Back to Journals » OncoTargets and Therapy » Volume 13
AZIN1-AS1, A Novel Oncogenic LncRNA, Promotes the Progression of Non-Small Cell Lung Cancer by Regulating MiR-513b-5p and DUSP11
Authors Cai Y, Wu Q, Liu Y, Wang J
Received 26 May 2020
Accepted for publication 11 August 2020
Published 9 October 2020 Volume 2020:13 Pages 9667—9678
DOI https://doi.org/10.2147/OTT.S261497
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Federico Perche
Yong Cai,1,* Qiongya Wu,1,* Yu Liu,1,* Jiying Wang2
1Department of Radiotherapy, Shanghai Pulmonary Hospital, Tongji University School of Medicine, Shanghai 200433, People’s Republic of China; 2Department of Oncology, Shanghai Pulmonary Hospital, Tongji University School of Medicine, Shanghai 200433, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Jiying Wang
Department of Oncology, Shanghai Pulmonary Hospital, Tongji University School of Medicine, Shanghai 200433, People’s Republic of China
Email [email protected]
Background: Emerging researches have demonstrated that aberrantly expressed long non-coding RNAs (lncRNAs) have great significance in non-small cell lung cancer (NSCLC) progression. The aim of this study was to explore the role of lncRNA AZIN1 antisense RNA 1 (AZIN1-AS1) in NSCLC and the related mechanism.
Methods: Expressions of AZIN1-AS1 and miR-513b-5p in NSCLC samples were detected by qRT-PCR. NSCLC cell lines (H1299 and HCC827) were used in vitro assays. CCK-8 assay, EdU assay, wound healing test and Transwell assay were carried out to test the biological influence of AZIN1-AS1 on NSCLC cells. Subcutaneous xenotransplanted tumor model and tail vein injection model were established to test the role of AZIN1-AS1 in vivo. Interactions between AZIN1-AS1 and miR-513b-5p, miR-513b-5p and dual-specificity phosphatase 11 (DUSP11) were determined by bioinformatic analysis, qRT-PCR, Western blot, and luciferase reporter assay.
Results: AZIN1-AS1 was up-regulated in NSCLC cells and tissues, while miR-513b-5p was significantly down-regulated. Silencing of AZIN1-AS1 or overexpression of miR-513b-5p markedly inhibited proliferation, migration and invasion of NSCLC cells, while overexpression of AZIN1-AS1 or inhibition of miR-513b-5p functioned oppositely. Importantly, AZIN1-AS1 mediated the promotion of malignancy of NSCLC cells was reversed by miR-513b-5p mimics. What’s more, AZIN1-AS1 could down-regulate miR-513b-5p via sponging it, and there existed a negative correlation between AZIN1-AS1 expression and miR-513b-5p expression in NSCLC samples. AZIN1-AS1 also enhanced the expression levels of DUSP11, which was proved as a target gene of miR-513b-5p. Further in vivo experiments showed that silencing of AZIN1-AS1 decreased tumor growth and metastasis, which was accompanied by overexpression of miR-513b-5p and inhibition of DUSP11 in tumor tissues.
Conclusion: AZIN1-AS1 acts as a tumor promoter in NSCLC, which is ascribed to the regulation of miR-513b-5p and DUSP11.
Keywords: non-small cell lung cancer, AZIN1-AS1, miR-513b-5p, DUSP11
Introduction
Lung cancer is with the highest morbidity and mortality among cancers worldwide.1–4 As the dominant pathologic type, non-small cell lung cancer (NSCLC) accounts for 80% of lung cancer.5 Although conventional treatment strategies (such as surgery, radiotherapy or chemotherapy) prolong the survival time of patients, the prognosis of NSCLC is still unsatisfactory, and the 5-year survival rate is less than 20%.6,7 It is necessary to figure out the molecular mechanism and look for new therapeutic targets of NSCLC.
Long non-coding RNAs are transcripts with length exceeding 200 nucleotides and without the ability to encode proteins. Previous studies have found that lncRNAs can control gene expression at epigenetic, transcriptional and post-transcriptional levels. LncRNAs are key regulators in various biological activities, such as chromatin modification, intranuclear transport, genomic imprinting, X chromosome silencing, transcriptional interference and transcriptional activation.8 Recent studies have shown that abnormal expression of lncRNAs is associated with many human diseases, and they are involved in the growth, invasion, metastasis and relapse of cancers.9 LncRNA AZIN1 antisense RNA 1 (AZIN1-AS1) was located on human chromosome 8q22.3, and its role and mechanism in cancer are still unclear.
MicroRNAs (miRNAs) are small non-coding RNAs with a length of about 18–24 nucleotides. They regulate the post-transcriptional process via binding to the 3ʹUTR of target gene, thus resulting in inhibition of translation or degradation of the mRNA and being involved in the regulation of cellular proliferation, differentiation and apoptosis.10 Accumulating studies show that miRNAs participate in the tumor progression.11 MiR-513b-5p origins from the precursor RNA transcribed from human chromosome Xq27.3. The expression of miR-513b-5p is remarkably down-regulated in testicular embryonic cancer, and it could impede the proliferation and metastasis of cancer cells by repressing interferon regulatory factor 2.12 Our previous study demonstrated that miR-513b-5p was down-regulated in NSCLC tissues and it could significantly inhibit the proliferation, invasion, migration, and promote apoptosis of NSCLC cells by regulating mTOR signaling pathway.13 However, the mechanism of miR-513b-5p dysregulation in NSCLC remains unclear.
In this study, we demonstrated that the expression of AZIN1-AS1 was significantly up-regulated in NSCLC tissues. Gain- and loss-of-function experiments confirmed that AZIN1-AS1 could promote the proliferation, migration and invasion of NSCLC cells, suggesting that AZIN1-AS1 could play a carcinogenic role in the progression of NSCLC. To further explore the downstream mechanism, we found that overexpression of AZIN1-AS1 could up-regulate the expression of dual-specificity phosphatase 11 (DUSP11), playing the role of competitive endogenous RNA (ceRNA) to regulate the expression of miR-513b-5p. This study explored the role of AZIN1-AS1/miR-513b-5p/DUSP11 axis in controlling NSCLC progression, which provided new potential targets for NSCLC treatment.
Materials and Methods
Tissue Samples
This study obtained the approval of the Ethics Committee of Shanghai Pulmonary Hospital. Written informed consents were acquired from each participating patient and all protocols were conducted in accordance with the principles of the Declaration of Helsinki. Thirty-six patients with primary NSCLC underwent NSCLC resection in our hospital from May 2017 to May 2018 were selected in this study. In the control group, the specimens were from adjacent tissues of the same patient (at least 3 cm away from the surgical margin), and no cancer cells were found by pathological examination after the operation. All specimens were stored in liquid nitrogen at −196°C.
Cell Culture and Cell Transfection
Human normal lung epithelial cell lines BEAS-2B and NSCLC cell lines (HCC827, NCI-H460, A549, H1299) were purchased from American Type Culture Collection (Rockville, USA). These cells were cultured in RPMI-1640 medium (ThermoFisher Scientific, Shanghai, China) supplemented with 10% fetal bovine serum (FBS) (Invitrogen, Carlsbad, CA, USA), 100U/mL penicillin and 100mg/mL streptomycin (Invitrogen, Carlsbad, CA, USA), and cultured in an incubator at 37°C with 5% CO2. AZIN1-AS1 overexpression plasmid (pcDNA3.1-AZIN1-AS1), knockdown plasmid (carrying AZIN1-AS1 shRNA), DUSP11 overexpression plasmid (pcDNA3.1-DUPS11), hsa-miR-153b-5p mimics and inhibitors, and corresponding control plasmids and miRNAs were obtained from GenePharma (Shanghai, China). The above vectors or miRNAs were transfected into cells using Lipofectamine 2000 reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions. 1µg/mL puromycin (Beyotime, Shanghai, China) was added into the medium to obtain stable cell lines transfected with plasmids.
Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
Total RNA in tissues and cells was extracted by TRIzol reagent (Invitrogen, Shanghai, China). MiRNA was extracted with mirPremier® microRNA Isolation Kit (Sigma, St. Louis, MO, USA). Total RNA was retrieved by First Strand cDNA Synthesis Kit (Thermo Fisher Scientific Inc., Rockford, IL, USA). In line with the manufacturer’s instructions, qRT-PCR was conducted with Light Cycler Fast Start DNA MasterPlus SYBR Green I kit (Roche Diagnostics, Burgess Hill, UK) to analyze the expression of AZIN1-AS1 and GAPDH. TaqMan microRNA assay and TaqMan Universal Master Mix II (Applied Biosystems, Foster City, CA, USA) were used to analyze the expression of miR-513b-5p and U6. The primers were designed and synthesized by RiboBio Co.,Ltd. (Guangzhou, China).
Cell Counting Kit-8 (CCK-8) Assay
HCC827 and H1299 cells in logarithmic growth phase were trypsinized, centrifuged and resuspended. After adjusting the cell density to 2×104/mL, the cells were inoculated into a 96-well plate with 100 μL cell suspension per well. After that, the 96-well plate was placed in an incubator for further culture. The absorbance of cells was measured in 24, 48, 72 and 96 hour, respectively. In brief, each well was incubated in the incubator for 1 hour with 10 µL CCK-8 solution (Dojindo Molecular Technologies, Kumamoto, Japan). After the termination of the culture, the 96-well plate was placed in a microplate reader to determine the absorbance (OD value) of each well at 450 nm wavelength.
Ethynyl-Deoxyuridine (EdU) Assay
EdU kit (RiboBio Co., Ltd., Guangzhou, China) was also used to evaluate the proliferation of NSCLC cells. HCC827 and H1299 cell lines were seeded into 24-well plates (containing a cover slip in each well), respectively, and cultured for 24 hours, and then incubated with EdU (final concentration of 50 mmol/L) for 2 hours. Then, the medium was discarded and the cover slips were washed gently with PBS for 3 times. After that, the cells were fixed in 4% paraformaldehyde for 10 min. Then, the cells were incubated with fluorescent staining solution for 30 minutes, washed with methanol twice, incubated with 1× Hoechst 33,342 for 30 minutes. After the cover slips were washed with PBS for three times and sealed, the images were obtained under fluorescence microscope (Olympus, Tokyo, Japan). The percentage of EdU-positive cells was calculated.
Transwell Assay
Cells were collected and resuspended in serum-free RPMI-1640 medium. Then, cells were added into the upper compartment of Transwell chamber (8 μm pore diameter, BD Biosciences, CA, USA) coated with Matrigel (BD, San Jose, CA, USA), while the 10% FBS-containing medium was added in the lower compartment. After 24 h’s culture, cells remaining in the upper compartment were wiped off with a cotton swab. Cells passing through the membrane were fixed with 4% polyformaldehyde and stained with crystal violet solution. At last, the cells were counted under a microscope.
Subcutaneous Xenotransplanted Tumor Model
Nude mice (6-week old) were purchased from the Laboratory Animal Center of Nanjing University. Cells in logarithmic growth phase from AZIN1-AS1 overexpression group or vector group, were trypsinized to prepare single-cell suspension. The cell density was adjusted to 2×l07/mL. One hundred μL cell suspension was subcutaneously inoculated to the left side (AZIN1-AS1 overexpression group) and right side (vector group) of each mouse. The longest diameter (L) and shortest diameter (W) of the tumors were measured every 3 days, and the volume of tumors (V) =0.5×L×W2 was calculated. In the lung metastasis study, there were 10 mice in each group. 1×107 cells of AZIN1-AS1 overexpression group or vector group were injected into the tail vein of the mice. Two weeks later, the mice were killed by cervical decapitation. Pathological examination was used to quantify lung metastasis. All procedures were reviewed and approved by the Animal Study Committee of Shanghai Pulmonary Hospital, and in compliance with the National Institutes of Health (NIH) Guide for the Care and Use of Laboratory Animals and Animal Welfare Act.
Dual-Luciferase Reporter Assay
The wild-type (WT) AZIN1-AS1 sequence or the WT DUSP11 3ʹ-UTR fragment containing predicted binding sites with miR-513b-5p was amplified and inserted into the pmirGLO dual-luciferase miRNA target expression vector (Promega Corp., Madison, WI, USA) to construct the report vector pmirGLO-AZIN1-AS1-WT or pmirGLO-DUSP11-WT. GeneArt Site-Directed Mutagenesis PLUS System (Thermo Fisher Scientific Inc., Rockford, IL, USA) was used to mutate the presumed binding site of miR-513b-5p with AZIN1-AS1 or DUSP11 3ʹ-UTR. Mutant (MUT) AZIN1-AS1 or DUSP11 3ʹ-UTR were inserted into pmirGLO dual-luciferase miRNA target expression vector to construct report vector pmirGLO-AZIN1-AS1-MUT or pmirGLO-DUSP11-MUT. The corresponding reporter vectors and miR-513b-5p or NC mimics were co-transfected into HEK293 cells and the cells were cultured for 48 h. After that, the luciferase activity of the cells in each group was measured using the dual-luciferase reporter assay system (Promega, Madison, WI, USA).
Western Blot
RIPA lysate (containing 1% PMSF) was used to extract total proteins from different groups of cells. Protein concentration was determined by BCA method. Protein samples were separated by SDS-PAGE, and transferred to polyvinylidene fluoride (PVDF) membranes (Millipore, Bedford, MA, USA). The membranes were blocked with 5% skimmed milk for 1 h at room temperature, and then the primary antibody (anti-DUSP11, ab127299, 1:1000, abcam) was used to incubate the membrane at 4°C overnight. After washing the membranes with TBST buffer, horseradish peroxidase (HRP)-labelled secondary antibody (Beyotime, Shanghai, China) was added and the membranes were incubated at 37°C for 1 h. After washing the membranes with TBST buffer again, the signals were detected by ChemiLucent ECL Detection system (Millipore, Bedford, MA, USA).
Statistical Methods
Statistical software SPSS 17.0 (SPSS Inc., Chicago, IL, USA) was used to analyze all the experimental data in this study. The data were expressed as mean ±standard deviation (x±s). t-test was used to compare the differences between two groups. P < 0.05 was statistically significant.
Results
Expression and Clinical Significance of AZIN1-AS1 in NSCLC
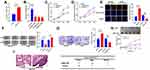
To preliminarily detect the expression level of AZIN1-AS1 in cancers, we searched the expressions of AZIN1-AS1 in multiple cancer tissues using GEPIA database (http://gepia.cancer-pku.cn/) and in NSCLC tissues using StarBase databases (http://starbase.sysu.edu.cn/). As shown, AZIN1-AS1 expression was up-regulated in NSCLC tissues (Figure 1A and B). To further validate the high expression of AZIN1-AS1 in NSCLC, we detected the expression level of AZIN1-AS1 in NSCLC tissues and cell lines using qRT-PCR. The results showed that the expression levels of AZIN1-AS1 in NSCLC tissues and cell lines were increased significantly compared with the control group (Figure 1C and D). Additionally, we examined the correlation between the expression level of AZIN1-AS1 and clinicopathological parameters of NSCLC patients. Patients were separated into AZIN1-AS1 high expression group and AZIN1-AS1 low expression group. The results suggested that the high expression of AZIN1-AS1 was significantly associated with an increase in tumor volume and the higher T stage (Table 1). These results hinted that AZIN1-AS1 was involved in the progression of NSCLC, and probably functioned as an oncogenic lncRNA.
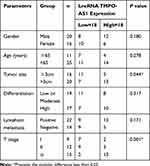 |
Table 1 Correlation Between LncRNA AZIN1-AS1 Expression and Clinical Features (n=36) |
The Effects of AZIN1-AS1 on Proliferation and Metastasis of NSCLC Cells in vitro and in vivo
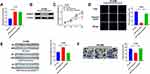
To explore the function of AZIN1-AS1 in NSCLC, we successfully constructed AZIN1-AS1 knockdown and overexpression cell lines with NSCLC cell lines H1299 and HCC827, respectively (Figure 2A and B). CCK-8 and EdU experiments showed that compared with the control group, the proliferation of NSCLC cells was, respectively, enhanced and repressed after overexpression and knockdown of AZIN1-AS1 (Figure 2C–E). Wound healing test and Transwell test were used to detect cell migration and invasion. Compared with the control group, the migration and invasion abilities of tumor cells were significantly increased after up-regulating of AZIN1-AS1 in cells, while the knockdown of AZIN1-AS1 in cells had the opposite effect (Figure 2F and G). In addition, in vivo experiments with nude mice showed that overexpression of AZIN1-AS1 significantly promoted the growth of transplanted tumors (Figure 2H); pulmonary metastasis experiment showed that 7 out of 10 mice in AZIN1-AS1 overexpression group of had severe pulmonary metastasis, and the incidence was markedly higher than that in the control group (1 out of 10) (Figure 2I).
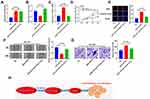
AZIN1-AS1 Can Target MiR-513b-5p
To explore the downstream molecular mechanism of AZIN1-AS1, we used StarBase to search for the potential targets of AZIN1-AS1. As shown in, AZIN1-AS1 contains a potential binding site of miR-513b-5p (Figure 3A). The levels of miR-513b-5p in NSCLC tissues and cells were further detected. The results of qRT-PCR indicated that the expression of miR-513b-5p was down-regulated in NSCLC tissues and cells (Figure 3B and C). Further experiments indicated that the expression levels of AZIN1-AS1 and miR-513b-5p in NSCLC tissues were negatively correlated (Figure 3D). Moreover, the luciferase reporter assay showed that miR-513b-5p notably suppressed the luciferase activity of AZIN1-AS1-WT reporter, but had no significant effect on AZIN1-AS1-MUT reporter (Figure 3E). Next, we explored the regulatory effect of AZIN1-AS1 on miR-513b-5p. As shown, the expression of miR-513b-5p in the AZIN1-AS1 overexpression group was significantly lower than that in the control group, while the expression of AZIN1-AS1 knockdown could increase the expression level of miR-513b-5p in NSCLC cells (Figure 3F). In conclusion, these data demonstrated that AZIN1-AS1 interacted with miR-513b-5p and inhibited the expression of miR-513b-5p.
Regulatory Effects of MiR-513b-5p on Proliferation and Metastasis of NSCLC Cells in vitro
In our previous study, miR-513b-5p was proved to suppress the malignant phenotypes of NSCLC A549 and H460 cell lines.13 To further validate the tumor-suppressive function of miR-513b-5p in NSCLC, in this study, we used H1299 and HCC827 cells to study the function of miR-513b-5p in NSCLC (Figure 4A and B). CCK-8 and EdU experiments showed that compared with the control group, the proliferation of H1299 cells transfected with miR-513b-5p mimics was significantly reduced, and the proliferation of HCC827 cells transfected with miR-513b-5p inhibitors was remarkably increased (Figure 4C–E). After investigating the effect of miR-513b-5p on the proliferation of cancer cells, we explored the effects of miR-513b-5p on the metastasis. As shown, the migration and invasion of the cells were reduced after transfection with miR-513b-5p mimics, while the transfection of miR-513b-5p inhibitors had the opposite effect (Figure 4F and G). These data further clarified that miR-513b-5p was associated with the malignant phenotypes of NSCLC and functioned as a tumor suppressor.
DUSP11 is the Functional Target of MiR-513b-5p
To explore the mechanism of the role of miR-513b-5p in NSCLC, we selected a candidate target for miR-513b-5p in TargetScan database (http://www.targetscan.org/vert_72/). As shown, miR-513b-5p was complementary to the 3ʹUTR of DUSP11, suggesting that DUSP11 might be a target gene for miR-513b-5p (Figure 5A). We further examined the expression of DUSP11 in NSCLC tissues and cell lines and demonstrated that the expression levels of DUSP11 in NSCLC tissues and cells were significantly higher than those of control group (Figure 5B and C). Dual-luciferase reporter assay showed that miR-513b-5p mimics could decrease the luciferase activity of luciferase reporter containing DUSP11 3ʹUTR-WT, but had no significant influence on the luciferase activity of DUSP11 3ʹUTR-MUT reporter (Figure 5D). In addition, qRT-PCR displayed that the expression levels of miR-513b-5p and DUSP11 in NSCLC tissues were negatively correlated, while the expression levels of AZIN1-AS1 and DUSP11 in NSCLC tissues were positively correlated (P < 0.05, Figure 5E and F). Western blot showed that the expression levels of DUSP11 were down-regulated and up-regulated, respectively, after transfection of mimics and inhibitors of miR-513b-5p in NSCLC cells. What’s more, as we expected, after overexpression and knockdown of AZIN1-AS1 in NSCLC cells, DUSP11 levels were up-regulated and down-regulated, respectively (Figure 5G and H). This indicated that AZIN1-AS1 could indirectly modulate the expression of DUSP11.
The Tumor-Suppressive Function of MiR-513b-5p Was Partly Dependent on Its Regulatory Function on DUSP11
To investigate the functional relationship between miR-513b-5p and DUSP11, DUSP11 overexpression plasmids were co-transfected with miR-513b-5p into H1299 cells. As shown, the co-transfection of DUSP11 overexpression plasmids did not affect the expression of miR-513b-5p in H1299 cells but significantly up-regulated the expression of DUSP11 (Figure 6A and B). Next, CCK-8 assay, EdU assay, wound healing assay and Transwell assay were performed, and as expected, these functional experiments indicated that miR-513b-5p suppressed the malignant phenotypes of H1299 cells, which could be partially reversed by the co-transfection of DUSP11 overexpression plasmids (Figure 6C–F). Collectively, these data validated that the tumor-suppressive function of miR-513b-5p was partly dependent on its regulatory function on DUSP11.
AZIN1-AS1 Affected the Proliferation and Metastasis of NSCLC Cells Through MiR-513-5p/DUSP11 Axis
To further verify the role of AZIN1-AS1 in NSCLC by regulating the expression of miR-513b-5p/DUSP11, we transfected miR-513b-5p mimics into HCC827 cells overexpressing AZIN1-AS1. We found that compared with AZIN1-AS1/NC group, AZIN1-AS1/miR-513b-5p mimics group had no significant change in AZIN1-AS1 level, while the expression of DUSP11 was significantly decreased (Figure 7A–C). The results of CCK-8 and EdU assays indicated that the proliferation of HCC827 cells was significantly increased by AZIN1-AS1 overexpression, while this was partly reversed by the co-transfection of miR-513b-5p mimics (Figure 7D and E). Wound healing and Transwell assays showed that the migration and invasion abilities of cancer cells were enhanced by AZIN1-AS1 overexpression, but reversed by the co-transfection of miR-513b-5p mimics (Figure 7F and G). Collectively, the above results further validated the role of AZIN1-AS1/miR-513b-5p/DUSP11 axis in NSCLC progression (Figure 7H).
Discussion
Accumulating studies show that lncRNA can affect the progression of tumors.14–17 For example, in NSCLC, lncRNA XIST could enhance the chemoresistance of cancer cells by regulating autophagy;16 lncRNA PVT1 promoted the proliferation and inhibits apoptosis of NSCLC cells by targeting miR-195.17 In this study, we found that AZIN1-AS1 was up-regulated in NSCLC tissues and cells compared with the control group. Additionally, its high expression in tumor tissues was associated with unfavorable pathological parameters. Furthermore, the biological function of AZIN1-AS1 in NSCLC was investigated. We observed that overexpression of AZIN1-AS1 could promote the proliferation and metastasis of NSCLC cells, whereas knockdown of AZIN1-AS1 could suppress these phenotypes. These results suggested that AZIN1-AS1 was an oncogenic lncRNA in NSCLC and promote the progression of NSCLC.
It has also been confirmed that the abnormal expression of miRNAs was related to NSCLC.18–20 For example, in NSCLC, the expression of miR-221 is significantly up-regulated, which promotes the malignant phenotype of NSCLC by targeting TIMP2;19 miR-486-5p is a tumor suppressor in NSCLC, which targets PIK3R1 to inhibit the progression of cancer.20 It has been known that the expression of miR-513b-5p was down-regulated in testicular embryonic carcinoma.12 Our previous study indicates that miR‐513b-5p could regulate the pathogenesis of NSCLC via regulating HMGB3.13 Herein, we further validated that the expression of miR-513b-5p in NSCLC tissues and cells was significantly down-regulated, and the proliferation and metastasis of NSCLC cells were inhibited by it.
More and more reports have found that lncRNAs can function as ceRNAs of miRNAs, and inhibit the expression of miRNAs as molecular sponges. For example, in breast cancer, MIR210HG acts as ceRNA of miR-1226-3p to regulate the expression of mucin-1c, promoting the occurrence of metastasis;21 in hepatocellular carcinoma, XIST regulates PDCD4 by targeting miR-497-5p to regulate the proliferation and migration of cancer cells.22 In this study, we identified the interaction between miR-513b-5p with AZIN1-AS1. We observed that the expression level of miR-513b-5p increased significantly after AZIN1-AS1 knockdown, while the overexpression of AZIN1-AS1 increased the expression of miR-513b-5p. And the luciferase activity assay confirmed the direct binding relationship between AZIN1-AS1 and miR-513b-5p. Analysis of clinical samples indicated a negative correlation between the expression levels of AZIN1-AS1 and miR-513b-5p. To some extent, this study explained the mechanism of miR-513b-5p dysregulation in NSCLC.
DUSP11 was located on human chromosome 2p13.1. The protein encoded by this gene is a member of the dual-specificity protein phosphatase subfamily. These phosphatases inactivate their targets by dephosphorylation of phosphoserine, threonine and phosphotyrosine residues, and negatively regulate the members of the mitogen-activated protein (MAP) kinase superfamily (MAPK/ERK, SAPK/JNK, p38), which was related to cellular proliferation and differentiation.23 In colonic cancer, high expression of DUSP11 is involved in the formation of multicellular spheroids, which is regarded as a typical characteristic of cellular malignant transformation.24 In this study, we firstly identified the DUSP11 as a target gene of miR-513b-5p. In addition, we observed that knockdown of AZIN1-AS1 could decrease the expression of DUSP11. These data showed that there existed an “AZIN1-AS1/miR-513b-5p/DUSP11” axis in NSCLC tumorigenesis.
In the following studies, other downstream miRNAs of AZIN1-AS1 needs to be identified. And it is necessary to explore whether the dysregulation of AZIN1-AS1 is associated with the overall survival time and disease-free survival time of NSCLC patients, which is quite important to evaluate its potential to be used as a biomarker. It is worth noting that DUSP11 is reported to be a target of p53, and contributes to the proliferation inhibition of osteosarcoma cells line U2OS.25 This study implies that the function of DUSP11 is specific in different cancers, and its role in NSCLC requires further investigation.
In conclusion, our study demonstrates that AZIN1-AS1 is up-regulated in NSCLC tissues and cells, and it promotes the proliferation and metastasis of NSCLC cells by regulating the expression of miR-513b-5p/DUSP11. To our best knowledge, this is the first study to investigate the role of AZIN1-AS1 in cancer biology and it is expected to provide new therapeutic targets for NSCLC patients.
Data Sharing Statement
All data generated or analyzed during this study are included in this published article.
Ethics Approval and Consent to Participate
The protocols of human samples collection were approved by the Ethics Committee of Shanghai Pulmonary Hospital. All participants provided written informed consent and the procedures were conducted in accordance with the principles of the Declaration of Helsinki. As for animal experiments, ethical and legal approval was obtained from Animal Study Committee of Shanghai Pulmonary Hospital prior to experiments. The protocols were in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals and Animal Welfare Act.
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Authors’ Information
Department of Radiation Oncology, Shanghai Pulmonary Hospital, Tongji University School of Medicine, Shanghai 200433, China.
Funding
There is no funding to report.
Disclosure
The authors declare that they have no competing interests for this work.
References
1. Ferlay J, Soerjomataram I, Dikshit R, et al. Cancer incidence and mortality worldwide: sources,methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–E386. doi:10.1002/ijc.29210
2. Wang L, Yu C, Liu Y, et al. Lung cancer mortality trends in China from 1988 to 2013: new challenges and opportunities for the government. Int J Environ Res Public Health. 2016;13(11):1052. doi:10.3390/ijerph13111052
3. Yap YS, Kwok LL, Syn N, et al. Predictors of hand-foot syndrome and pyridoxine for prevention of capecitabine-induced hand-foot syndrome: a randomized clinical trial. JAMA Oncol. 2017;3(11):1538–1545. doi:10.1001/jamaoncol.2017.1269
4. Chen W, Zheng R, Zhang S, et al. Cancer incidence and mortality in China in 2013: an analysis based on urbanization level. Chin J Cancer Res. 2017;29(1):1–10. doi:10.21147/j.issn.1000-9604.2017.01.01
5. Siegel R, Naishadham D, Jemal A. Cancer statistics, 2013. CA Cancer J Clin. 2013;63(1):11–30. doi:10.3322/caac.21166
6. Allemani C, Weir HK, Carreira H, et al. CONCORD working group. Global surveillance of cancer survival 1995–2009: analysis of individual data for 25,676,887 patients from 279 population-based registries in 67 countries (CONCORD-2). Lancet. 2015;385(9972):977–1010. doi:10.1016/S0140-6736(14)62038-9
7. Sheng J, Wang L, Han Y, et al. Dual roles of protein as a template and a sulfur provider: a general approach to metal sulfides for efficient photothermal therapy of cancer. Small. 2018;14(1):1702529. doi:10.1002/smll.201702529
8. Wang KC, Yang YW, Liu B, et al. A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature. 2011;472(7341):120–124. doi:10.1038/nature09819
9. Bhan A, Soleimani M, Mandal SS. Long noncoding RNA and cancer: a new paradigm. Cancer Res. 2017;77(15):3965–3981. doi:10.1158/0008-5472.CAN-16-2634
10. Guarnieri DJ, Dileone RJ. MicmRNAs: a new class of gene regulators. Ann Med. 2008;40(3):197–208. doi:10.1080/07853890701771823
11. Tutar Y. miRNA and cancer; computational and experimental approaches. Curr Pharm Biotechnol. 2014;15(5):429. doi:10.2174/138920101505140828161335
12. Wang X, Zhang X, Wang G, Wang L, Lin Y, Sun F. Hsa-miR-513b-5p suppresses cell proliferation and promotes P53 expression by targeting IRF2 in testicular embryonal carcinoma cells. Gene. 2017;626:344–353. doi:10.1016/j.gene.2017.05.033
13. Wang J, Sheng Z, Cai Y. Effects of microRNA-513b on cell proliferation, apoptosis, invasion, and migration by targeting HMGB3 through regulation of mTOR signaling pathway in non-small-cell lung cancer. J Cell Physiol. 2019;234(7):10934–10941. doi:10.1002/jcp.27921
14. Zeng Z, Xu FY, Zheng H, et al. LncRNA-MTA2TR functions as a promoter in pancreatic cancer via driving deacetylation-dependent accumulation of HIF-1α. Theranostics. 2019;9(18):5298–5314. doi:10.7150/thno.34559
15. Zhou F, Shen F, Zheng Z, Ruan J. The LncRNA XIRP2-AS1 predicts favorable prognosis in colon cancer. Onco Targets Ther. 2019;12:5767–5778. doi:10.2147/OTT.S215419
16. Sun W, Zu Y, Fu X, Deng Y. Knockdown of lncRNA-XIST enhances the chemosensitivity of NSCLC cells via suppression of autophagy. Oncol Rep. 2017;38(6):3347–3354. doi:10.3892/or.2017.6056
17. Wu D, Li Y, Zhang H, Hu X. Knockdown of Lncrna PVT1 enhances radiosensitivity in non-small cell lung cancer by sponging Mir-195. Cell Physiol Biochem. 2017;42(6):2453–2466. doi:10.1159/000480209
18. Yang D, Du G, Xu A, Xi X, Li D. Expression of miR-149-3p inhibits proliferation, migration, and invasion of bladder cancer by targeting S100A4. Am J Cancer Res. 2017;7(11):2209–2219.
19. Yin Z, Xu M, Li P. miRNA-221 acts as an oncogenic role by directly targeting TIMP2 in non-small-cell lung carcinoma. Gene. 2017;620:46–53. doi:10.1016/j.gene.2017.04.007
20. Tian F, Wang J, Ouyang T, et al. MiR-486-5p serves as a good biomarker in nonsmall cell lung cancer and suppresses cell growth with the involvement of a target PIK3R1. Front Genet. 2019;10:688. doi:10.3389/fgene.2019.00688
21. Li XY, Zhou LY, Luo H, et al. The long noncoding RNA MIR210HG promotes tumor metastasis by acting as a ceRNA of miR-1226-3p to regulate mucin-1c expression in invasive breast cancer. Aging (Albany NY). 2019:11.
22. Zhang Y, Zhu Z, Huang S, et al. lncRNA XIST regulates proliferation and migration of hepatocellular carcinoma cells by acting as miR-497-5p molecular sponge and targeting PDCD4. Cancer Cell Int. 2019;19(1):198. doi:10.1186/s12935-019-0909-8
23. Burke JM, Sullivan CS. DUSP11- an RNA phosphatase that regulates host and viral non-coding RNAs in mammalian cells. RNA Biol. 2017;14(11):1457–1465. doi:10.1080/15476286.2017.1306169
24. Dardousis K, Voolstra C, Roengvoraphoj M, et al. Identification of differentially expressed genes involved in the formation of multicellular tumor spheroids by HT-29 colon carcinoma cells. Mol Ther. 2007;15(1):94–102. doi:10.1038/sj.mt.6300003
25. Caprara G, Zamponi R, Melixetian M, Helin K. Isolation and characterization of DUSP11, a novel p53 target gene. J Cell Mol Med. 2009;13(8B):2158–2170. doi:10.1111/j.1582-4934.2008.00616.x
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.