Back to Journals » Clinical Ophthalmology » Volume 11
Associations between CRYBA4 gene variants and high myopia in a Japanese population
Authors Kawagoe T, Ota M, Meguro A , Takeuchi M, Yamane T, Shimazaki H, Takeuchi M , Okada E, Teshigawara T , Mizuki N
Received 11 July 2017
Accepted for publication 9 October 2017
Published 7 December 2017 Volume 2017:11 Pages 2151—2156
DOI https://doi.org/10.2147/OPTH.S146038
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Scott Fraser
Tatsukata Kawagoe,1 Masao Ota,1,2 Akira Meguro,1 Masaki Takeuchi,1 Takahiro Yamane,1 Haruna Shimazaki,3 Masaru Takeuchi,3 Eiichi Okada,4 Takeshi Teshigawara,1,5,6 Nobuhisa Mizuki1
1Department of Ophthalmology and Visual Science, Yokohama City University Graduate School of Medicine, Yokohama, 2Department of Medicine, Division of Gastroenterology and Hepatology, Shinshu University School of Medicine, Matsumoto, 3Department of Ophthalmology, National Defense Medical College, Tokorozawa, 4Okada Eye Clinic, Yokohama, 5Yokosuka Chuoh Eye Clinic, Yokosuka, 6Tsurumi Chuoh Eye Clinic, Yokohama, Japan
Purpose: The crystallin beta A4 (CRYBA4) gene variant, rs2009066, was previously reported to be associated with high myopia in a southern Chinese population. In the present study, we investigated whether CRYBA4 variants were associated with high myopia in a Japanese population.
Methods: We recruited 1,063 Japanese patients with high myopia (spherical equivalent [SE] ≤–9.00 D in both eyes) and 1,009 healthy Japanese subjects (SE >–1.00 D). We genotyped rs2009066 and three tagging single-nucleotide polymorphisms (SNPs), rs16982456, rs2071861, and rs4276, in the CRYBA4 region.
Results: We did not find any significant association between these four SNPs and high myopia in an allele analysis. However, rs2009066 and rs2071861, which were in strong linkage disequilibrium (LD; r2=0.86), showed a marginal association with high myopia in the recessive genotype model of risk alleles (rs2009066 G allele: P=0.032, odds ratio [OR] =1.31; rs2071861 A allele: P=0.037, OR =1.31). Nevertheless, this association became insignificant after correcting for multiple testing (Pc >0.05).
Conclusion: This study showed no significant association between CRYBA4 variants and high myopia in a Japanese population. Our findings did not correspond with a previous study. Further genetic studies with other populations are needed to elucidate a potential contribution of the CRYBA4 region in the development of high myopia.
Keywords: high myopia, CRYBA4, association study, polymorphism
Introduction
Myopia is the most frequent ocular disorder worldwide in the modern world.1 High myopia is distinguished from common myopia, based on a spherical equivalent (SE) refractive error of <−6 D or an axial length (AL) of >26 mm. It is well known that high myopia is associated with an increased risk of various ocular diseases, including retinal detachment, glaucoma, and cataract.2 The prevalence of high myopia in East Asian and Southeast Asian populations is higher than in Caucasian populations.3–7
Although the pathogenesis of high myopia remains uncertain, previous studies have indicated that genetic and environmental factors, such as education, are involved in the progression of high myopia.8–13 Familial linkage studies and twin studies have identified 25 myopia loci (MYP1 to MYP25) (OMIM; http://omim.org/),14 and many genome-wide association studies have dealt with several candidate loci/genes for myopia, refractive error, and/or elongation of the AL in several ethnic populations.15–20 Most of the identified genes were not confirmed in follow-up studies on either common or high myopia, and it is not yet apparent whether the same genetic background is associated with both myopias.
Family-based linkage analyses have identified a large number of chromosomal regions, for example, in the MYP loci (MYP1–3, MYP5–6, MYP15–17, MYP18–19, MYP21–22, MYP24–25), associated with high myopia (OMIM; http://omim.org/).21 Among these, we focused on the MYP6 locus on chromosome 22q13.33 containing the SCO2 cytochrome c oxidase assembly protein (SCO2) gene, which encodes a copper chaperone required for cytochrome c oxidase assembly.22 Tran-Viet et al showed that heterozygous mutations in the SCO2 gene cause high-grade myopia in patients in the USA and that SCO2 expression was significantly downregulated in myopic mouse retinae.23 Another study also identified other heterozygous mutations in SCO2 causing high myopia in the People’s Republic of China.24 These findings suggest SCO2 mutations are an important risk factor for the development of high myopia. However, Wakazono et al suggested that SCO2 mutations may play a limited role in extreme myopia among Japanese patients.25 Furthermore, the SCO2 E140K mutation identified by Tran-Viet et al was not associated with high myopia in a Poland population.26 Thus, in the MYP6 locus, the contribution of SCO2 to myopia development is still unclear.
Recently, Ho et al performed a case–control association study with 26 potential candidate genes in a southern Chinese population to identify myopia susceptibility genes in the MYP6 locus. That study identified a novel gene, crystallin beta A4 (CRYBA4), which conferred susceptibility to high myopia.27 CRYBA4 is part of a four-gene cluster of β-crystallin spanning approximately 1.4 Mbp on chromosome 22. Previous studies28,29 have shown that missense mutations in the CRYBA4 gene are involved in cataractogenesis and microphthalmia and indicated that CRYBA4 plays an important role in eye development. These evidences suggest that CRYBA4 variants may be a risk factor for the development of high myopia.
To date, no replication study for the findings on CRYBA4 has been performed in other ethnic populations. The aim of the current study was to investigate whether CRYBA4 variants were associated with high myopia in a Japanese population. Here, we assessed multiple single-nucleotide polymorphisms (SNPs) in the CRYBA4 gene to determine their association with the risk of disease in Japanese patients with high myopia.
Methods
Subjects
We recruited 1,063 unrelated Japanese individuals with high myopia (SE ≤−9.00 D in both eyes) and 1,009 unrelated healthy Japanese control subjects (SE >−1.00 D in both eyes) at Yokohama City University, Okada Eye Clinic, and Aoto Eye Clinic in Japan. All participants were unrelated to each other, but had similar social backgrounds and resided in the same urban area. They were diagnosed with comprehensive ophthalmologic tests, which included an AL measurement, a fundus examination, and determinations of spherical power and corneal curvature (Autorefractor; NIDEK ARK-730A, ARK-700A TOPCON KP-8100P, BIO & PACHY Meter AL-2000; Tomey Corporation). The subjects with high myopia had no known genetic diseases associated with myopia and/or high myopia, including glaucoma, keratoconus, or Marfan syndrome. We excluded individuals younger than 20 years old from the control cohort to exclude individuals with potential myopia. Written informed consent was obtained from all participants. The study methodology adhered to the tenets of the Declaration of Helsinki, and the study was approved by the relevant Ethics Committees at Yokohama City University, Okada Eye Clinic, and Aoto Eye Clinic.
SNP genotyping of the CRYBA4 gene region
Genomic DNA was extracted from peripheral blood samples with the QIAamp DNA Blood Mini Kit (Qiagen, Hilden, Germany). Procedures were performed under standardized conditions to prevent variation in DNA quality.
We evaluated rs2009066, which is located 3 kb downstream of the CRYBA4 gene. rs2009066 was previously reported to be the gene most significantly associated with high myopia.25 We also selected three tagging SNPs (rs16982456, rs2071861, and rs4276) that covered the entire CRYBA4 gene, including 10 kb upstream and downstream (minor allele frequency ≥5%, pairwise r2≥0.8), with the LD tag SNP selection tool in the SNPinfo web server (https://snpinfo.niehs.nih.gov/). Genotyping was performed with the polymerase chain reaction (PCR) method; we used the TaqMan 5′ exonuclease assay with validated TaqMan primer-probe sets supplied by Applied Biosystems (Foster City, CA, USA). The PCR mixture (total volume, 10 μL) contained 1× TaqMan Universal PCR Master Mix (Applied Biosystems), 24 nM of each primer-probe set, and 3 ng genomic DNA. The PCR conditions were as follows: 95°C for 10 min, followed by 40 cycles of denaturation at 92°C for 15 s and annealing/extension at 60°C for 1 min. Probe fluorescence was detected with the StepOne Plus Real-Time PCR System (Applied Biosystems).
Statistical analysis
Allele frequencies, genotype frequencies, Hardy–Weinberg equilibrium (HWE), and LD were assessed with SNP & Variation Suite 8.4.0 software (Golden Helix, Inc., Bozeman, MT, USA; http://www.goldenhelix.com), SNPStats software (Catalan Institute of Oncology, Barcelona, Spain; http://bioinfo.iconcologia.net/SNPstats),30 and Haploview 4.1 software.31 The obtained P-values were corrected for multiple hypothesis testing with Bonferroni’s method. A corrected P (Pc) value <0.05 was considered significant. Multiple inheritance models were used to analyze genotype data in assessing each risk allele. These inheritance models were additive (R/R vs R/nR vs nR/nR), dominant (R/R + R/nR vs nR/nR), and recessive (R/R vs R/nR + nR/nR), where R was the risk allele and nR was the non-risk allele. P-values and odds ratios (ORs) in genotype models were adjusted for age and sex (Padj and ORadj, respectively).
Results
The clinical characteristics of the study populations are shown in Table 1. Patient age ranged from 13 to 72 years (mean 38.7±11.8 years), and 43.1% of patients were male. The average SEs of the patients were −11.21±1.92 D in the right eye and −11.13±1.95 D in the left eye. The average ALs of the patients were 27.64±1.18 mm (range 23.92–33.85 mm) for the right eye and 27.60±1.16 mm (range 23.99–33.17 mm) for the left eye. The ages of controls ranged from 20 to 87 years (mean 58.3±12.2 years), and 44.3% were male. The average SEs were 0.49±0.64 D (range −0.75 to 3.50 D) in the right eye and 0.49±0.63 D (range −0.75 to 3.00 D) in the left eye. The average ALs were 23.22±0.80 mm (range 18.76–25.96 mm) and 23.20±0.79 mm (range 18.99–26.05 mm) for the right and left eyes, respectively.
Among the controls, the genotype frequencies of all four SNPs analyzed were in HWE (P>0.05). Figure 1 shows the overall LD patterns for the four SNPs. These four SNPs were located in one haplotype block, and three SNPs (rs2071861, rs4276, and rs2009066) were in strong LD with each other (r2≥0.41).
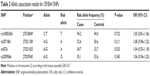
Table 2 shows the allelic association results for the four SNPs. None of the four SNPs showed any significant association with high myopia. Table 3 shows the genotype association results of the four SNPs, calculated for each of the three inheritance models (additive, dominant, and recessive models). No significant association was found for any of the four SNPs in the additive or dominant models. On the other hand, rs2009066 and rs2071861, which were in almost complete LD (r2=0.86), showed significant differences between cases and controls in the recessive model of risk alleles (P=0.032, OR =1.31 and P=0.037, OR =1.31, respectively). However, the differences did not remain significant after applying the Bonferroni correction for multiple testing (Pc >0.05). We did not find any significant association between the other two SNPs and high myopia in the recessive model.
Discussion
High myopia is characterized by AL elongation and thinning of the sclera,32–34 but there are several distinct clinical phenotypes.35–37 High myopia is thought to be caused by multiple genes interacting with the environment. Although many candidate gene studies4–9 and genome-wide association studies10–16 have been conducted, the genetic basis for susceptibility to high myopia has not been fully elucidated.
Ho et al recently identified CRYBA4 as a new gene associated with susceptibility to high myopia. They performed a case–control association study using southern Chinese subjects in Hong Kong.27 They reported that rs2009066 was the SNP most significantly associated with high myopia and its risk allele G had a dominant inheritance effect and a high OR of 1.74.27 In the current study, we found a lack of association between CRYBA4 variants and high myopia in our Japanese population, although the rs2009066 allele G had a possible effect (OR =1.31, but not significant) on disease risk in the recessive inheritance model which is inconsistent with observations from the previous study. This discrepancy may result from complex differences between the two studies. Indeed, these two studies focused on different ethnic groups and had different sample sizes; moreover, there may have been differences in environmental factors, and possibly, genotyping errors.
One of the possible limitations in the present and previous studies is the medium sample size (1,063 patients and 1,009 controls from Japan, 658 patients and 655 controls from Hong Kong). Limited sample sizes can sometimes lead to false-positive or false-negative results in a case–control association study. Various complex diseases often involve the integration of multiple genetic and environmental factors, and risk alleles for such diseases confer small effect sizes (OR <1.5).38 Therefore, it is suggested that the sample sizes in these two studies may have not enough statistical power to accurately detect the association between CRYBA4 variants and high myopia (a common disease with small effect sizes).
The structural stability of crystallins is important for maintaining transparency and refractive error in the lens.39 In the human lens, there are three major crystallin classes, α-crystallin, β-crystallin, and γ-crystallin, which contain 40%, 35%, and 25% of total crystallin proteins, respectively.28 β-crystallins are the major constituents of human lens.40 CRYBA4 is a 196-amino-acid protein and constitutes 5% of the total crystallins in the young human lens.40 The expression of CRYBA4 is limited to the lens fiber cells; it is not found in the retina, cornea, iris, choroid, sclera, etc.41 CRYBA4 mutations have been reported to be involved in cataractogenesis and microphthalmia,28,29 and it has been suggested that CRYBA4 variants may affect eye development. Considering the functional role of CRYBA4, it is not surprising that it has been associated with ocular abnormalities, including myopia. However, only one study has reported that rs2009066, which is located near the CRYBA4 gene, was a marker for susceptibility to high myopia.27 In contrast, the present study showed no association between rs2009066 and high myopia in the Japanese population. Although our results do not support those of previous study, we could not still exclude the possibility that CRYBA4 variants may affect the risk of high myopia. To clarify the role of CRYBA4 gene variants in the risk of high myopia, further genetic analyses should be performed in different populations.
Acknowledgments
The authors thank all the subjects for their participation in this study. They also thank Dr Yasuhito Iijima and the medical staff at the Aoto Eye Clinic for their assistance with sample collection and diagnosis.
Disclosure
The authors report no conflicts of interest in this work.
References
Pararajasegaram R. VISION 2020-the right to sight: from strategies to action. Am J Ophthalmol. 1999;128(3):359–360. | ||
Saw SM, Gazzard G, Shih-Yen EC, Chua WH. Myopia and associated pathological complications. Ophthalmic Physiol Opt. 2005;25(5):381–391. | ||
Lam CS, Goldschmidt E, Edwards MH. Prevalence of myopia in local and international schools in Hong Kong. Optom Vis Sci. 2004;81(5):317–322. | ||
Pan CW, Zheng YF, Anuar AR, et al. Prevalence of refractive errors in a multiethnic Asian population: the Singapore epidemiology of eye disease study. Invest Ophthalmol Vis Sci. 2013;54(4):2590–2598. | ||
Fledelius HC. Myopia prevalence in Scandinavia. A survey, with emphasis on factors of relevance for epidemiological refraction studies in general. Acta Ophthalmol Suppl. 1988;185:44–50. | ||
Wensor M, McCarty CA, Taylor HR. Prevalence and risk factors of myopia in Victoria, Australia. Arch Ophthalmol. 1999;117(5):658–663. | ||
Kempen JH, Mitchell P, Lee KE, et al; Eye Diseases Prevalence Research Group. The prevalence of refractive errors among adults in the United States, Western Europe, and Australia. Arch Ophthalmol. 2004;122(4):495–505. | ||
Baird PN, Schäche M, Dirani M. The GEnes in Myopia (GEM) study in understanding the aetiology of refractive errors. Prog Retin Eye Res. 2010;29(6):520–542. | ||
Au Eong KG, Tay TH, Lim MK. Education and myopia in 110,236 young Singaporean males. Singapore Med J. 1993;34(6):489–492. | ||
Au Eong KG, Tay TH, Lim MK. Race, culture and myopia in 110,236 young Singaporean males. Singapore Med J. 1993;34(1):29–32. | ||
Morgan IG, Rose KA. Myopia and international educational performance. Ophthalmic Physiol Opt. 2013;33(3):329–338. | ||
French AN, Ashby RS, Morgan IG, Rose KA. Time outdoors and the prevention of myopia. Exp Eye Res. 2013;114:56–68. | ||
Fan Q, Verhoeven VJ, Wojciechowski R, et al. Meta-analysis of gene-environment-wide association scans accounting for education level identifies additional loci for refractive error. Nat Commun. 2016;7:11008. | ||
Jacobi FK, Pusch CM. A decade in search of myopia genes. Front Biosci (Landmark Ed). 2010;15:359–372. | ||
Hysi PG, Young TL, Mackey DA, et al. A genome-wide association study for myopia and refractive error identifies a susceptibility locus at 15q25. Nat Genet. 2010;42(10):902–905. | ||
Solouki AM, Verhoeven VJ, van Duijn CM, et al. A genome-wide association study identifies a susceptibility locus for refractive errors and myopia at 15q14. Nat Genet. 2010;42(10):897–901. | ||
Fan Q, Barathi VA, Cheng CY, et al. Genetic variants on chromosome 1q41 influence ocular axial length and high myopia. PLoS Genet. 2012;8(6):e1002753. | ||
Nakanishi H, Yamada R, Gotoh N, et al. A genome-wide association analysis identified a novel susceptible locus for pathological myopia at 11q24.1. PLoS Genet. 2009;5(9):e1000660. | ||
Cheng CY, Schache M, Ikram MK, et al. Nine loci for ocular axial length identified through genome-wide association studies, including shared loci with refractive error. Am J Hum Genet. 2013;93(2):264–277. | ||
Simpson CL, Wojciechowski R, Oexle K, et al. Genome-wide meta-analysis of myopia and hyperopia provides evidence for replication of 11 loci. PLoS One. 2014;9(9):e107110. | ||
Hawthorne FA, Young TL. Genetic contributions to myopic refractive error: insights from human studies and supporting evidence from animal models. Exp Eye Res. 2013;114:141–149. | ||
Robinson NJ, Winge DR. Copper metallochaperones. Annu Rev Biochem. 2010;79:537–562. | ||
Tran-Viet KN, Powell C, Barathi VA, et al. Mutations in SCO2 are associated with autosomal-dominant high-grade myopia. Am J Hum Genet. 2013;92(5):820–826. | ||
Jiang D, Li J, Xiao X, et al. Detection of mutations in LRPAP1, CTSH, LEPREL1, ZNF644, SLC39A5, and SCO2 in 298 families with early-onset high myopia by exome sequencing. Invest Ophthalmol Vis Sci. 2014;56(1):339–345. | ||
Wakazono T, Miyake M, Yamashiro K, Yoshikawa M, Yoshimura N. Association between SCO2 mutation and extreme myopia in Japanese patients. Jpn J Ophthalmol. 2016;60(4):319–325. | ||
Piekutowska-Abramczuk D, Kocyła-Karczmarewicz B, Małkowska M, et al. No evidence for association of SCO2 heterozygosity with high-grade myopia or other diseases with possible mitochondrial dysfunction. JIMD Rep. 2016;27:63–68. | ||
Ho DW, Yap MK, Ng PW, Fung WY, Yip SP. Association of high myopia with crystallin beta A4 (CRYBA4) gene polymorphisms in the linkage-identified MYP6 locus. PLoS One. 2012;7(6):e40238. | ||
Billingsley G, Santhiya ST, Paterson AD, et al. CRYBA4, a novel human cataract gene, is also involved in microphthalmia. Am J Hum Genet. 2006;79(4):702–709. | ||
Zhou G, Zhou N, Hu S, Zhao L, Zhang C, Qi Y. A missense mutation in CRYBA4 associated with congenital cataract and microcornea. Mol Vis. 2010;16:1019–1024. | ||
Solé X, Guinó E, Valls J, Iniesta R, Moreno V. SNPStats: a web tool for the analysis of association studies. Bioinformatics. 2006;22(15):1928–1929. | ||
Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21(2):263–265. | ||
Xu L, Li Y, Wang S, Wang Y, Jonas JB. Characteristics of highly myopic eyes: the Beijing Eye Study. Ophthalmology. 2007;114(1):121–126. | ||
Ohno-Matsui K, Shimada N, Yasuzumi K, et al. Long-term development of significant visual field defects in highly myopic eyes. Am J Ophthalmol. 2011;152(2):256–265.e1. | ||
Rada JA, Shelton S, Norton TT. The sclera and myopia. Exp Eye Res. 2006;82(2):185–200. | ||
Samarawickrama C, Mitchell P, Tong L, et al. Myopia-related optic disc and retinal changes in adolescent children from Singapore. Ophthalmology. 2011;118(10):2050–2057. | ||
Ohno-Matsui K, Akiba M, Moriyama M, et al. Acquired optic nerve and peripapillary pits in pathologic myopia. Ophthalmology. 2012;119(8):1685–1692. | ||
Hwang YH, Kim YY. Myopic optic disc changes in adolescents. Ophthalmology. 2012;119(4):885–886. | ||
Ku CS, Loy EY, Pawitan Y, Chia KS. The pursuit of genome-wide association studies: where are we now? J Hum Genet. 2010;55:195–206. | ||
Hejtmancik JF. The genetics of cataract: our vision becomes clearer. Am J Hum Genet. 1998;62(3):520–525. | ||
Lampi KJ, Ma Z, Shih M, et al. Sequence analysis of betaA3, betaB3, and betaA4 crystallins completes the identification of the major proteins in young human lens. J Biol Chem. 1997;272(4):2268–2275. | ||
Bloemendal H, de Jong W, Jaenicke R, Lubsen NH, Slingsby C, Tardieu A. Ageing and vision: structure, stability and function of lens crystallins. Prog Biophys Mol Biol. 2004;86(3):407–485. |
 © 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2017 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.