest in this work.
References
1. Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi:10.3322/caac.21262
2. Coory M, Gkolia P, Yang IA, Bowman RV, Fong KM. Systematic review of multidisciplinary teams in the management of lung cancer. Cancer. 2008;60(1):14–21. doi:10.1016/j.lungcan.2008.01.008
3. Debevec L, Debeljak A. Multidisciplinary management of lung cancer. J Thorac Oncol. 2007;2(6):577. doi:10.1097/JTO.0b013e318060f16d
4. Shiraishi N, Natsume A, Togayachi A, et al. Identification and characterization of three novel beta 1,3-N-acetylglucosaminyltransferases structurally related to the beta 1,3-galactosyltransferase family. J Biol Chem. 2001;276(5):3498–3507. doi:10.1074/jbc.M004800200
5. Yeh JC, Hiraoka N, Petryniak B, et al. Novel sulfated lymphocyte homing receptors and their control by a Core1 extension beta 1,3-N-acetylglucosaminyltransferase. Cell. 2001;105(7):957–969. doi:10.1016/S0092-8674(01)00394-4
6. Gromova I, Gromov P, Celis JE. A novel member of the glycosyltransferase family, beta 3 Gn-T2, highly downregulated in invasive human bladder transitional cell carcinomas. Mol Carcinog. 2001;32(2):61–72. doi:10.1002/mc.1065
7. Lu C-H, Wu W-Y, Lai Y-J, Yang C-M, Yu L-C. Suppression of B3GNT7 gene expression in colon adenocarcinoma and its potential effect in the metastasis of colon cancer cells. Glycobiology. 2014;24(4):359–367. doi:10.1093/glycob/cwu002
8. Liu Z, Shen L, Xu L, Sun X, Zhou J, Wu S. Down-regulation of beta-1,3-N-acetylglucosaminyltransferase-8 by siRNA inhibits the growth of human gastric cancer. Mol Med Rep. 2011;4(3):497–503. doi:10.3892/mmr.2011.448
9. Ho WL, Che MI, Chou CH, et al. B3GNT3 expression suppresses cell migration and invasion and predicts favorable outcomes in neuroblastoma. Cancer Sci. 2013;104(12):1600–1608. doi:10.1111/cas.12294
10. Barkeer S, Chugh S, Karmakar S, et al. Novel role of O-glycosyltransferases GALNT3 and B3GNT3 in the self-renewal of pancreatic cancer stem cells. BMC Cancer. 2018;18(1):1157. doi:10.1186/s12885-018-5074-2
11. Zhang W, Hou T, Niu C, Song L, Zhang Y. B3GNT3 expression is a novel marker correlated with pelvic lymph node metastasis and poor clinical outcome in early-stage cervical cancer. PLoS One. 2015;10(12):e0144360. doi:10.1371/journal.pone.0144360
12. Gao L, Zhang H, Zhang B, Zhu J, Chen C, Liu W. B3GNT3 overexpression is associated with unfavourable survival in non-small cell lung cancer. J Clin Pathol. 2018;71(7):642–647. doi:10.1136/jclinpath-2017-204860
13. Treiber T, Treiber N, Meister G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat Rev Mol Cell Biol. 2019;20(1):5–20. doi:10.1038/s41580-018-0059-1
14. Peng Y, Croce CM. The role of MicroRNAs in human cancer. Signal Transduct Target Ther. 2016;1:15004. doi:10.1038/sigtrans.2015.4
15. Castro D, Moreira M, Gouveia AM, Pozza DH, De Mello RA. MicroRNAs in lung cancer. Oncotarget. 2017;8(46):81679–81685. doi:10.18632/oncotarget.v8i46
16. Mao G, Liu Y, Fang X, et al. Tumor-derived microRNA-494 promotes angiogenesis in non-small cell lung cancer. Angiogenesis. 2015;18(3):373–382. doi:10.1007/s10456-015-9474-5
17. Yuan Y, Du W, Wang Y, et al. Suppression of AKT expression by miR-153 produced anti-tumor activity in lung cancer. Int J Cancer. 2015;136(6):1333–1340. doi:10.1002/ijc.29103
18. Shi ZM, Wang L, Shen H, et al. Downregulation of miR-218 contributes to epithelial-mesenchymal transition and tumor metastasis in lung cancer by targeting Slug/ZEB2 signaling. Oncogene. 2017;36(18):2577–2588. doi:10.1038/onc.2016.414
19. Luo Y, Sun R, Zhang J, Sun T, Liu X, Yang B. miR-506 inhibits the proliferation and invasion by targeting IGF2BP1 in glioblastoma. Am J Transl Res. 2015;7(10):2007–2014.
20. Zhou Y, Jin Z, Wang C. Glycogen phosphorylase B promotes ovarian cancer progression via Wnt/beta-catenin signaling and is regulated by miR-133a-3p. Biomed Pharmacother. 2019;120:109449. doi:10.1016/j.biopha.2019.109449
21. Peng S, Geng J, Sun R, Tian Z, Wei H. Polyinosinic-polycytidylic acid liposome induces human hepatoma cells apoptosis which correlates to the up-regulation of RIG-I like receptors. Cancer Sci. 2009;100(3):529–536. doi:10.1111/j.1349-7006.2008.01062.x
22. J C, Y Y, C X, et al. Low microRNA-139 expression associates with poor prognosis in patients with tumors: a meta-analysis. Hepatobiliary Pancreat Dis Int. 2018;undefined(undefined):undefined.
23. He Y, Yu D, Zhu L, Zhong S, Zhao J, Tang J. miR-149 in human cancer: a systemic review. J Cancer. 2018;9(2):375–388. doi:10.7150/jca.21044
24. Hua D, Qin F, Shen L, et al. beta3GnT8 regulates laryngeal carcinoma cell proliferation via targeting MMPs/TIMPs and TGF-beta1. Asian Pac J Cancer Prev. 2012;13(5):2087–2093. doi:10.7314/APJCP.2012.13.5.2087
25. Liu J, Shen L, Yang L, Hu S, Xu L, Wu S. High expression of beta3GnT8 is associated with the metastatic potential of human glioma. Int J Mol Med. 2014;33(6):1459–1468. doi:10.3892/ijmm.2014.1736
26. Witz IP. The selectin-selectin ligand axis in tumor progression. Cancer Metastasis Rev. 2008;27(1):19–30. doi:10.1007/s10555-007-9101-z
27. Li CW, Lim SO, Chung EM, et al. Eradication of triple-negative breast cancer cells by targeting glycosylated PD-L1. Cancer Cell. 2018;33(2):187–201 e110. doi:10.1016/j.ccell.2018.01.009
28. Tian P, Yan L. Inhibition of MicroRNA-149-5p induces apoptosis of acute myeloid leukemia cell line THP-1 by targeting Fas Ligand (FASLG). Med Sci Monit. 2016;22:5116–5123. doi:10.12659/MSM.899114
29. Shimada K, Nakamura M, De Velasco MA, Tanaka M, Ouji Y, Konishi N. Syndecan-1, a new target molecule involved in progression of androgen-independent prostate cancer. Cancer Sci. 2009;100(7):1248–1254. doi:10.1111/j.1349-7006.2009.01174.x
30. Pan S-J, Zhan S-K, Pei B-G, Sun Q-F, Bian L-G, Sun B-M. MicroRNA-149 inhibits proliferation and invasion of glioma cells via blockade of AKT1 signaling. Int J Immunopathol Pharmacol. 2012;25(4):871–881. doi:10.1177/039463201202500405
31. Wang Y, Zheng X, Zhang Z, et al. MicroRNA-149 inhibits proliferation and cell cycle progression through the targeting of ZBTB2 in human gastric cancer. PLoS One. 2012;7(10):e41693. doi:10.1371/journal.pone.0041693
32. Zhang Y, Guo X, Xiong L, et al. Comprehensive analysis of microRNA-regulated protein interaction network reveals the tumor suppressive role of microRNA-149 in human hepatocellular carcinoma via targeting AKT-mTOR pathway. Mol Cancer. 2014;13:253. doi:10.1186/1476-4598-13-253
33. Xu Y, Chen X, Lin L, Chen H, Yu S, Li D. MicroRNA-149 is associated with clinical outcome in human neuroblastoma and modulates cancer cell proliferation through Rap1 independent of MYCN amplification. Biochimie. 2017;139:1–8. doi:10.1016/j.biochi.2017.04.011
34. Ke Y, Zhao W, Xiong J, Cao R. miR-149 inhibits non-small-cell lung cancer cells EMT by targeting FOXM1. Biochem Res Int. 2013;2013:506731. doi:10.1155/2013/506731
35. Zhang X, Wang S, Wang H, et al. Circular RNA circNRIP1 acts as a microRNA-149-5p sponge to promote gastric cancer progression via the AKT1/mTOR pathway. Mol Cancer. 2019;18(1):20. doi:10.1186/s12943-018-0935-5
36. Rui-Da X, F F, Xs Y, Zd L, Lf L. miR-149-5p inhibits cell growth by regulating TWEAK/Fn14/PI3K/AKT pathway and predicts favorable survival in human osteosarcoma. Int J Immunopathol Pharmacol. 2018;32:2058738418786656.
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.