Back to Journals » Pharmacogenomics and Personalized Medicine » Volume 13
Association Study of MAP3K1 SNPs and Risk Factors with Susceptibility to Esophageal Squamous Cell Carcinoma in a Chinese Population: A Case–Control Study
Authors Yang Y , Zhou Q, Pan H, Wang L, Qian C
Received 8 April 2020
Accepted for publication 4 June 2020
Published 3 July 2020 Volume 2020:13 Pages 189—197
DOI https://doi.org/10.2147/PGPM.S256230
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Martin H Bluth
Yiling Yang,1,* Qiang Zhou,2,* Huiwen Pan,2 Liming Wang,1,3 Cheng Qian4
1Department of Oncology, Affiliated People’s Hospital of Jiangsu University, Zhenjiang, Jiangsu 212002, People’s Republic of China; 2Department of Cardiothoracic Surgery, Affiliated People’s Hospital of Jiangsu University, Zhenjiang, Jiangsu 212002, People’s Republic of China; 3Department of Respiratory, Xuhui Hospital-Affiliated Hospital of Zhongshan Hospital of Fudan University, Shanghai 200032, People’s Republic of China; 4Department of Thoracic Surgery, Zhongshan Hospital of Fudan University, Shanghai 200032, People’s Republic of China
*These authors contributed equally to this work
Correspondence: Liming Wang; Cheng Qian Email [email protected]; [email protected]
Purpose: The aim of this study was to screen the predisposed population and explore possible interactions between genetic polymorphisms and risk factors involved in the tumorigenesis and progression of ESCC (esophageal squamous cell carcinoma), in hope of identifying possible therapeutic targets along the way.
Patients and Methods: Cases (1043) and controls (1315) were enrolled to evaluate the possible association between MAP3K1 SNPs and ESCC risk. Subgroup analyses include MAP3K1 variants, gender, age, smoking and drinking status.
Results: Among all three single locus polymorphisms of MAP3K1, only the heterozygote genotype of rs702689 AG is shown to be associated with increased risk for developing ESCC (OR=1.272, 95% confidence interval=1.061– 1.525, p=0.009). Moreover, stratified analysis results observed altered susceptibility among patients with exposure to risk factors combined with certain genetic variant to ESCC.
Conclusion: This study reveals that MAP3K1 rs702689 AG genotype might facilitate the tumorigenesis in ESCC, particularly among women, patients who were over 63y and those who never drink nor smoke.
Keywords: single nucleotide polymorphism, esophageal cancer
Introduction
Esophageal cancer is one of the most rapidly-growing cancer diagnoses around the world, with it being the 7th most common cancer worldwide and 5th in China,1 among which, esophageal squamous cell carcinoma (ESCC) is the dominant pathological type which accounts for more than 90% of all the cases. In the high-risk regions, there is a strong tendency observed towards familial aggregation.2 Previous studies also reported that genetic susceptibility and other risk factors (environmental, demographic, lifestyle or dietary factors) may interact in ways to influence ESCC risk,3–7 so it’s reasonable to speculate that genetic susceptibility, in conjunction with demographic and lifestyle factors (age, gender, smoking and drinking habit), plays a role in the etiology of ESCC. In terms of treatment, the multimodality approach remains the first-line treatment as has been confirmed by a series of landmark clinical trials.8,9 However, the prognosis and 5-year survival rate remain poor, as well as no driver mutation has been detected, emphasizing the urgency to better understand the tumorigenesis and progression mechanisms of ESCC.
Mitogen-activated protein kinase (MAPK) signaling pathway is critical for human cancer cell survival, apoptosis, migration, tumor-immune system interactions and resistance to drug therapy.10,11 It is composed of three signaling families: extracellular signal-regulated kinases (ERKs), c-Jun N-terminal kinases (JNKs) and p38 MAPKs. Cross-communications between the MAPK/ERK pathway and parallel pathways, such as the PI3K-AKT and Wnt-Ca2+ ones, are found to be critical for abnormal proliferation and therapy resistance.12,13 Mitogen-activated protein kinases kinase kinase 1(MAP3K1) is a 196-kDa serine-threonine kinase that belongs to the MAP3K family.14 MAP3K1 activates JNK and ERK pathway, respectively.15 Recently, MAP3K1 was found to regulate both proteolytic degradation and migration of tumor cells.16 Moreover, MAP3K1 gene was identified in several GWAS researches of Desmoplastic melanoma and Breast cancer.17,18 Nevertheless, the contribution of MAP3K1 gene variants to ESCC is unknown.
Here, in order to explore the possible association between MAP3K1 single nucleotide polymorphisms (SNPs) and ESCC risk, a hospital-based case–control study was conducted in this high-risk region of China where the incidence rate of esophageal cancer can go as high as 1‰–1.3‰. Meanwhile, Jiangsu province has the second highest mortality rate related to esophageal cancer around China.19 Moreover, we hypothesized that there may be possible interactions between gene variations and risk factors leading to the abnormally high incidence of esophageal cancer in China, so stratification analyses were performed to validate if there is synergistic effect between genetic mutations and risk factors in the development of ESCC.
Patients and Methods
Study Populations
A total of 1043 histologically diagnosed ESCC patients and 1315 healthy, well-matched (in terms of age and gender, p = 0.121 and 0.880, respectively) controls were recruited from the Affiliated People’s Hospital of Jiangsu University (Zhenjiang, China) for a duration from October 2008 to January 2017 for this study. No patients had ever been diagnosed with cancer or received chemotherapy or radiotherapy at any point prior to this study.
Written informed consent was obtained from each individual before sample and personal information collection. With consent, two milliliters of venous blood was collected from each subject. Prospectively collected demographic and related risk factors information included age, race, sex, smoking and drinking history. To better define the smoking and drinking status, the “Smokers” cohort included individuals who smoked at least one cigarette per day for more than one year, whereas subjects who had more than three alcoholic drinks a week for more than six months were included in the “Alcohol drinkers” cohort.
Ethical Approval
We have complied with the World Medical Association Declaration of Helsinki regarding ethical conduct of research involving human subjects and/or animals. This study was approved by the Review Board of Jiangsu University.
Genomic DNA Extraction, SNP Selection and Genotyping
Blood samples collected from individuals using Vacutainers were transferred to tubes lined with ethylenediamine tetra-acetic acid (EDTA). Genomic DNA was extracted from the sample with the QIAamp DNA Blood Mini Kit (Qiagen, Berlin, Germany) following the protocol provided. Sample DNA was amplified by PCR according to the manufacturer’s protocol. The samples were genotyped using the Ligation Detection Reaction (LDR) method with technical support from Biotechnology Inc. (Shanghai, China). For quality control, 10% of the total samples were randomly selected for repeated analyses. Pilot linkage disequilibrium analyses were performed in the Chinese Han population to choose the SNP loci with moderate correlation, and tagging SNPs were selected for further analyses.
Statistical Analysis
All statistical analyses were carried out with SPSS 23.0 statistical package (SPCC Inc., Chicago, IL). Tests about Hardy–Weinberg equilibrium (HWE) for genotypes were conducted with a goodness-of-fit χ2 test based on the calculated genotype frequencies between case and control groups. Those who did not meet HWE were excluded from further study. ORs with 95% CIs were calculated to assess the strength of association between the three MAP3K1 SNPs and ESCC risk. Further to estimate the overall risk of ESCC imparted by the synergistic effect between MAP3K1 SNPs and other exogenous factors such as smoking and drinking, stratified analyses were performed to examine the statistical differences. The associations between three SNPs and the risk of ESCC were analyzed by PLINK software (v1.07, available at http://zzz.bwh.harvard.edu/plink/download.shtml). Crude ORs and adjusted ORs when adjusted for age, sex, smoking and alcohol drinking status were also calculated using unconditional logistic regression analyses. Bilateral probability tests were taken, p value < 0.05 was considered statistically significant.
Results
Characteristics of the Study Population
The characteristics of study subjects, including demographics and lifestyle factors, were presented in Table 1. The controls and cases cohorts were well matched in terms of age and gender (χ2 test, p = 0.121 and 0.880, respectively). In this case, lower incidence rate of ESCC was observed in female compared to male cohort (27.33% vs.72.67%); subjects over 63y suffered higher incidence of ESCC compared to people <63y (54.84% vs 45.16%). Moreover, the cigarette-smoking rate (43.53% vs 26.70%, p < 0.001) and alcohol-drinking rate (31.54% vs 7.07%, p < 0.001) were both significantly higher among ESCC patients.
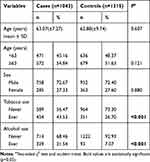 |
Table 1 Distribution of Selected Demographic Variables and Risk Factors in ESCC Cases and Controls |
As shown in Table 2, the genotyping successful rates were all beyond 98.5%. Genotype frequencies for the three chosen polymorphism loci in control subjects all met Hardy–Weinberg equilibrium (p value for HWE, all p > 0.05) (Table 2).
 |
Table 2 Primary Information for rs702688, rs702689, rs72644086 Polymorphisms |
Association Between Risk of ESCC and Three Polymorphisms
As shown in Table 3, in single marker analysis, statistically significant difference in genotype frequencies of rs702689 were observed between the cases and controls (p =0.009). Also, the differences among frequencies of the three genotypes were statistically significant (p =0.012). In all, as assessed by the allelic, dominant, co-dominant, recessive and Cochran-Armitage trend tests, only rs702689 was shown to be associated with increased risk for developing ESCC (Table 3).
 |
Table 3 Main Effects of MAP3K1 SNPs on ESCC Risk |
Stratification Analyses on Three Polymorphisms and Risk of ESCC
To further evaluate the effects of MAP3K1 rs702688, rs702689, rs72644086 variants on ESCC risk combined with different gender, age, smoking and alcohol drinking status, stratification analyses were performed as demonstrated in Tables 4–6. MAP3K1 rs702688 AG+GG genotype increased male susceptibility to ESCC, while rs702689 AG and AA+AG genotype increased susceptibility to ESCC in female cohort. rs702689 AG and AA+AG genotype increased the susceptibility to ESCC among people over 63y, contrarily increased risk for ESCC was observed in rs72644086 AG and AG+GG genotype combined with age<63. Meanwhile, rs702689 AG and AA+AG genotype increased susceptibility to ESCC in non-smoking subgroup. In drinking subgroup, rs72644086 AG and AG+GG genotype doubled the risk for ESCC. Interestingly, rs702689 AG and AA+AG genotype were found to be associated with higher risk for ESCC in non-alcohol consumption cohort.
 |
Table 4 Stratified Analyses Between MAP3K1 rs702688 Polymorphism and ESCC Risk by Sex, Age, Smoking Status and Alcohol Consumption |
 |
Table 5 Stratified Analyses Between MAP3K1 rs702689 Polymorphism and ESCC Risk by Sex, Age, Smoking Status and Alcohol Consumption |
 |
Table 6 Stratified Analyses Between MAP3K1 rs72644086 Polymorphism and ESCC Risk by Sex, Age, Smoking Status and Alcohol Consumption |
Linkage Disequilibrium Analyses and Association Tests
Linkage disequilibrium analyses in cases were performed as shown in Table 7. There were strong correlations between the three loci. Strong associations between these 3 loci were detected in association test with help from Haploview software (version 4.2). Especially between rs702688 and rs702689, stronger associations were detected.
 |
Table 7 Linkage Disequilibrium Analyses of MAP3K1 rs702688, rs702689, rs72644086 in Case Group |
Haplotype Analyses of MAP3K1 Polymorphisms and Susceptibility to ESCC
As demonstrated in Table 8, Haplotype analyses showed that MAP3K1 Ars702688Grs702689Ars72644086 was the protective haplotype against ESCC (crude OR=0.844, 95% CI=0.751–0.949, p=0.004). Meanwhile, MAP3K1 Grs702688Ars702689Ars72644086 was associated with increased risk for ESCC (crude OR=1.230, 95% CI=1.020–1.482, p=0.029).
 |
Table 8 MAP3K1 Haplotype Frequencies in Cases and Controls and Risk of ESCC |
Power Calculation
The power calculation was performed by “Power and Sample Size Calculation” Software (http://biostat.mc.vanderbilt.edu/wiki/Main/PowerSampleSize). Based on the assumption that the type I error probability for a two-sided test α equals 0.05, the probability of exposure in controls P0 is 0.295. In the current study, with ligation detection reaction method, the successful rates of genotyping exceed 98%. There were 1315 controls and 1043 cases successfully genotyped. The ratio of control/case (m) equals 1.08, and the correlation coefficient for exposure between matched case and controls (f) is 1.30. The power value is 0.814.
Discussion
In this current study, we investigated the possible interactions (and association) of genetic polymorphisms in MAP3K1 with risk factors and the susceptibility to ESCC in the at-risk Chinese population. Our study demonstrated that MAP3K1 rs702689 polymorphism seemed to play a substantial role in conditioning the risk for developing ESCC. To better understand the interactions between gene and chosen risk factors, stratification studies were carried out, indicating MAP3K1 rs702688, rs702689, rs72644086 polymorphisms altered susceptibility to ESCC according to different age, sex, smoking and alcohol consumption status.
MAP3K1 (also known as MEK1) is a serine/threonine kinase, an important member of the highly conserved MAPK signaling pathway. Of all the 19 MAP3Ks, only MAP3K1 contains a plant homeodomain (PHD) motif and an E3 ubiquitin (Ub) ligase which means it could regulate both protein phosphorylation and ubiquitin proteasome system.20 Due to its unique biological structure, MAP3K1 has complex roles in the regulation of cell death, survival, migration and differentiation. MAP3K1 was initially considered to be more of a pro-apoptotic factor, although recent studies have proposed a more complicated role for it to play in deciding cell fate. The general consensus based on various researches is that full-length MAP3K1 promotes cell survival while the caspase 3-cleavaged c-terminal fragment containing kinase domain induces cell apoptosis.
There is a growing body of studies suggesting that MAP3K1 is widely involved in a variety of diseases. Besides being the primary mutated gene in breast cancer,21,22 MAP3K1 has been shown to be the target of various stimuli in the tumorigenesis, progression and invasion in other types of cancer. In colorectal cancer, Salem et al discovered that MAP3K1 is the direct target of MiR-375 which leads to apoptosis via NF-kB and PI3K/AKT pathways.23 Guo et al observed miR-196b suppressed proliferation, migration and invasion of human choriocarcinoma cells by inhibiting its transcriptional target MAP3K1, making both of them potential targets for clinical treatment.24 Similar study suggests MiR-451 inhibited the proliferation of EC9706 by targeting CDKN2D and MAP3K1.25
MAPK pathway is considered to be one of the most deregulated pathway in esophageal cancer.26 Hu et al discovered FAT1 promotes epithelial mesenchymal transition (EMT) via MAPK/ERK signaling pathway thus contributes to tumorigenesis in esophageal squamous cell cancer.27 Similarly, MAPK pathway was down-regulated by PPARγ-activated TLA4 pathway which leads to inhibited proliferation and induced apoptosis in EC cells.28,29 O’Callaghan et al identified the function and mechanism underlying the rarely reported isoform of p38, the p38δ, loss of which could promote ESCC proliferation, migration and anchorage-independent growth.30 Further to prove the point, over-expression of miR302a inhibited the viability and invasion of esophageal cancer cells via the MAPK and PI3K/AKT signaling pathways.31 Several studies also reported that the inhibition of MAPK pathway could enhance target sensitivity to tumor Ag–specific CTL lysis, therefore improving the prognosis in esophageal and gastric cancer patients.32
To the best of our knowledge, this is the first ever report to study the association between MAP3K1 SNPs and ESCC with or without exposure to risk factors. Among all three loci we examined, only rs702689 was studied before in terms of pediatric age asthma, familial colorectal tumors and 46, XY disorder.33–36 Based on our results, the heterozygous genotype of MAP3K1 rs702689 increased the susceptibility to ESCC. Especially, female, age ≥63, non-smoking and non-alcoholic characteristics may interact with the heterozygous genotype of MAP3K1 rs702689 in ways which were associated with an uptick in susceptibility to ESCC. Smoking and alcohol consumption are widely accepted as high-risk factors of ESCC. Interestingly, rs702689 SNP was implicated in the increased risk of ESCC among non-smokers and non-alcohol consumers, which suggests strong genetic correlation between MAP3K1 rs702689 SNP and ESCC risk. Although rs72644086 polymorphism was not associated with the susceptibility to ESCC in single marker analyses, alcohol consumption significantly increased ESCC risk in MAP3K1 rs72644086 AG, AG+GG genotypes. The findings exemplified the significance of interactions between genetic susceptibility and risk factors which both contribute to the carcinogenesis in esophageal epithelial.
However, the mechanisms underlying the altered susceptibility towards ESCC remain unknown. That is identified rs702689 could cause the missense mutation of MAP3K1 gene in exon 14.37 MAP3K1 mutation increases the expression of translated protein, consequently activating the MAPK signaling pathway to enhance the growth and proliferation of malignant cells. Still, further functional experiments are warranted.
Several limitations of our study must be acknowledged. First, our sample study population is pretty restricted which means replicative studies in different areas might appear challenging. So to solve this problem, multi-center collaborations should be encouraged. Second, further studies will be necessary to decipher the pathologies underlying the altered susceptibility that the MAP3K1 SNPs conferred. What is more, MAP3K1 SNPs might also be correlated with the survival of ESCC patients. However, since our follow-up studies are still in process, so the outcome is not available right now. Also, in stratified analysis, some other confounding factors (educational level, income, BMI, physical exercise record) fail to be included; thus, the analysis results stood the chance of being hampered. Last but not least, as ESCC is the dominant subtype of EC in China which accounts for over 90% of the cases, so our case cohorts are all ESCC. Nevertheless, esophageal adenocarcinoma is the increasingly common subtype in Western Europe and the US, and the etiologies of these two malignancies are totally different; thus, further research are necessary to verify this genetic association in EAC subgroup.
In conclusion, we found that the heterozygous genotype of MAP3K1 rs702689 AG was associated with increased risk for ESCC. Synergistic interactions between gene variants and risk factors were observed among female, age ≥63 and patients with no previous smoking or drinking history. However, further functional studies are in dire need to elucidate the underlying mechanisms for the altered susceptibility.
Acknowledgments
We acknowledge the technical assistance from Genesky Biotechnologies Inc. (Shanghai, China). Yiling Yang and Qiang Zhou are co-first authors for this study.
Disclosure
The authors declare no conflicts of interest in this work.
References
1. World cancer research fund. Available from: https://www.wcrf.org/dietandcancer/cancer-trends/oesophageal-cancer-statistics.
2. Pan D, Su M, Huang G, et al. MTHFR C677T genetic polymorphism in combination with serum vitamin B2, B12 and aberrant DNA methylation of P16 and P53 genes in esophageal squamous cell carcinoma and esophageal precancerous lesions: a case-control study. Cancer Cell Int. 2019;19:288. doi:10.1186/s12935-019-1012-x
3. Chen J, Huang ZJ, Duan YQ, Xiao XR, Jiang JQ, Zhang R. Aberrant DNA methylation of P16, MGMT, and hMLH1 genes in combination with MTHFR C677T genetic polymorphism and folate intake in esophageal squamous cell carcinoma. Asian Pac J Cancer Prev. 2012;13(10):
4. Yokoyama A, Kakiuchi N, Yoshizato T, et al. Age-related remodelling of oesophageal epithelia by mutated cancer drivers. Nature. 2019;565(7739):
5. Suo C, Yang Y, Yuan Z, et al. Alcohol intake interacts with functional genetic polymorphisms of aldehyde dehydrogenase (ALDH2) and alcohol dehydrogenase (ADH) to increase esophageal squamous cell cancer risk. J Thorac Oncol. 2019;14(4):
6. Dumitrescu RG. Alcohol-induced epigenetic changes in cancer. Methods Mol Biol. 2018;1856:157–172. doi:10.1007/978-1-4939-8751-1_9
7. Liang P, Zhang W, Wang W, et al. PLCE1 polymorphisms and risk of esophageal and gastric cancer in a northwestern Chinese population. Biomed Res Int. 2019;2019:9765191. doi:10.1155/2019/9765191
8. Batra R, Malhotra GK, Singh S, Are C. Managing squamous cell esophageal cancer. Surg Clin North Am. 2019;99(3):
9. Egyud MR, Tseng JF, Suzuki K. Multidisciplinary therapy of esophageal cancer. Surg Clin North Am. 2019;99(3):419–437. doi:10.1016/j.suc.2019.02.002
10. Pham TT, Angus SP, Johnson GL. MAP3K1: genomic alterations in cancer and function in promoting cell survival or apoptosis. Genes Cancer. 2013;4(11–12):
11. Burotto M, Chiou VL, Lee JM, Kohn EC. The MAPK pathway across different malignancies: a new perspective. Cancer. 2014;120(22):3446–3456. doi:10.1002/cncr.28864
12. Mendoza MC, Er EE, Blenis J. The Ras-ERK and PI3K-mTOR pathways: cross-talk and compensation. Trends Biochem Sci. 2011;36(6):
13. Cammareri P, Vincent DF, Hodder MC, et al. TGFβ pathway limits dedifferentiation following WNT and MAPK pathway activation to suppress intestinal tumourigenesis. Cell Death Differ. 2017;24(10):1681–1693. doi:10.1038/cdd.2017.92
14. Uhlik MT, Abell AN, Cuevas BD, Nakamura K, Johnson GL. Wiring diagrams of MAPK regulation by MEKK1, 2, and 3. Biochem Cell Biol. 2004;82(6):658–663. doi:10.1139/o04-114
15. Hirai S, Noda K, Moriguchi T, et al. Differential activation of two JNK activators, MKK7 and SEK1, by MKN28-derived nonreceptor serine/threonine kinase/mixed lineage kinase 2. J Biol Chem. 1998;273(13):7406–7412. doi:10.1074/jbc.273.13.7406
16. Suddason T, Gallagher E. A RING to rule them all? Insights into the Map3k1 PHD motif provide a new mechanistic understanding into the diverse roles of Map3k1. Cell Death Differ. 2015;22(4):540–548. doi:10.1038/cdd.2014.239
17. Shain AH, Garrido M, Botton T, et al. Exome sequencing of desmoplastic melanoma identifies recurrent NFKBIE promoter mutations and diverse activating mutations in the MAPK pathway. Nat Genet. 2015;47(10):1194–1199. doi:10.1038/ng.3382
18. Jara L, Gonzalez-Hormazabal P, Cerceño K, et al. Genetic variants in FGFR2 and MAP3K1 are associated with the risk of familial and early-onset breast cancer in a South-American population. Breast Cancer Res Treat. 2013;137(2):559–569. doi:10.1007/s10549-012-2359-z
19. Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi:10.3322/caac.21338
20. Charlaftis N, Suddason T, Wu X, Anwar S, Karin M, Gallagher E. The MEKK1 PHD ubiquitinates TAB1 to activate MAPKs in response to cytokines. EMBO J. 2014;33(21):2581–2596. doi:10.15252/embj.201488351
21. Kuo SH, Yang SY, You SL, et al. Polymorphisms of ESR1, UGT1A1, HCN1, MAP3K1 and CYP2B6 are associated with the prognosis of hormone receptor-positive early breast cancer. Oncotarget. 2017;8(13):20925–20938. doi:10.18632/oncotarget.14995
22. Glubb DM, Maranian MJ, Michailidou K, et al. Fine-scale mapping of the 5q11.2 breast cancer locus reveals at least three independent risk variants regulating MAP3K1. Am J Hum Genet. 2015;96(1):5–20. doi:10.1016/j.ajhg.2014.11.009
23. Salem SM, Hamed AR, Mosaad RM. MTDH and MAP3K1 are direct targets of apoptosis-regulating miRNAs in colorectal carcinoma. Biomed Pharmacother. 2017;94:767–773. doi:10.1016/j.biopha.2017.07.153
24. Guo Z, Sui L, Qi J, et al. miR-196b inhibits cell migration and invasion through targeting MAP3K1 in hydatidiform mole. Biomed Pharmacother. 2019;113:108760. doi:10.1016/j.biopha.2019.108760
25. Zang WQ, Yang X, Wang T, et al. MiR-451 inhibits proliferation of esophageal carcinoma cell line EC9706 by targeting CDKN2D and MAP3K1. World J Gastroenterol. 2015;21(19):5867–5876. doi:10.3748/wjg.v21.i19.5867
26. Santarpia L, Lippman SM, El-Naggar AK. Targeting the MAPK-RAS-RAF signaling pathway in cancer therapy. Expert Opin Ther Targets. 2012;16(1):103–119. doi:10.1517/14728222.2011.645805
27. Hu X, Zhai Y, Kong P, et al. FAT1 prevents epithelial mesenchymal transition (EMT) via MAPK/ERK signaling pathway in esophageal squamous cell cancer. Cancer Lett. 2017;397:83–93. doi:10.1016/j.canlet.2017.03.033
28. Sutter AP, Maaser K, Gerst B, Krahn A, Zeitz M, Scherübl H. Enhancement of peripheral benzodiazepine receptor ligand-induced apoptosis and cell cycle arrest of esophageal cancer cells by simultaneous inhibition of MAPK/ERK kinase. Biochem Pharmacol. 2004;67(9):1701–1710. doi:10.1016/j.bcp.2004.01.009
29. Wu K, Yang Y, Liu D, et al. Activation of PPARγ suppresses proliferation and induces apoptosis of esophageal cancer cells by inhibiting TLR4-dependent MAPK pathway. Oncotarget. 2016;7(28):44572–44582. doi:10.18632/oncotarget.10067
30. O’Callaghan C, Fanning LJ, Houston A, Barry OP. Loss of p38δ mitogen-activated protein kinase expression promotes oesophageal squamous cell carcinoma proliferation, migration and anchorage-independent growth. Int J Oncol. 2013;43(2):405–415. doi:10.3892/ijo.2013.1968
31. Xia D, Tian S, Chen Z, Qin W, Liu Q. miR302a inhibits the proliferation of esophageal cancer cells through the MAPK and PI3K/Akt signaling pathways. Oncol Lett. 2018;15(3):3937–3943. doi:10.3892/ol.2018.7782
32. Mimura K, Shiraishi K, Mueller A, et al. The MAPK pathway is a predominant regulator of HLA-A expression in esophageal and gastric cancer. J Immunol. 2013;191(12):6261–6272. doi:10.4049/jimmunol.1301597
33. Szczepankiewicz A, Sobkowiak P, Rachel M, et al. Multilocus analysis of candidate genes involved in neurogenic inflammation in pediatric asthma and related phenotypes: a case-control study. J Asthma. 2012;49(4):329–335. doi:10.3109/02770903.2012.669442
34. Dominguez-Valentin M, Nakken S, Tubeuf H, et al. Identification of genetic variants for clinical management of familial colorectal tumors. BMC Med Genet. 2018;19(1):26. doi:10.1186/s12881-018-0533-9
35. Loke J, Pearlman A, Radi O, et al. Mutations in MAP3K1 tilt the balance from SOX9/FGF9 to WNT/β-catenin signaling. Hum Mol Genet. 2014;23(4):1073–1083. doi:10.1093/hmg/ddt502
36. Esposito S, Ierardi V, Daleno C, et al. Genetic polymorphisms and risk of recurrent wheezing in pediatric age. BMC Pulm Med. 2014;14:162. doi:10.1186/1471-2466-14-162
37. Das DK, Rahate SG, Mehta BP, Gawde HM, Tamhankar PM. Mutation analysis of mitogen activated protein kinase 1 gene in Indian cases of 46,XY disorder of sex development. Indian J Hum Genet. 2013;19(4):437–442. doi:10.4103/0971-6866.124372
 © 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2020 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
