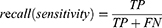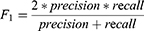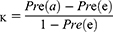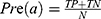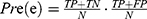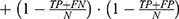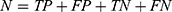Back to Journals » Nature and Science of Sleep » Volume 13
Automatic Sleep Stage Classification of Children with Sleep-Disordered Breathing Using the Modularized Network
Authors Wang H, Lin G, Li Y, Zhang X, Xu W, Wang X, Han D
Received 27 August 2021
Accepted for publication 12 October 2021
Published 30 November 2021 Volume 2021:13 Pages 2101—2112
DOI https://doi.org/10.2147/NSS.S336344
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Professor Ahmed BaHammam
Huijun Wang,1– 3,* Guodong Lin,4,* Yanru Li,1– 3 Xiaoqing Zhang,1– 3 Wen Xu,1– 3 Xingjun Wang,4 Demin Han1– 3
1Department of Otorhinolaryngology Head and Neck Surgery, Beijing Tongren Hospital, Capital Medical University, Beijing, People’s Republic of China; 2Obstructive Sleep Apnea-Hypopnea Syndrome Clinical Diagnosis and Therapy and Research Centre, Capital Medical University, Beijing, People’s Republic of China; 3Key Laboratory of Otolaryngology-Head and Neck Surgery, Ministry of Education, Capital Medical University, Beijing, People’s Republic of China; 4Department of Electronic Engineering, Tsinghua Shenzhen International Graduate School, Tsinghua University, Shenzhen, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Demin Han
Beijing Tongren Hospital, Capital Medical University, No. 1 Dongjiaominxiang Street, Dongcheng District, Beijing, 100730, People’s Republic of China
Tel +86-010-58269335
Fax +86-010-58269331
Email [email protected]
Xingjun Wang
Tsinghua Shenzhen International Graduate School, University Town of Shenzhen, Nanshan District, Shenzhen, 518055, People’s Republic of China
Tel +86-18038153071
Email [email protected]
Purpose: To develop an automatic sleep stage analysis model for children and evaluate the effect of the model on the diagnosis of sleep-disordered breathing (SDB).
Patients and Methods: Three hundred and forty-four SDB patients aged between 2 to 18 years who completed polysomnography (PSG) to assess the severity of the disease were enrolled in this study. We developed deep neural networks to stage sleep from electroencephalography (EEG), electrooculography (EOG) and electromyogram (EMG). The model performance was estimated by accuracy, precision, recall, F1-score, and Cohen’s Kappa coefficient (ĸ). And we compared the difference in calculation of sleep parameters among the technicians, the model ensemble, and the single-channel EEG model.
Results: The numbers of raw data divided into training, validation, and testing were 240, 36, and 68, respectively. The best performance appeared in the model ensemble of which the accuracy was 83.36% (ĸ=0.7817) in 5-stages, and the accuracy was 96.76% (ĸ=0.8236) in 2-stages. The single-channel EEG model showed the classification satisfyingly as well. There was no significant difference in TST, SE, SOL, time in W, time in N1+N2, time in N3, and OAHI between technician and the model (P> 0.05). On the datasets from sleep-EDF-13 and sleep-EDF-18, the average classification accuracies achieved were 92.76% and 91.94% in 5-stages by using the proposed method, respectively.
Conclusion: This research established the model for pediatric automatic sleep stage classification with satisfying reliability and generalizability. In addition, it could be applied for calculating quantitative sleep parameters and evaluating the severity of SDB.
Keywords: sleep-disordered breathing, SDB, deep learning, sleep stage, children
Introduction
Sleep-disordered breathing (SDB) represents a spectrum of breathing disorders ranging from primary snoring (PS) to obstructive sleep apnea (OSA) that disrupts nocturnal respiration and architecture of sleep, which is highly prevalent in children who are at the critical stage of growth and development.1 It is meaningful for early diagnosis and intervention because SDB was verified to be associated with the functioning of various organs, including immune responses, cardiovascular function, and neurocognitive function.2
The mobility of SDB in children who suffer from snoring, mouth breathing, or apnea ranged from 7.9 to 13.4% as measured according to the Pediatric Sleep Questionnaire (PSQ).3,4 Based on guidelines provided by the American Academy of Paediatrics in 2012, the morbidity due to OSA in children is 1 to 5%.5 Since overnight polysomnography (PSG) remains the gold standard for diagnosing the severity of SDB, diagnostic efficiency is largely dependent on the availability and accessibility of this technique.
Sleep stage classification is the first step for data analysis in PSG, according to the strict criteria proposed by the American Academy of Sleep Medicine (AASM).6 It takes nearly 1 to 2 hours for technicians to identify sleep stages manually. However, intra-rater reliability (IRR) of classification is also known to be the subject of considerable variability. The SIESTA database arising from an EU-funded project found that the overall agreement for the AASM standard was 82.0% (ĸ = 0.76) based on an epoch-by-epoch comparison.7 The accurate and user-friendly sleep staging system would assist sleep experts and provide critical clinical utility.
Because of time consuming nature and labor intensiveness of the manual method, several methods for automatic scoring of long-term sleep data have been researched in the past decades. The accuracy of the models mostly ranges from 78–90%.8–12 Development and evaluation of software dedicated to automatic sleep staging (AS) face several issues, among which are: (1) EEG has a low signal-to-noise ratio (SNR), as the brain activity measured is often covered by multiple sources of environmental, physiological, and activity-specific noise called “artifacts”; (2) the generalization capabilities of models need to be further verified, for patients of different ages, pathophysiological, and treatment in the real world.13,14
Behavioral and physiological characteristics of sleep in normal children vary significantly from sleep in adults.1 There is a dynamic changing process in the frequency and amplitude of the characteristic waves in different ages. Considerable differences occur within and between individual children. How to classify the sleep stages in children of different ages correctly concerns the evaluation of sleep efficiency and management of pediatric sleep-related disorders.
An automated deep neural network which has achieved human-level annotation performance with an average accuracy of 81.81% was proposed by using a multi-model integration strategy with multiple signals in our laboratory.8 Based on unfiltered EEG in a large sample of children with sleep-disordered breathing, our study was devoted to developing an automatic sleep stage analysis model with good generalizability in the clinical setting.
Patients and Methods
Study Datasets
The study was conducted under the principles stated in the Declaration of Helsinki and approved by the Institutional Review Board (IRB) of the Beijing Tongren Hospital (TRECKY2017-032). Written informed consent was obtained from each parent of the children for inclusion in the study and the use of their medical records. According to the IRB’s decision, this study used public datasets of sleep-EDF for model verification without obtaining patients’ informed consent.
Clinical Data
We recruited children in the Beijing Tongren Hospital between January 2017 and June 2021. The inclusion criteria were as follows: (1) 2 to 18 years old; (2) snoring more than 3 days every week; (3) total sleep time more than 6 hours; (4) children’s parents voluntarily participated in the study and signed informed consent forms. The exclusion criteria were as follows: (1) cannot cooperate to complete overnight polysomnography; (2) PSG recording integration failed; (3) PSG recordings could not be analyzed by technicians because of a large number of artifacts.
Sleep-EDF Database
For validating the generalization of the model, we used expanded version of the sleep-EDF database in which PSG recordings lasting approximately 20 hours or 9 hours each were collected from healthy subjects and some who had mild difficulty falling asleep after temazepam intake but were otherwise healthy, including sleep-EDF-13 (61 PSG recordings) and sleep-EDF-18 (197 PSG recordings).15 We combined the N3 and N4 according to the R&K rule into N3 with the AASM guidelines. For comparison with other articles, we input the integral PSG data with long periods of wake and only included 30 minutes of the wake before and after sleep.
Overnight Polysomnography (PSG)
The PSG was performed using the different computerized data collection systems of Compumedics S series (Compumedics Inc, Australia) and Alice 6 (Phillips Inc, America), including EEG(C3/A2, C4/A1), EOG (ROC, LOC), EMG, electrocardiogram (ECG), nasal and oral cannula pressure, recording of respiratory (thoracic and abdominal) movements, and pulse oximetry for oxygen saturation (SpO2).
The highly trained, experienced (more than 10 years) PSG technician scored sleep stages and respiratory events in a 30s epoch following the AASM guidelines (2012).6 It is classified into one of the following five categories: (1) Wake (W), (2) Non-rapid eye movement stage 1 (N1), (3) Non-rapid eye movement stage 2 (N2), (4) Non-rapid eye movement stage 3 (N3) or (5) Rapid eye movement stage (REM). Sleep stages of N1 and N2 are also called shallow sleep, and N3 is called deep sleep. In our study, we additionally classified sleep stages into 4 stages (W vs shallow sleep vs deep sleep vs REM), 3 stages (W vs non-rapid eye movement stage vs REM), and 2 stages (W vs Sleep).
The obstructive apnea-hypopnea index (OAHI) was defined as the number of apnea and hypopnea events per hour of sleep and was used to indicate the severity of SDB: PS (OAHI<1); mild OSA (1≤OAHI<5); moderate OSA (5≤OAHI<10); severe OSA (OAHI≥10).16,17
Data Processing
Signal Preprocessing
The frequency of the PSG data in the A6 dataset was 200 Hz while it is 256 Hz in the Compumedics dataset. To facilitate subsequent processing and retain necessary information of the signal at the same time, we filtered the signal at 50Hz and then downsampled it to 50 Hz. Based on this operation, we could eliminate high-frequency noise without spectral aliasing. Moreover, the amplitude of the signal was scaled to 100.
Label Smoothing
In the multi-classification task, the neural network would output the degree of confidence of each category. The probability vector could be obtained after the softmax processing. The cross-entropy loss of the network was computed as:
Where p is the ground truth probability vector and q is the network predicted probability vector, and K is the total number of classes. When the ground truth probability vector is in the form of one-hot (hard label), pi=1 if i equals to the ground truth class c, otherwise pi = 0. In this way, it may induce overfitting. To solve this problem, label smoothing converts hard label to soft label:
Where ɛ is constant and 0<ɛ<1. Label smoothing can improve the generalization of neural networks and prevent the network from becoming over-confident and overfitting.
One-to-Many Label
When doctors classify the sleep stage, it is not only based on the current epoch but also the adjacent ones. To make sure that the neural network can learn the features of the adjacent epoch, we used the one-to-many label, which means that the label of the current epoch corresponds to the signal of the current epoch and the adjacent ones. In our work, we took the signal of epoch before and after into consideration, which means that each label corresponded to a 90 seconds signal.
Neural Network
In this work, we used the modularized network architecture which consists of convolution block and multi-branch convolution block. Modularized network architecture has been widely used in neural networks nowadays such as VGG-nets18 and ResNets,19 of which the effectiveness has been proven by a variety of tasks.20–23 The neural network can become deeper without the limitation of the growing number of hyper-parameters by stacking blocks of the same structure.
Convolution Block
The structure of the convolution block is shown in Figure 1. The convolution layers are used to perform channel number conversion and extract the feature’s potential mapping. The shortcut connection is used to concatenate the input and output of the second ReLU, which can make the network deeper without the problem of vanishing gradient and exploding gradient. The length of the feature is halved by the average pooling layer. Batch normalization (BN) and ReLU are used before and after the convolution layer respectively. BN normalizes the distribution of input and ensures the input of the convolution layer has the same distribution as far as possible, which can alleviate the problem of vanishing gradient in training and accelerate the training speech of the model.
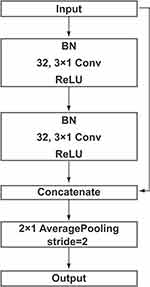 |
Figure 1 An overview of convolution block. Note: Batch normalization (BN) and ReLU were used before and after the convolution layer respectively. |
Multi-Branch Convolution Block
The structure of the multi-branch convolution block is shown in Figure 2. Multi-branch architecture is widely used by the family of Inception models24–26 and ResNeXt.27 The first convolution layer is used to perform channel number conversion. The channel of feature is split into g group and as input of the convolution layer with a kernel size of 3×1 respectively, then concatenate them together. The third convolution layer with a kernel size of 1×1 guarantees that the number of channels is the same as the input. We also used the shortcut and average pooling as in convolution block. Multi-branch convolution block can reduce the number of parameters of the network and prevent the model from overfitting.
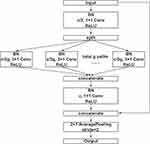 |
Figure 2 An overview of multi-branch convolution block. Notes: The channel of feature was split into g group and as input of the convolution layer respectively, then we concatenated them together. The specific values of c and g are shown in Figure 3. |
Overall Architecture and Training
The proposed overall network architecture is shown in Figure 3, which is mainly composed of convolution block and multi-branch convolution block. After the feature was extracted by the last multi-branch convolution block, we used the global average pooling layer to reduce the parameter amount of the model. Our model had great effectiveness even though it only had 0.16 million parameters, which is fewer parameters than most models. In other words, the model was easier to train and obtain higher training speed. The optimizer used in model training was Adabound28 (learning rate=0.0001, beta1=0.9, beta2=0.999, gamma=0.001) and batch size was 32. Moreover, early stopping callback on the validation loss with the patience of 5 epochs. All experiments in this study were performed on an NVIDIA GeForce RTX 3090 GPU.
 |
Figure 3 An overview of our overall network architecture based on convolution block and multi-branch convolution block. |
Model Evaluation and Statistical Analysis
The performance of the neural network model was evaluated by the overall accuracy, precision, recall, weighted F1 score, and Cohen’s Kappa coefficient. Their calculation formula was as follows:
where TP, TN, FP, and FN are the true positive, the true negative, the false positive, and the false negative, respectively.
Statistical analysis was performed using SPSS 22 software (SPSS Inc., Chicago, IL). We used Shapiro–Wilk test for normal data distribution test. Data were presented as mean±standard deviation or median (P25, P75). Variables with normal distribution were analyzed by t-test. Variables with nonnormal distribution were analyzed by Wilcoxon rank-sum test. The limit of agreement between technicians and the models was also analyzed using Bland-Altman plots.
Results
Study Population
The numbers of raw data collected by Alice 6 and Compumedics S series were 201 and 143, respectively. All the epochs were randomly split into training, validation, and testing sets at a ratio of 7:1:2. There was no significant difference in the demographic and polysomnographic parameters among the different datasets (Table 1). The distribution of W, N1, N2, N3, and REM was imbalanced at the radio of 1.74:1:7.33:3.94.
 |
Table 1 The Demographic and Polysomnographic Data of Children in Different Datasets |
Model Performance
In Table 2, we showed the accuracy and Cohen’s kappa of different models using channel combinations. The best performance with an average accuracy of 83.36% (ĸ=0.7817) in 5-stages appeared in the model ensemble which included the C3/A2+C4/A1+LOC+ROC+EMG, C3/A2, C4/A1, LOC, and ROC. And this model could also classify the stage of wake and sleep successfully with an average accuracy of 96.76% (ĸ=0.8236). In the single-channel EEG model, the performance of C3 was better than LOC and EMG. The confusion matrix for displaying the five sleep stage classification between the network prediction and technician was pictured in Figure 4. Except for single-channel EMG, N1 had precision (45.24–53.48%) and sensitivity (14.93–29.82%), which were lower than others. Most of them were classified as N2. We also compared the sleep stage classification performance of various ages of children, different data collection systems, and severities of OSA. The results were listed in the supplemental material (Table S1–S3).
 |
Table 2 Comparison of Testing Performances Using Different Input Channels |
Quantitative Sleep Parameters
There was no significant difference in total sleep time (TST), sleep efficiency (SE), sleep onset latency (SOL), time of wake, time of shallow sleep, and time of deep sleep between the network prediction using the model ensemble and technicians (P>0.05). The model would overestimate the sleep time in REM (P<0.001) by about 7.73min (Figure 5). Using the single-channel EEG model, we found that it underestimated SOL by about 1.63min (Supplemental Material, Figure S1).
Analysis of Respiratory Parameters
Using the stage classification of the model ensemble, we reviewed the analysis of respiratory events in the testing dataset. In Table 3, there was good consistency between the model and technicians (P=0.303). And there was no mistake in the diagnosis of OSA severity between the two sleep stage analyses.
 |
Table 3 Comparison of the Sleep Parameters and Respiratory Parameters Between Network Prediction and Analysis of Technicians |
Comparative Experiment on the Public Dataset
We conducted 4-fold cross-validation to evaluate the performance of the model which used channels of Fpz-Cz, Pz-Oz, and EOG. For 5-stages, the testing achieved accuracy of 92.76%(ĸ=0.8778) and 91.94% (ĸ=0.8521) of sleep-EDF-13 and sleep-EDF-18 with integral data, respectively. When removing large amounts of stage of wake, model performance dropped with the accuracy of 85.75% (ĸ=0.8015) and 84.58% (ĸ=0.7862) of sleep-EDF-13 and sleep-EDF-18, respectively. The comparison of similar research was shown in Table 4.
 |
Table 4 Performance Comparison of Various Research on the Public Dataset of Sleep-EDF |
Discussion
This study was the first to use unfiltered raw data of PSG based on large clinical samples in children with SDB for sleep staging model training, and compared the quantitative sleep parameters under sleep stage classification by experts and the model, such as total sleep time, sleep efficiency, sleep onset latency, and the time of each sleep stage. We used the modularized network which had lower number of parameters to propose an automatic sleep stage analysis model for children. For the 5-stages, the accuracy was 83.36%, and the cohen’s Kappa coefficient was 0.7881. This model could accurately distinguish between wake and sleep stages and showed no significant difference in diagnosis of the severity of children with SDB. Compared with similar studies, using sleep-EDF for verification, this model performed well.
Previous studies have trained models based on public datasets (collected from healthy adults and insomnia patients without other diseases), which have achieved good accuracy (78–92%). However, the original PSG data may have electrodes falling off, unstable baseline, high impedance, etc., and the accuracy of the model is affected by factors such as sleep fragmentation and arousal.8 Before this, other studies established sleep staging models based on pediatric PSG data,10,11 but its sample size, using more channels, and larger model load might limit its clinical application. Due to the different EEG patterns of children of different ages, we used the 2–18 years old to train children’s PSG, and the model had better generalization. The accuracy of sleep staging for children aged 2–6, 7–13, and 14–18 were 82.68%, 83.86%, and 84.71%, respectively. Similar to other research, the accuracy decreased as the severity of SDB increased.8,32 The testing set of this study included 10 children, whose AHI ranged from 5.03 to 39.42. The accuracy of automatic sleep stage classification was 82.74% in 5-stages, which was a little less than the performance in children with primary snoring and mild OSA. However, there was less influence on the calculation of classification in the severity of SDB.
Considering the clinical application of the automatic sleep staging model, we analyzed the performance of different channel combinations and single-channel EEG. Except for the single-channel EMG, all models had an accuracy of more than 95% for 2-stages, and the performance of 5-stages was equivalent to that of experienced sleep technicians. The lower accuracy of single-channel EEG model might be related to the smaller differences in different sleep stages. Affected by sweating and electrode shedding, many artifacts in EMG and small differences in EMG signals in N1, N2, and N3 phases were the explanation for the poor effect of the single-channel EMG model. Similar to other studies, the specificity and sensitivity of N1 stage classification were at a low level due to the small proportion of N1 in this database. A large number of N1 was divided into N2 and REM, which may be related to N1 which is mostly in the transition of sleep stage and the characteristics are not obvious. Some studies have performed separate data amplification for the N1-35-, which can increase the diagnostic efficiency of N1. We will use it in the optimization of the model in the future. The N3 stage has a higher proportion in children’s sleep, and the slow-wave has the characteristics of high amplitude. Compared with the adult EEG staging model,8,36,37 the accuracy of the N3 in this study had been greatly improved.
Comparing the ability of sleep stage classification and quantitative sleep parameters illustrates the practicality of the neural network model. The difference between model ensemble and technician analysis was small. The model underestimated the total sleep time by 1.00min, time in slight sleep (N1+N2) by 5.21min, time in N3 by 3.52min, and overestimated sleep onset latency by 0.44min, time in W by 0.56min, and time in REM by 7.73min. This inconsistency is tolerable. Similarly, the single-channel EEG model obtained a similar performance. Others based on non-contact radar technology, wearable devices, electrocardiograms, and respiratory dynamics, etc., had lower accuracy in judging different sleep stages than the single-channel EEG model of this study.38–40 Using the automated sleep stage, we re-analyzed the respiratory events, and the calculated OAHI was not different from the manual analysis. In the future, adding a single-channel EEG module to wearable devices and performing automatic sleep stage classification will be more efficient to screen children with sleep-disordered breathing.
As shown in Table 4, a variety of deep learning networks achieved good performance.10,29–34 With combined channels of EEG and EOG, our results were at a leading level. Recently, researchers have begun to consider how to design algorithms to be small, efficient, and robust. Based on previous research, we have improved data preprocessing and algorithms: (1) we filtered the signal at 50Hz, which would preserve EEG characteristics, remove high-frequency noise while reducing the amount of calculation. (2) Label smoothing mainly uses soft label, which modifies the weight of the real sample label category when calculating the loss function, and finally has the effect of suppressing overfitting. (3) Compared with other classifiers,30,41–43 our model achieved a huge performance improvement, meanwhile the number of parameters (number of trainable parameters in a single model was 0.15 million) was much smaller than in other similar studies (Table 5).
 |
Table 5 Overall Accuracy in 5-Stages and Training Model Number of Trainable Parameters of Similar Classifiers |
There are still several limitations in our study. Even though our study population involved children aged 2–18, there is still a lack of validation for children under 2 years of age. SDB patients have mainly primary snoring and mild OSA, and the proportion of N1 in the children’s clinical sample database was very small, which could affect its performance.
Conclusion
In our study, we created an automatic sleep stage analysis model with the modularized network with satisfying reliability and generalizability, which could be applied to calculating quantitative sleep parameters and evaluating the severity of SDB.
Acknowledgments
This study could not have been conducted without the help of the study participants, technologists, and physicians at the Department of Otolaryngology-Head and Neck Surgery, Beijing Tongren Hospital and Department of Electronic Engineering, Tsinghua Shenzhen International Graduate School, Tsinghua University.
Funding
This work was financially supported by the Science and Technology Innovation Committee of Shenzhen (WDZC20200818121348001), the National Natural Science Foundation of China (81970866, 81800894), the Consulting research project of Chinese Academy of Engineering (2019-XZ-29), Shenzhen Municipal Natural Science Foundation, and the Shenzhen Science and Technology Innovation Committee (KCXFZ202002011010487).
Disclosure
The authors report no conflicts of interest in this work.
References
1. Sheldon SH, Ferber R, Kryger MH, Gozal D. Principles and Practice of Pediatric Sleep Medicine. Elsevier Inc; 2014.
2. Zandieh SO, Cespedes A, Ciarleglio A, et al. Asthma and subjective sleep disordered breathing in a large cohort of urban adolescents. J Asthma. 2017;54(1):62–68. doi:10.1080/02770903.2016.1188942
3. Abazi Y, Cenko F, Cardella M, et al. Sleep Disordered Breathing: an Epidemiological Study among Albanian Children and Adolescents. Int J Environ Res Public Health. 2020;17(22):8586. doi:10.3390/ijerph17228586
4. Guo Y, Pan Z, Gao F, et al. Characteristics and risk factors of children with sleep-disordered breathing in Wuxi, China. BMC Pediatr. 2020;20(1):310. doi:10.1186/s12887-020-02207-5
5. Marcus CL, Brooks LJ, Draper KA, et al. Diagnosis and management of childhood obstructive sleep apnea syndrome. Pediatrics. 2012;130(3):576–584. doi:10.1542/peds.2012-1671
6. Berry RB, Budhiraja R, Gottlieb DJ, et al. Rules for scoring respiratory events in sleep: update of the 2007 AASM Manual for the Scoring of Sleep and Associated Events. Deliberations of the Sleep Apnea Definitions Task Force of the American Academy of Sleep Medicine. J Clin Sleep Med. 2012;8(5):597–619. doi:10.5664/jcsm.2172
7. Danker-Hopfe H, Anderer P, Zeitlhofer J, et al. Interrater reliability for sleep scoring according to the Rechtschaffen & Kales and the new AASM standard. J Sleep Res. 2009;18(1):74–84. doi:10.1111/j.1365-2869.2008.00700.x
8. Zhang X, Xu M, Li Y, et al. Automated multi-model deep neural network for sleep stage scoring with unfiltered clinical data. Sleep Breath. 2020;24(2):581–590. doi:10.1007/s11325-019-02008-w
9. Peter-Derex L, Berthomier C, Taillard J, et al. Automatic analysis of single-channel sleep EEG in a large spectrum of sleep disorders. J Clin Sleep Med. 2021;17(3):393–402. doi:10.5664/jcsm.8864
10. Huang X, Shirahama K, Li F, et al. Sleep stage classification for child patients using DeConvolutional Neural Network. Artif Intell Med. 2020;110:101981. doi:10.1016/j.artmed.2020.101981
11. Venkatesh K, Poonguzhali S, Mohanavelu K, et al. Sleep Stages Classification Using Neural Network with Single Channel EEG. IEEE Access. 2019;7:96495–96505. doi:10.1109/ACCESS.2019.2928129
12. Sharma M, Tiwari J, Acharya UR. Automatic Sleep-Stage Scoring in Healthy and Sleep Disorder Patients Using Optimal Wavelet Filter Bank Technique with EEG Signals. Int J Environ Res Public Health. 2021;18(6):3087. doi:10.3390/ijerph18063087
13. Younes M, Raneri J, Hanly P. Staging Sleep in Polysomnograms: analysis of Inter-Scorer Variability. J Clin Sleep Med. 2016;12(6):885–894. doi:10.5664/jcsm.5894
14. Roy Y, Banville H, Albuquerque I, et al. Deep learning-based electroencephalography analysis: a systematic review. J Neural Eng. 2019;16(5):051001. doi:10.1088/1741-2552/ab260c
15. Kemp B, Zwinderman AH, Tuk B, et al. Analysis of a sleep-dependent neuronal feedback loop: the slow-wave microcontinuity of the EEG. IEEE Trans Biomed Eng. 2000;47(9):1185–1194. doi:10.1109/10.867928
16. Sateia MJ. International classification of sleep disorders-third edition: highlights and modifications. Chest. 2014;146(5):1387–1394. doi:10.1378/chest.14-0970
17. Society of Pediatric Surgery CM. Chinese guideline for the diagnosis and treatment of childhood obstructive sleep apnea (2020). Zhonghua Er Bi Yan Hou Tou Jing Wai Ke Za Zhi. 2020;55(8):729–747.
18. Simonyan K, Zisserman A. Very Deep Convolutional Networks for Large-Scale Image Recognition. Computer Sci. 2014.
19. He K, Zhang X, Ren S, et al. Deep Residual Learning for Image Recognition.
20. Shelhamer E, Long J, Darrell T. Fully Convolutional Networks for Semantic Segmentation. IEEE Trans Pattern Anal Mach Intell. 2017;39(4):640–651. doi:10.1109/TPAMI.2016.2572683
21. Oord A, Dieleman S, Zen H, et al. WaveNet: a Generative Model for Raw Audio. arXiv:1609.03499. 2016.
22. Pinheiro P, Collobert R, Dollar P. Learning to Segments Objects Candidates. Adv Neural Inf Process Syst. 2015;2:1547.
23. Xiong W, Droppo J, Huang X, et al. The Microsoft 2016 Conversational Speech Recognition System.
24. Szegedy C, Liu W, Jia Y, et al. Going Deeper with Convolutions. IEEE Computer Soc. 2014.
25. Szegedy C, Vanhoucke V, Ioffe S, et al. Rethinking the Inception Architecture for Computer Vision.2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR).
26. Szegedy C, Ioffe S, Vanhoucke V, et al. Inception-v4, Inception-ResNet and the Impact of Residual Connections on Learning.
27. Xie S, Girshick R, Dollár P, et al. Aggregated Residual Transformations for Deep Neural Networks.2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR).
28. Luo L, Xiong Y, Liu Y, et al. Adaptive Gradient Methods with Dynamic Bound of Learning Rate.
29. Delimayanti MK, Purnama B, Nguyen NG, et al. Classification of Brainwaves for Sleep Stages by High-Dimensional FFT Features from EEG Signals. Applied Sciences. 2020;10:5. doi:10.3390/app10051797
30. Yildirim O, Baloglu UB, Acharya UR. A Deep Learning Model for Automated Sleep Stages Classification Using PSG Signals. Int J Environ Res Public Health. 2019;16(4):599. doi:10.3390/ijerph16040599
31. da Silveira TLT, Kozakevicius AJ, Rodrigues CR. Single-channel EEG sleep stage classification based on a streamlined set of statistical features in wavelet domain. Med Biol Eng Comput. 2017;55(2):343–352. doi:10.1007/s11517-016-1519-4
32. Korkalainen H, Aakko J, Nikkonen S, et al. Accurate Deep Learning-Based Sleep Staging in a Clinical Population With Suspected Obstructive Sleep Apnea. IEEE J Biomed Health Inform. 2020;24(7):2073–2081.
33. Mousavi S, Afghah F, Acharya UR. SleepEEGNet: automated sleep stage scoring with sequence to sequence deep learning approach. PLoS One. 2019;14(5):e0216456. doi:10.1371/journal.pone.0216456
34. Khalili E, Mohammadzadeh Asl B. Automatic Sleep Stage Classification Using Temporal Convolutional Neural Network and New Data Augmentation Technique from Raw Single-Channel EEG. Comput Methods Programs Biomed. 2021;204:106063. doi:10.1016/j.cmpb.2021.106063
35. Chriskos P, Frantzidis CA, Gkivogkli PT. Automatic Sleep Staging Employing Convolutional Neural Networks and Cortical Connectivity Images. IEEE Trans Neural Netw Learn Syst. 2020;31(1):113–123. doi:10.1109/TNNLS.2019.2899781
36. Guillot A, Sauvet F, During EH, et al. Dreem Open Datasets: multi-Scored Sleep Datasets to Compare Human and Automated Sleep Staging. IEEE Trans Neural Syst Rehabil Eng. 2020;28(9):1955–1965. doi:10.1109/TNSRE.2020.3011181
37. Hassan AR, Bhuiyan MI. A decision support system for automatic sleep staging from EEG signals using tunable Q-factor wavelet transform and spectral features. J Neurosci Methods. 2016;271:107–118. doi:10.1016/j.jneumeth.2016.07.012
38. Scott H, Lovato N, Lack L. The Development and Accuracy of the THIM Wearable Device for Estimating Sleep and Wakefulness. Nat Sci Sleep. 2021;13:39–53. doi:10.2147/NSS.S287048
39. Toften S, Pallesen S, Hrozanova M, et al. Validation of sleep stage classification using non-contact radar technology and machine learning (Somnofy®). Sleep Med. 2020;75:54–61. doi:10.1016/j.sleep.2020.02.022
40. Sun H, Ganglberger W, Panneerselvam E, et al. Sleep staging from electrocardiography and respiration with deep learning. Sleep. 2020;43(7):zsz306. doi:10.1093/sleep/zsz306
41. Supratak A, Dong H, Wu C, et al. DeepSleepNet: a Model for Automatic Sleep Stage Scoring Based on Raw Single-Channel EEG. IEEE Trans Neural Syst Rehabil Eng. 2017;25(11):1998–2008. doi:10.1109/TNSRE.2017.2721116
42. Sors A, Bonnet S, Mirek S, et al. A convolutional neural network for sleep stage scoring from raw single-channel EEG. Biomed Signal Process Control. 2018;42:107–114. doi:10.1016/j.bspc.2017.12.001
43. Tsinalis O, Matthews PM, Guo Y. Automatic Sleep Stage Scoring Using Time-Frequency Analysis and Stacked Sparse Autoencoders. Ann Biomed Eng. 2016;44(5):1587–1597. doi:10.1007/s10439-015-1444-y
 © 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2021 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.