Back to Journals » OncoTargets and Therapy » Volume 9
ATG7 promotes the tumorigenesis of lung cancer but might be dispensable for prognosis predication: a clinicopathologic study
Authors Sun S, Wang Z, Tang F, Hu P, Yang Z, Xue C, Gong J, Shi L, Xie C
Received 5 March 2016
Accepted for publication 15 June 2016
Published 11 August 2016 Volume 2016:9 Pages 4975—4981
DOI https://doi.org/10.2147/OTT.S107876
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr William C. Cho
Shaoxing Sun,1,2,* Zhihao Wang,1,2,* Fang Tang,1,2 Pengchao Hu,3 Zetian Yang,4 Chao Xue,4 Jun Gong,1,2 Liu Shi,1,2 Conghua Xie,1,2
1Department of Radiation and Medical Oncology, 2Hubei Key Laboratory of Tumor Biological Behaviors, Zhongnan Hospital of Wuhan University, 3Department of Pathology and Pathophysiology, School of Basic Medical Sciences, Wuhan University, 4Department of Thoracic and Cardiovascular Surgery, Zhongnan Hospital of Wuhan University, Wuhan, People’s Republic of China
*These authors contributed equally to this work
Abstract: Lung cancer is the most frequent cause of cancer-related death worldwide. Dysregulated autophagy is often observed in lung cancer. Autophagy-related 7 (ATG7) is an autophagy gene that is essential for the biogenesis of autophagosomes. Although ATG7-deficient mouse models have demonstrated that ATG7-dependent autophagy is required for lung cancer tumorigenesis, the relationship between ATG7 expression levels and human lung cancer is unclear. Here, we demonstrate that ATG7 was overexpressed in human lung cancer tissues compared with normal tissues. However, ATG7 expression was not associated with tumor differentiation, tumor size, or TNM stage. Moreover, the overexpression of ATG7 did not influence the overall survival of the lung cancer patients. Therefore, our results indicate that ATG7 might be dispensable for tumor growth and chemotherapy efficacy in human lung cancer.
Keywords: ATG7, autophagy, lung cancer, prognosis, clinicopathologic study
Introduction
Lung cancer is the leading cause of cancer-related death worldwide. Surgery, chemotherapy, and radiotherapy are the main approaches to lung cancer treatment.1 Despite therapeutic advances, the overall 5-year survival of lung cancer remains very low.2 Carcinogenesis and the progression of lung cancer are very complicated processes that are accompanied by complex cell signal network alterations, which contribute to cell biology behavioral changes.3 As a fundamentally important mechanism of substance recycling, autophagy has recently sparked great interest because it is now recognized to be involved in various developmental processes and various diseases, including lung cancer.4
Autophagy is an essential, conserved lysosomal degradation pathway that controls the quality of the cytoplasm by eliminating protein aggregates and damaged organelles.4,5 If not specified, autophagy means macroautophagy, which is the major type of autophagy. Autophagy allows cells to degrade their own proteins and organelles to maintain the cellular homoeostasis that is required for normal growth and development, short-term adaptations to stress, and the long-term survival of optimally fit cells.4,5 Dysregulation of autophagy is often observed in cancer.6 However, the role of autophagy in cancer is context-dependent. In some settings, autophagy may limit tumorigenesis by preserving protein and organelle quality control, whereas in other contexts, autophagy promotes metabolism, adaptations to stress, and tumor growth.7 The functions of autophagy are conserved from yeast through mammals and are controlled by the autophagy-related genes (ATG).8 Among these genes, the common ubiquitin E1-like activating enzyme autophagy-related 7 (ATG7) is essential for autophagy via its control of the assembly and expansion of autophagosomal membranes. ATG7 deficiency reduces KRAS-driven lung tumorigenesis but also causes robust early tumor establishment in BrafV600E-driven mouse lung cancer models.9,10 Recently, Eng et al demonstrated that ATG7-dependent autophagy is dispensable to the growth of KRAS-mutant tumors and their chemosensitivity.11 Inconsistent results drawn from different models have caused confusion regarding the role of ATG7-dependent autophagy in tumorigenesis. Although numerous studies of the role of ATG7 deficiency in mouse models have been conducted, the relationship between ATG7 expression levels and the clinicopathologic characteristics of human lung cancer remains unclear.
With this issue in mind, the present study examined ATG7 expression in human lung cancer samples and evaluated its association with clinicopathological parameters and its prognostic significance. We first compared ATG7 expression in lung cancer tissues with that in normal lung tissues. We found that ATG7 was overexpressed in human lung cancer tissues compared to the normal tissues. However, ATG7 expression was not associated with tumor differentiation, tumor size, TNM stage, or prognosis.
Materials and methods
Ethics statement
This study was approved by the Ethics Committee of Zhongnan Hospital of Wuhan University. Written informed consent was obtained from all participants.
Patients and tissue samples
Cohort 1
Forty-one pairs of fresh tissue samples from lung cancer patients with no prior treatment were collected during the second half of 2013. The tumor tissues were sampled by the surgeon, and normal samples were taken from the grossly normal lung at least 5 cm away from the tumor in the resected lung specimens. All tissues were fixed in formalin at more than ten times the volume of the tissue within 10 minutes following resection from the body and for at least 12 hours. Paraffin embedding was performed within 48 hours, and to confirm the pathological characteristics, hematoxylin and eosin (HE) staining was performed prior to immunohistochemistry. The HE slices were examined by a well-trained pathologist, and those HE slices that coincided with official reports were submitted for further analysis. Meanwhile, baseline data were collected. The International Association for Study of Lung Cancer staging system (7th edition) was used for the staging.
Cohort 2
To investigate the relationships between ATG7 expression and the clinical outcomes of the non-small-cell lung cancer patients, we screened the surgery sample library of the Department of Pathology, Zhongnan Hospital of Wuhan University for non-small-cell lung cancer (NSCLC) patient samples from 2010 through 2011. None of the patients underwent radiotherapy or chemotherapy before surgery. Tissue samples from 76 included patients were obtained with the assistance of the work staff of that department. The baseline characteristics were primarily collected by consulting archived medical records, and the same staging system was used to reclassify the stages. The follow-ups were completed primarily via telephone contact within 2 months. Thirteen patients were excluded from the study due to loss of contact. The remaining samples were from patients with a median age of 60 (range 37–79) and were submitted for further immunohistochemical analysis, and survival data analyses were performed.
Immunohistochemistry analysis
After the confirmation of suitable tissue structure based on the HE staining, paraffin-embedded tumor and normal tissues from cohort 1 were cut at 4 μm and mounted on glass slides. Along with the samples from cohort 2, which were prepared in the same manner, all slides were dewaxed in xylene and then rehydrated in ethanol. For antigen exposure, all slides were placed in 0.01 mol/L citrate buffer (pH =6), heated in a microwave oven for 15 minutes, and then left at room temperature for 20 minutes. An UltraSensitive™ SP (Goat) immunohistochemistry kit (Fuzhou Maixin Biotech. Co. Ltd., Fuzhou, Fujian, People’s Republic of China) was used for the immunohistochemistry detection. The procedures were performed strictly according to the manufacturer’s instructions. 3,3′-Diaminobenzidine was used as the chromogen. Anti-ATG7 primary antibody ab53255 (Abcam, San Francisco, CA, USA) at a concentration of 1:100 was applied. A reddish brown precipitate located in the cytoplasm of the cells indicated a positive reaction. In each immunohistochemistry run, a hepatocellular carcinoma section served as a positive control, and the omission of the primary antibody served as a negative control. The scorings were calculated by two independent investigators with no access to the clinical data. Positive staining area (0, none; 1, <10%; 2, 10%–50%; 3, 50%–80%; and 4, >80%) and staining strength (0, negative; 1, weak; 2, moderate; and 3, strong) were considered. The final scores were determined by the combined staining score (area score plus strength score). Immunoreactive scores equal or >3 were used to the classify tumors with positive ATG7 expression, and staining scores below 3 were indicated negative ATG7 expression.
Statistical analysis
The positive staining rates of the tumor and normal tissue slides from cohort 1 were calculated and compared using a chi-square test. The relationships between the characteristics of the patients from cohort 2 and the ATG7 expression levels in the tumor tissues were examined with Spearman correlation analyses. Survival curves were plotted using Kaplan–Meier methods and compared using the log-rank test. Multivariate Cox proportional hazard models were used to identify the potential prognostic significance of each parameter. Trend tests were also performed. All statistical work was performed with licensed SPSS 20.0 software (IBM Corporation, Armonk, NY, USA). A P-value <0.05 was considered statistically significant.
Results
ATG7 is overexpressed in lung cancer tissues compared to normal tissues
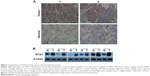
Although numerous studies have demonstrated the role of ATG7 in the tumorigenesis of lung cancer, the relationships between ATG7 expression levels and human lung cancer clinicopathologic characteristics remain unclear.9–11 We first compared the expression of ATG7 in the lung cancer tissues and normal lung tissues. As presented in Table 1, 41 paired tissues were collected, and immunohistochemistry was performed by a well-trained pathological technician. Thirty-four (82.9%) males and seven (17.1%) females with a mean age of 60.6 (40–76) years were diagnosed with adenocarcinomas (51.2%), squamous carcinomas (43.6%), and small-cell carcinomas (2.4%). The majority of the patients (65.8%) were in the local advanced stage, which coincides with the epidemiology data. The ATG7 protein was primarily detected in the cytoplasm of the tumor cells (Figure 1A). Positive staining was identified in 26 (63.4%) tumor tissue samples, whereas only nine (9.8%) normal lung tissue samples were considered positive. A chi-square test revealed a significant difference (P=0.00). We also confirmed this finding with Western blotting (Figure 1B).
  | Table 1 Characteristics of lung cancer patients with paired tissues (total N=41) |
ATG7 expression levels are not correlated with the clinicopathological parameters or prognoses of lung cancer patients
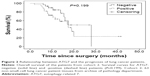
Because ATG7 is overexpressed in lung cancer tissues, and previous studies have demonstrated that ATG7 is required for efficient tumorigenesis in lung cancer, we speculated that ATG7 might be negatively correlated with the prognoses of lung cancer patients.10,12 Surprisingly however, in cohort 1, ATG7 expression was not associated with tumor differentiation (P=0.799), tumor size (P=0.986), or TNM stage (T stage, P=0.781; metastases of the lymph nodes, P=0.923) (Table 2). Because the ATG7 expression level was not associated with the clinicopathological parameters, we next sought to determine whether ATG7 expression was correlated with the prognoses of lung cancer patients. As presented in Table 3, 63 patients with a mean age of 59.4 (37–79) were included. Forty-seven males and 16 females had been diagnosed with lung cancer and selected surgery as the initial treatment. The majority of the patients (92.1%) had pathologically clean residuals. Seven patients in stage IV underwent operations mainly due to insufficient examinations and consequent incorrect staging. The relationships between ATG7 expression and the patients’ features were also analyzed, and similar to cohort 1, no significant results were obtained (Table 4). The patients with no ATG7 expression had a median survival time of 17.5 months (95% confidence interval, 11.9–23.1 months), and the patients with positive ATG7 expression had the same survival time of 17.5 months (95% confidence interval, 11.3–23.7 months). No significant difference was noticed (P=0.199, Figure 2). In cohort 2, ATG7 expression was also not associated with tumor differentiation (P=0.860), tumor size (P=0.942), or T stage (P=0.151, Table 4).
Discussion
Lung cancer is the leading cause of cancer-related mortality worldwide. Despite recent therapeutic advances, traditional surgical resection, chemotherapy, and radiotherapy have not satisfactorily improved the long-term survival of patients with NSCLC.13 Approximately 40% of patients with NSCLC die of recurrence or metastasis within 5 years after potentially curative resection.1 Autophagy is a catabolic process that regulates the lysosomal turnover of damaged proteins and organelles to maintain cellular homeostasis.14 As a fundamentally important mechanism of substance recycling, autophagy has recently sparked great interest because it is now recognized to be involved in various diseases, including lung cancer.15 Recent data have suggested that lung cancers in particular may be selectively dependent on autophagy.12
The functions of autophagy are conserved from yeast through mammals and are controlled by the autophagy-related genes (ATG).16 One particular member of the ATG protein family, ATG7, has been the focus of recent research. ATG7 acts as an E1-like activating enzyme that facilitates both microtubule-associated protein light chain 3-phosphatidylethanolamine and ATG12 conjugation. Thus, ATG7 stands at the hub of these two ubiquitin-like systems to involve light chain 3 and ATG12 in autophagic vesicle expansion.17 Because ATG7 is such an important autophagy-related gene, numerous studies have utilized ATG7 knockout to generate autophagy deficiency mouse models to illuminate the relationship between autophagy and cancer. However, the inconsistent results from different mouse models motivated us to examine the relationships between ATG7 expression and clinicopathological parameters. Here, our results indicated that ATG7 promotes the tumorigenesis of lung cancer but is dispensable for prognosis predication. Guo et al demonstrated that although ATG7 deficiency in KRAS-induced lung tumors reduces the tumor burden by half, no association with an overall survival advantage has been observed in mouse models.18 Recently, Eng et al demonstrated that ATG7 is dispensable to the growth of KRAS-mutant tumors and chemotherapy efficacy.11 Our results partly confirmed their conclusion. ATG7 is essential for autophagy and knockout of ATG7 is often used to generate autophagy-deficient models. The role of autophagy in cancer is context-dependent.19 p53 Status has been demonstrated to determine the role of autophagy in tumor development.20 Therefore, our results suggest that ATG7-dependent autophagy might not regulate overall survival in unselected patients with lung cancer.
At present, there are no validated biomarkers that are indicative of autophagy inhibitors. The discovery of EGFR mutations that confer sensitivity to tyrosine kinase inhibitors in lung adenocarcinomas in 2004 heralded the beginning of the era of precision medicine for lung cancer.3,13 Efforts have been made to identify novel therapeutic targets and define the mechanisms of sensitivity and resistance to targeted therapies. The definition of molecular subgroups of lung cancer with prognoses that are closely correlated with autophagy and the characterizations of the genomic landscape of these lung cancer subtypes seem urgent. Autophagy dysregulation is often observed in cancer, and autophagy has been demonstrated to confer resistance to several first-line lung cancer chemotherapy agents.21–23 Therefore, the inhibition of autophagy is a promising strategy for improving the overall survival of lung cancer patients. A number of clinical trials are now revealing the promising role of the autophagy inhibitor chloroquine as a novel antitumor drug.24 However, several studies have reported that chloroquine sensitizes cancer cells to chemotherapy independently of autophagy inhibition.11,25 Autophagy inhibitors with higher specificity need to be discovered and tested in clinical trials. Moreover, ATG7-independent autophagy exists in cells.26 Future studies are needed to clarify the role of ATG7-independent autophagy in cancer.
Acknowledgment
This work was supported by the Natural Sciences Foundation of Hubei Province (number 2013CFA006).
Disclosure
The authors report no conflicts of interest in this work.
References
Ettinger DS, Wood DE, Akerley W, et al. Non-small cell lung cancer, version 6.2015. J Natl Compr Canc Netw. 2015;13(5):515–524. | ||
DeSantis CE, Lin CC, Mariotto AB, et al. Cancer treatment and survivorship statistics, 2014. CA Cancer J Clin. 2014;64(4):252–271. | ||
Politi K, Herbst RS. Lung cancer in the era of precision medicine. Clin Cancer Res. 2015;21(10):2213–2220. | ||
Jiang P, Mizushima N. Autophagy and human diseases. Cell Res. 2014;24(1):69–79. | ||
Rubinsztein DC, Gestwicki JE, Murphy LO, Klionsky DJ. Potential therapeutic applications of autophagy. Nat Rev Drug Discov. 2007;6(4):304–312. | ||
Guo JY, Xia B, White E. Autophagy-mediated tumor promotion. Cell. 2013;155(6):1216–1219. | ||
Duffy A, Le J, Sausville E, Emadi A. Autophagy modulation: a target for cancer treatment development. Cancer Chemother Pharmacol. 2015;75(3):439–447. | ||
Xiong J. Atg7 in development and disease: panacea or Pandora’s box? Protein Cell. 2015;6(10):722–734. | ||
Guo JY, Karsli-Uzunbas G, Mathew R, et al. Autophagy suppresses progression of K-ras-induced lung tumors to oncocytomas and maintains lipid homeostasis. Genes Dev. 2013;27(13):1447–1461. | ||
Strohecker AM, Guo JY, Karsli-Uzunbas G, et al. Autophagy sustains mitochondrial glutamine metabolism and growth of BrafV600E-driven lung tumors. Cancer Discov. 2013;3(11):1272–1285. | ||
Eng CH, Wang Z, Tkach D, et al. Macroautophagy is dispensable for growth of KRAS mutant tumors and chloroquine efficacy. Proc Natl Acad Sci U S A. 2016;113(1):182–187. | ||
Karsli-Uzunbas G, Guo JY, Price S, et al. Autophagy is required for glucose homeostasis and lung tumor maintenance. Cancer Discov. 2014;4(8):914–927. | ||
Patel JN, Ersek JL, Kim ES. Lung cancer biomarkers, targeted therapies and clinical assays. Transl Lung Cancer Res. 2015;4(5):503–514. | ||
Rubinsztein DC, Codogno P, Levine B. Autophagy modulation as a potential therapeutic target for diverse diseases. Nat Rev Drug Discov. 2012;11(9):709–730. | ||
Rao S, Tortola L, Perlot T, et al. A dual role for autophagy in a murine model of lung cancer. Nat Commun. 2014;5:3056. | ||
Bento CF, Renna M, Ghislat G, et al. Mammalian autophagy: how does it work? Annu Rev Biochem. 2016;85:685–713. | ||
Noda NN, Inagaki F. Mechanisms of autophagy. Annu Rev Biophys. 2015;44:101–122. | ||
Guo JY, Chen HY, Mathew R, et al. Activated Ras requires autophagy to maintain oxidative metabolism and tumorigenesis. Genes Dev. 2011;25(5):460–470. | ||
White E. Deconvoluting the context-dependent role for autophagy in cancer. Nat Rev Cancer. 2012;12(6):401–410. | ||
Rosenfeldt MT, O’Prey J, Morton JP, et al. p53 status determines the role of autophagy in pancreatic tumour development. Nature. 2013;504(7479):296–300. | ||
Xu Z, Hang J, Hu J, Gao B. Gefitinib, an EGFR tyrosine kinase inhibitor, activates autophagy through AMPK in human lung cancer cells. J Buon. 2014;19(2):466–473. | ||
Zou Y, Ling YH, Sironi J, Schwartz EL, Perez-Soler R, Piperdi B. The autophagy inhibitor chloroquine overcomes the innate resistance of wild-type EGFR non-small-cell lung cancer cells to erlotinib. J Thorac Oncol. 2013;8(6):693–702. | ||
Sui X, Kong N, Zhu M, et al. Cotargeting EGFR and autophagy signaling: A novel therapeutic strategy for non-small-cell lung cancer. Mol Clin Oncol. 2014;2(1):8–12. | ||
Pascolo S. Time to use a dose of chloroquine as an adjuvant to anti-cancer chemotherapies. Eur J Pharmacol. 2016;771:139–144. | ||
Maycotte P, Aryal S, Cummings CT, Thorburn J, Morgan MJ, Thorburn A. Chloroquine sensitizes breast cancer cells to chemotherapy independent of autophagy. Autophagy. 2012;8(2):200–212. | ||
Nishida Y, Arakawa S, Fujitani K, et al. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature. 2009;461(7264):654–658. |
 © 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2016 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.