Back to Journals » Cancer Management and Research » Volume 10
Association study of genetic variation of lncRNA MALAT1 with carcinogenesis of colorectal cancer
Authors Zhao K, Jin S, Wei B, Cao S, Xiong Z
Received 14 June 2018
Accepted for publication 6 August 2018
Published 23 November 2018 Volume 2018:10 Pages 6257—6261
DOI https://doi.org/10.2147/CMAR.S177244
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 2
Editor who approved publication: Dr Antonella D'Anneo
Kexin Zhao,1 Si Jin,2 Bo Wei,3 Shiqiong Cao,1 Zhifan Xiong1
1Division of Gastroenterology, Liyuan Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan 430077, People’s Republic of China; 2Department of Endocrinology, Liyuan Hospital, Tongji Medical College, Huazhong University of Science, Wuhan 430077, People’s Republic of China; 3Department of Oncology, Liyuan Hospital, Tongji Medical College, Huazhong University of Science, Wuhan 430077, People’s Republic of China
Introduction: Colorectal cancer (CRC) remains a major public health concern worldwide. However, the detailed molecular mechanisms of CRC remain poorly understood.
Methods: In the current study, we evaluated associations of four genetic variants located in the promoter and gene region of long noncoding RNAs metastasis-associated lung adenocarcinoma transcript 1 (lncRNA MALAT1) with CRC susceptibility among a Chinese population with 966 CRC cases and 988 healthy controls, using a two-stage, case–control study design (400 CRC cases and 400 controls in stage 1, and 566 CRC cases and 588 controls in stage 2).
Results: We found that the minor alleles of rs619586 (OR=0.73; 95% CI=0.60–0.88; P=0.001) and rs1194338 (OR=0.80; 95% CI=0.70–0.92; P=0.001) were significantly associated with decreased CRC susceptibility. Compared with those with rs619586 −AA genotype, the risk of CRC was significantly lower in individuals with AG genotype (OR=0.76; 95% CI=0.61–0.95) and GG genotype (OR=0.46; 95% CI=0.23–0.90). Compared with those with rs1194338 −CC genotype, the risk of CRC was significantly lower in individuals with AC genotype (OR=0.79; 95% CI=0.65–0.95) and AA genotype (OR=0.68; 95% CI=0.51–0.89).
Conclusion: Taken together, our findings provided strong evidence for the hypothesis that genetic variants in lncRNA MALAT1 might contribute to the carcinogenesis of CRC.
Keywords: colorectal cancer, genetic, lncRNA, susceptibility, MALAT1
Introduction
Colorectal cancer (CRC) ranks the third most prevalent cancer type in both men and women worldwide.1 According to the Colorectal Cancer Statistics in 2017, there were about 135,430 newly diagnosed CRC cases and 50,260 deaths from the disease in United States in 2017.2 It was reported that 376,300 new CRC cases and 191,000 CRC death occurred per year in China.3 CRC has a complicated carcinogenesis involving multiple risk factors for its initiation, promotion, and progression, as well as its relapse and metastasis.2,4 In addition to some well-known risk factors, such as older age, male gender, obesity, lack of physical exercise, adenomas, and smoking, genetic factors also play an important role in CRC carcinogenesis.2,4–7 In spite of great effort for the research of CRC carcinogenesis, the detailed molecular mechanisms of CRC remain poorly understood.8,9
Recently, the long noncoding RNAs (lncRNAs), which are defined as transcripts longer than 200 nucleotides that are not translated into protein, have gradually been reported to be an important class of RNAs with pivotal roles in regulation of gene expression, and involved in multitudinous human complex diseases.10,11 However, the biological functions and precise mechanisms of the majority of lncRNAs are still poorly understood. LncRNA-metastasis-associated lung adenocarcinoma transcript 1 (MALAT1), originally identified as a marker for predicting metastasis and prognosis of early-stage non-small-cell lung cancer patients, was reported to be dysregulated in multiple human cancer types,12 diabetic retinopathy,13 thoracic aortic aneurysm,14 etc. Studies also found that MALAT1 contributed to the cell proliferation/migration/invasion, malignancy, tumor growth, metastasis, and poor prognosis of CRC.15–19 Additionally, many epidemiologic studies have explored the link between genetic variants in MALAT1 and disease susceptibility.20–25 Meta-analyses showed that MALAT1 rs619586 was associated with overall cancer risk.26 However, the associations between genetic variants of lncRNA MALAT1 and carcinogenesis of CRC were less investigated. Only Li et al found rs1194338 in the promoter region of MALAT1 was significantly associated with decreased CRC risk.24 In this study, we genotyped the tag single-nucleotide polymorphisms (tagSNPs) located in lncRNA MALAT1 gene, as well as an SNP located in the promoter region of MALAT1, and aimed to investigate their associations with the CRC susceptibility among a Chinese population.
Patients and methods
Study subjects
In the current study, we adopted a two-stage, case–control study design, which aims to reduce the false discovery rate, and the subjects were divided into two groups according the recruitment time. A total of 966 CRC cases and 988 healthy controls were included in this study. The inclusion criterion for patients was histopathologically confirmed CRC without previous chemotherapy or radiotherapy. Cancer-free controls came from health check-up programs at the same hospital during the same time, which were matched to cases by gender and age group (±5 years). Face-to-face interviews were conducted by trained interviewers, who administered a structured questionnaire relating to demographic characteristics and lifestyle-related factors. Meanwhile, 5 mL peripheral blood from each individual was collected and stored in tubes containing sodium citrate anticoagulant, at –80°C, for DNA isolation. This study was approved by the Medical Ethical Committee of Liyuan Hospital, and all subjects enrolled were heritably unrelated ethnic Han Chinese. Written informed consent was signed by each participant.
SNP selection, DNA extraction, and genotyping
TagSNPs of lncRNA MALAT1 were selected using GVS online server (http://gvs.gs.washington.edu/GVS150/index.jsp). Linkage disequilibrium value r2 should be less than 0.8, and the minor allele frequency of the selected SNP should be above 0.05 in Chinese Han Beijing population. As a result three tagSNPs (rs11227209, rs619586, and rs3200401) were selected. Another SNP (rs1194338) in the promoter region of MALAT1, which was reported to be associated with CRC susceptibility,24 was also included. DNA was extracted from peripheral blood leukocytes with RelaxGene Blood System DP319-02 (Tiangen, Beijing, People’s Republic of China). A260/A280 ratio of the purified DNA samples was assessed to ensure purity. DNA samples were then stored at -80°C for further genotyping. Candidate SNPs were genotyped by the TaqMan SNP Genotyping Assay on an ABI PRISM 7900HT Fast Real-Time PCR platform (Applied Biosystems, Waltham, MA, USA). For quality control, we randomly selected 10% of these DNA samples for further validation, which were 100% consistent. Two researchers performed genotyping independently to cross-validate the results.
Statistical analysis
Categorical data were expressed as number (percentage) and analyzed using chi-squared test.
Continuous data were expressed as mean±SD and analyzed using Student’s t-test. Hardy–Weinberg equilibrium (HWE) was tested by using a chi-squared goodness-of-fit test. ORs and 95% CIs were selected to estimate the strength of association between SNPs and risk of CRC adjusted for age, gender, alcohol drinking and smoking status, and family history of cancer. The analyses were done first per allele and then per genotype. All statistics were performed using SPSS software 19.0 (IBM Corporation, Armonk, NY, USA), and P<0.05 was considered as statistically significant.
Results
Characteristics of study population
The descriptive characteristics of the study participants are provided in Table 1. No significant differences were found between the CRC patients and controls with regard to the distribution of age, gender, and smoking and drinking status in the discovery set and validation set, yet the distribution of family history of cancer in CRC patients was significantly higher than that in the controls in both stages (P<0.001). Overall, 52.5% of the tumor site was located at colon.
  | Table 1 The demographic characteristics between the colorectal cancer cases and controls |
Associations of candidate SNPs and CRC risk in stage 1
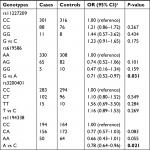
Table 2 shows the effects of four SNPs (rs11227209, rs619586, rs3200401, and rs1194338) on CRC susceptibility in stage 1. The genotypic distributions of four SNPs among the controls were in accordance with HWE (P>0.05). In the four loci, the per allele association of rs619586 (OR=0.71; 95% CI=0.52–0.97; P=0.031) and rs1194338 (OR=0.78; 95% CI=0.64–0.96; P=0.021) was statistically significant, after being adjusted for age, gender, alcohol drinking and smoking status, and family history of cancer. However, the genotypic distribution for rs11227209 and rs3200401 was not statistically significant, indicating no evidence of association with CRC risk.
Associations of candidate SNPs and CRC risk in stage 2
Further, we replicated the above finding in an independent stage II samples (Table 3). Both rs619586 and rs1194338 were validated for significant association (P<0.05). When pooled together, the minor alleles of rs619586 (OR=0.73; 95% CI=0.60–0.88; P=0.001) and rs1194338 (OR=0.80; 95% CI=0.70–0.92; P=0.001) were significantly associated with CRC susceptibility. Compared with those with rs619586 −AA genotype, the risk of CRC was significantly lower in individuals with AG genotype (OR=0.76; 95% CI=0.61–0.95) and GG genotype (OR=0.46; 95% CI=0.23–0.90). Compared with those with rs1194338 −CC genotype, the risk of CRC was significantly lower in individuals with AC genotype (OR=0.79; 95% CI=0.65–0.95) and AA genotype (OR=0.68; 95% CI=0.51–0.89).
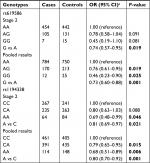  | Table 3 Association of MALAT1 variants with colorectal cancer risk in stage 2 and combined stage Notes: aAdjusted by age, gender, alcohol drinking and smoking status, and family history of cancer. |
Discussion
Up to date, CRC remains a major public health concern worldwide. The current study explored the associations between genetic variants located in the promoter and gene region of lncRNA MALAT1 and CRC susceptibility among a Chinese population, using a two-stage, case–control study design. We detected that minor alleles of both MALAT1 rs619586 and rs1194338 were significantly associated with decreased CRC susceptibility in two stages.
Increasing evidence shows that lncRNAs can be used as biomarkers for the prediction of susceptibility, prognosis, or as tumor therapeutic targets in human cancers.27–29 MALAT1, also known as NEAT2 (noncoding nuclear-enriched abundant transcript 2), was located at 11q13.1. Tripathi et al first identified that MALAT1 could regulate alternative splicing by modulating serine/arginine splicing factor phosphorylation.30 It was also found to be involved in multiple types of physiological processes, such as alternative splicing, nuclear organization, epigenetic modulating of gene expression, and various pathological processes, ranging from diabetes complications to cancers.12,31–33 Functional investigations indicated that G allele of rs619586 could trigger higher expression of MALAT1 in multiple tissues, which might affect the physiological processes consequently.3,29 Function of MALAT1 rs1194338 was only studied in 71 CRC tissues samples; however, no statistically significant difference of MALAT1 expression between CC genotype and AA genotype was detected.6
Genetic variants in lncRNA MALAT1 have also been linked to various kinds of diseases. Li et al found rs1194338 was significantly associated with decreased CRC risk.24 Wang et al identified that rs3200401 was associated with survival for advanced lung adenocarcinoma patients.23 Peng et al showed that individuals with genotype AG of rs619586 have a decreased risk of breast cancer.25 Additionally, Wang et al found that rs619586AG/GG genotypes might protect against the occurrence of coronary atherosclerotic heart disease.34 Li et al identified that MALAT1 rs619586 was significantly associated with lower risk of congenital heart disease.20 Zhuo et al showed rs619586 was associated with a decreased risk of pulmonary arterial hypertension.21 All the above findings were consistent with our results, which showed rs619586 and rs1194338 were significantly associated with decreased CRC susceptibility.
Strengths and limitations
A relatively small sample size for the stratification and interaction analyses is the limitation of this study that should be pointed out. However, for the main effect, the statistical power for rs619586 was 88.2%, and the statistical power for rs1194338 was 91.1%. The strength of this study is that we used a two-stage investigation to validate the association between the genetic variants and CRC susceptibility.
Conclusion
Conclusively, the current findings indicate that genetic variants in lncRNA MALAT1 may influence the risk of CRC. However, since the exact mechanism and function of these gene variants have not yet been fully defined, the present findings need to be confirmed in further studies with other populations in order to clarify the association between these polymorphisms and the risk of CRC.
Disclosure
The authors report no conflicts of interest in this work.
References
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin. 2018;68(1):7–30. | ||
Siegel RL, Miller KD, Fedewa SA, et al. Colorectal cancer statistics, 2017. CA Cancer J Clin. 2017;67(3):177–193. | ||
Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. | ||
Stoffel EM, Koeppe E, Everett J, et al. Germline Genetic Features of Young Individuals With Colorectal Cancer. Gastroenterology. 2018;154(4):897–905. | ||
Wang X, Fang H, Cheng Y, et al. The molecular landscape of synchronous colorectal cancer reveals genetic heterogeneity. Carcinogenesis. 2018;39(5):708–718. | ||
Schubert SA, Ruano D, Elsayed FA, et al. Evidence for genetic association between chromosome 1q loci and predisposition to colorectal neoplasia. Br J Cancer. 2018;118(2):e4. | ||
Witold K, Anna K, Maciej T, Jakub J. Adenomas - Genetic factors in colorectal cancer prevention. Rep Pract Oncol Radiother. 2018;23(2):75–83. | ||
Weigl K, Thomsen H, Balavarca Y, Hellwege JN, Shrubsole MJ, Brenner H. Genetic Risk Score Is Associated With Prevalence of Advanced Neoplasms in a Colorectal Cancer Screening Population. Gastroenterology. 2018;155(1):88–98. | ||
Jeon J, Du M, Schoen RE, et al; Colorectal Transdisciplinary Study and Genetics and Epidemiology of Colorectal Cancer Consortium. Determining Risk of Colorectal Cancer and Starting Age of Screening Based on Lifestyle, Environmental, and Genetic Factors. Gastroenterology. 2018;154(8):2152–2164. | ||
Zheng LL, Li JH, Wu J, et al. deepBase v2.0: identification, expression, evolution and function of small RNAs, LncRNAs and circular RNAs from deep-sequencing data. Nucleic Acids Res. 2016;44(D1):D196–D202. | ||
Xie W, Yuan S, Sun Z, Li Y. Long noncoding and circular RNAs in lung cancer: advances and perspectives. Epigenomics. 2016;8(9):1275–1287. | ||
Amodio N, Raimondi L, Juli G, et al. MALAT1: a druggable long non-coding RNA for targeted anti-cancer approaches. J Hematol Oncol. 2018;11(1):63. | ||
Biswas S, Thomas AA, Chen S, et al. MALAT1: An Epigenetic Regulator of Inflammation in Diabetic Retinopathy. Sci Rep. 2018;8(1):6526. | ||
Lino Cardenas CL, Kessinger CW, Cheng Y, et al. An HDAC9-MALAT1-BRG1 complex mediates smooth muscle dysfunction in thoracic aortic aneurysm. Nat Commun. 2018;9(1):1009. | ||
Yang MH, Hu ZY, Xu C, et al. MALAT1 promotes colorectal cancer cell proliferation/migration/invasion via PRKA kinase anchor protein 9. Biochim Biophys Acta. 2015;1852(1):166–174. | ||
Ji Q, Zhang L, Liu X, et al. Long non-coding RNA MALAT1 promotes tumour growth and metastasis in colorectal cancer through binding to SFPQ and releasing oncogene PTBP2 from SFPQ/PTBP2 complex. Br J Cancer. 2014;111(4):736–748. | ||
Xu C, Yang M, Tian J, Wang X, Li Z. MALAT-1: a long non-coding RNA and its important 3’ end functional motif in colorectal cancer metastasis. Int J Oncol. 2011;39(1):169–175. | ||
Zheng HT, Shi DB, Wang YW, et al. High expression of lncRNA MALAT1 suggests a biomarker of poor prognosis in colorectal cancer. Int J Clin Exp Pathol. 2014;7(6):3174–3181. | ||
Wu C, Zhu X, Tao K, et al. MALAT1 promotes the colorectal cancer malignancy by increasing DCP1A expression and miR203 downregulation. Mol Carcinog. 2018;57(10):1421–1431. | ||
Li Q, Zhu W, Zhang B, et al. The MALAT1 gene polymorphism and its relationship with the onset of congenital heart disease in Chinese. Biosci Rep. 2018;38(3):BSR20171381. | ||
Zhuo Y, Zeng Q, Zhang P, Li G, Xie Q, Cheng Y. Functional polymorphism of lncRNA MALAT1 contributes to pulmonary arterial hypertension susceptibility in Chinese people. Clin Chem Lab Med. 2017;55(1):38–46. | ||
Wang R, Sun Y, Li L, et al. Preclinical Study using Malat1 Small Interfering RNA or Androgen Receptor Splicing Variant 7 Degradation Enhancer ASC-J9® to Suppress Enzalutamide-resistant Prostate Cancer Progression. Eur Urol. 2017;72(5):835–844. | ||
Wang JZ, Xiang JJ, Wu LG, et al. A genetic variant in long non-coding RNA MALAT1 associated with survival outcome among patients with advanced lung adenocarcinoma: a survival cohort analysis. BMC Cancer. 2017;17(1):167. | ||
Li Y, Bao C, Gu S, et al. Associations between novel genetic variants in the promoter region of MALAT1 and risk of colorectal cancer. Oncotarget. 2017;8(54):92604–92614. | ||
Peng R, Luo C, Guo Q, et al. Association analyses of genetic variants in long non-coding RNA MALAT1 with breast cancer susceptibility and mRNA expression of MALAT1 in Chinese Han population. Gene. 2018;642:241–248. | ||
Huang X, Zhang W, Shao Z. Association between long non-coding RNA polymorphisms and cancer risk: a meta-analysis. Biosci Rep. 2018;38(4):BSR20180365. | ||
Zhang B, Mao YS, Diermeier SD, et al. Identification and Characterization of a Class of MALAT1-like Genomic Loci. Cell Rep. 2017;19(8):1723–1738. | ||
Xia H, Chen Q, Chen Y, et al. The lncRNA MALAT1 is a novel biomarker for gastric cancer metastasis. Oncotarget. 2016;7(35):56209–56218. | ||
Szcześniak MW, Makałowska I. lncRNA-RNA Interactions across the Human Transcriptome. PLoS One. 2016;11(3):e0150353. | ||
Tripathi V, Ellis JD, Shen Z, et al. The nuclear-retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell. 2010;39(6):925–938. | ||
Zhang B, Arun G, Mao YS, et al. The lncRNA Malat1 is dispensable for mouse development but its transcription plays a cis-regulatory role in the adult. Cell Rep. 2012;2(1):111–123. | ||
Uchida S, Dimmeler S. Long noncoding RNAs in cardiovascular diseases. Circ Res. 2015;116(4):737–750. | ||
Gutschner T, Hämmerle M, Diederichs S. MALAT1 – a paradigm for long noncoding RNA function in cancer. J Mol Med. 2013;91(7):791–801. | ||
Wang G, Li Y, Peng Y, Tang J, Li H. Association of polymorphisms in MALAT1 with risk of coronary atherosclerotic heart disease in a Chinese population. Lipids Health Dis. 2018;17(1):75. |
 © 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2018 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.