Back to Journals » ImmunoTargets and Therapy » Volume 12
Association of Toll-like Receptors 1, 2, 4, 6, 8, 9 and 10 Genes Polymorphisms and Susceptibility to Pulmonary Tuberculosis in Sudanese Patients
Authors Mhmoud NA
Received 15 January 2023
Accepted for publication 29 March 2023
Published 6 April 2023 Volume 2023:12 Pages 47—75
DOI https://doi.org/10.2147/ITT.S404915
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Michael Shurin
Najwa A Mhmoud
Faculty of Medical Laboratory Sciences, Department of Medical Microbiology and Immunology, University of Khartoum, Khartoum, Sudan
Correspondence: Najwa A Mhmoud, Faculty of Medical Laboratory Sciences, Department of Medical Microbiology and Immunology University of Khartoum, P.O. Box 102, Khartoum, Sudan, Fax +249-83-383590, Email [email protected]
Background: Genetic factors are important contributors to the development of a wide range of complex disease. Polymorphisms in genes encoding for toll-like receptors (TLRs) usually influence the efficiency of the immune response to infection and are associated with disease susceptibility and progression. Therefore, we aim to describe the first association between TLR1, TLR2, TLR4 TLR6, TLR8, TLR9 and TLR10 genes polymorphisms and susceptibility to pulmonary tuberculosis (PTB) in Sudanese patients.
Methodology: Here we performed a case study which included 160 tuberculosis patients and 220 healthy matched controls from Sudan. In the study population, we evaluated the possible association between 86 markers in TLR1, TLR2, TLR4 TLR6, TLR8, TLR9 and TLR10 genes polymorphisms and susceptibility to PTB disease in Sudanese population using polymerase chain reaction and restriction fragment length polymorphism (PCR-RFLP).
Results: From our results it appeared that in the PTB population the TLR1 (rs5743557, rs4833095, rs5743596), TLR2 (rs5743704, rs5743708, rs3804099), TLR4 (rs4986790, rs4986791), TLR6 (rs5743810), TLR8 (rs3764879, rs3764880), TLR9 (rs352165, rs352167, rs187084) and TLR10 (rs4129009) were significantly more often encountered (p< 0.0001) than in the control population and were associated with PTB in the Sudanese population. For the other polymorphisms tested, no association with PTB was found in the population tested.
Conclusion: The work describes novel mutations in TLR1, TLR2, TLR4, TLR6, TLR8, TLR9 and TLR10 genes and their association with PTB infection in Sudanese population. These results will enhance our ability to determine the risk of developing the disease by targeting specific TLR pathways to reduce the severity of the disease. Future studies are needed in a larger sample to replicate our findings and understand the mechanism of association of TLR polymorphism in PTB.
Keywords: tuberculosis, toll-like receptors, PCR-RFLP
Introduction
Tuberculosis (TB) has been declared a major public health problem worldwide, due to the emergence of almost untreatable strains of Mycobacterium tuberculosis (M. tuberculosis). In addition to that, the only existing vaccine against TB, the BCG vaccine, fails to protect against pulmonary TB, the most important form of disease manifestation.1 It kills approximately 10 million people and nine million new TB cases are reported worldwide annually.1,2 Moreover, in the last two decades, three major coronavirus epidemics have been reported worldwide.3 The burden of COVID-19, HIV, and TB is one of the major and persistent global health challenges of the twenty-first century.4–10
Genetic factors are important contributors to the development of a wide range of complex disease.11–21 A person who is susceptible to a particular infectious disease, such as TB, the risk of developing the disease is higher than one who has not inherited the genetic risk factor.11–21 These evidences on the influence of host genetic factors in TB susceptibility led to the development of strategies to identify candidate genes or susceptibility loci in the human genome. Identification of polymorphisms in genes has enabled linkage and association studies to be used in explaining individual variation in susceptibility to and severity of TB in humans.
In this context, toll-like receptors (TLRs) are a family of proteins that are expressed either on the extra cellular cell surface (TLR1, 2, 4, 5, 6) or in the cytosol or on endosomal membranes (TLR3, 7, 8, 9) of macrophages and dendritic cells. TLRs are essential for recognition of a broad repertoire of pathogen-associated molecular patterns (PAMPs) on macrophages and dendritic cells and play an important role in the innate responses against M. tuberculosis.22–26 Genetic variations of TLR1, TLR2, TLR4, TLR6, TLR8, TLR9 and TLR10 have been associated with the susceptibility to TB in different ethnic groups.27–33 In contrast, other studies have failed to demonstrate significant associations of TLRs polymorphisms with TB.34–37
However, to date, no TLR mutations or single-nucleotide polymorphisms (SNPs) have been established as accepted risk factors for PTB among different ethnic populations. Besides, only one study have been performed on Sudanese patients to understand the role of TLR2 polymorphism in PTB.31 Therefore, to confirm the role of TLR variants in the increased risk for PTB we conducted the candidate gene association study by investigating 86 SNPs present in TLR1, TLR2, TLR4 TLR6, TLR8, TLR9 and TLR10 genes in a sub-Saharan Sudanese pulmonary tuberculosis patients.
Materials and Methods
Study Population
A prospective, cross sectional, case-control study was carried out during the period between 2019 and 2020 at Abu-Angah Hospital, Khartoum, Sudan. 160 patients with active pulmonary TB and 220 healthy controls were included. EDTA blood samples were taken from all patients and healthy controls. All tuberculosis patients had microbiological (by culture and/or smear) or radiological evidence of M. tuberculosis disease (Table 1).
 |
Table 1 Characteristics of the Study Population |
The healthy controls had no evidence of tuberculosis disease by clinical examination, and were matched on age, gender and BCG status (Table 1).
The collected blood samples were tested for other infectious diseases and that included hepatitis B (HBsAg, InTec products, INC, China), hepatitis C (Rapid Anti-HCV Test, InTec products, INC, China), syphilis (RAPIDAN TESTER, product code: RTTP01, Turkey), and HIV (HIV1, 2 Cassette test, Clinotech Diagnostics & Pharmaceuticals, Canada). Blood samples were stored at −20 ° C until use. Nasopharyngeal swab for was used to define Corona Virus Disease-2019 (COVID-19).
Ethics Statement
The present study was approved by the Ethics Committee of University of Khartoum, Khartoum, Sudan (5/2018). This study adheres to the Declaration of Helsinki (1964). Written informed consents were obtained from all participants in the study or legally responsible guardians for participants less than 18 years old.
DNA Isolation
Genomic DNA was isolated from blood samples with the large volume kit for the MagNA Pure system (Roche, Almere, The Netherlands) according to the manufacturer’s descriptions. The isolated DNA was stored at −20°C.
Genotyping
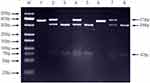
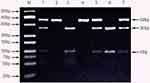
Genomic variants of TLR1 (rs5743604, rs5743611, rs5743618, rs76600635, rs5743551, rs5743557, rs5743565, rs5743566, rs5743580, rs5743594 rs4833095, rs5743595, rs5743596); TLR2 (rs1816702, rs5743704 rs5743708, rs7656411, rs11938228, rs893629, rs1898830, rs121917864, rs4696480, rs3804099, rs5743699 rs3804100); TLR4 (rs7869402, rs1927907, rs1927911, rs1927914, rs6478317, rs55912718, rs5030719, rs10759931, rs10759933, rs2770150, rs1554973, rs11536878, rs11536879, rs7873784, rs11536889, rs4986790, rs4986791, rs11536897, rs11536898); TLR6 (rs3796508, rs5743810, rs5743831, rs1039559, rs6531670, rs5743788, rs5743794); TLR8 (rs4830805, rs4830808, rs3747414, rs3761624, rs1548731, rs2109134, rs3788935, rs1013150, rs5744068, rs3764879, rs5744080, rs3764880, rs17256081, rs5741883, rs2407992, rs178998); TLR9 (rs5743836, rs164637, rs352139, rs352140, rs352143, rs352162, rs352165, rs352167, rs187084); and TLR10 (rs4129009, rs7694115, rs10856839, rs11466645, rs4274855, rs11096955, rs11096956, rs11096957, rs7698870, and rs10776483) genes were detected by PCR followed by restriction enzyme fragment analysis (PCR-RFLP) (Figures 1 and 2). All PCR primers are stated in Table 2. Each of the PCRs consisted of a pre-denaturation step of 4 minutes at 94°C and 40 cycles each of 30 seconds denaturation at 94°C, 30 seconds annealing at 55°C and 30 seconds elongation at 72°C. This was followed by a post-elongation step of 7 minutes at 72°C. Restriction endonucleases were selected using the NEBcutter software (http://nc2.neb.com/NEBcutter2/). Restriction endonucleases were obtained from Fermentas (st. Leon-rot, Germany), and Roche (Penzberg, Germany) and were used as described by the manufacturer. Restriction fragments were visualized by electrophoresis on 2% agarose gels (Hispanagar, Sphaero Q, Leiden, The Netherlands).
 |
Table 2 PCR Primers and Restriction Enzymes for genotyping the Different Single Nucleotide Polymorphisms |
The criteria for the selection of SNPs for this study were based on their previously reported to change the level or function of corresponding gene products and influence susceptibility/resistance to infections. Also, the selection was based on the publically available information on the polymorphisms of TLR genes available in the 1000 Genomes project for the South Asian population (http://www.internationalgenome.org).38
Statistical Analysis
The mean age of the patient population and the control population were compared by the unpaired t-test. Gender, occupation and BCG-vaccination status between the patient and control population were compared with the Fisher exact test. Verification of Hardy-Weinberg equilibrium (HWE) was performed with Pearson’s χ2 test. The effect of the TLR1; TLR2; TLR4; TLR6; TLR8; TLR9; and TLR10 polymorphisms on susceptibility to tuberculosis were assessed with the Fisher exact test. P-value of <0.05 was deemed statistically significant. All statistical analyses were performed using SPSS for Windows v11.0 statistical analysis software. This study was in accordance with the principles of the Helsinki Declaration (1964). This study was in accordance with the principles of the Helsinki Declaration (1964) This study was in accordance with the principles of the Helsinki Declaration (1964)
Results
Characteristics of Tuberculosis Patients and Healthy Control Subjects
One hundred and sixty Sudanese tuberculosis patients were included into the study. The diagnosis of tuberculosis was based on the presence of MTB in a positive Ziehl-Nielson (ZN) smear of a sputum specimen and/or by positive culture with tuberculosis and radiological evidence (chest X-ray) (Table 1). The control population comprised 220 healthy unrelated people from the same endemic area in Sudan, they were matched on gender and BCG-status (Table 1) and showed no signs of any lung disease. Unfortunately, the occupation of the control population differed from that of the patient population.
Distribution of TLR1, TLR2, TLR4, TLR6, TLR8, TLR9 and TLR10 Genes Polymorphisms
To detect the possible deficiencies in TLR1; TLR2; TLR4; TLR6; TLR8; TLR9; and TLR10 production among tuberculosis patients, genotype (Table 3) and allele frequencies (Table 4) in the promoters of the genes encoding for TLR1, TLR2, TLR4, and TLR6 were determined. To determine if the SNPs reached Hardy-Weinberg equilibrium (HWE), the Pearson’s χ2 test was performed. It appeared that in the patients and control population, all genotype distributions were in Hardy-Weinberg disequilibrium.
 |
Table 3 Genotype Distributions and Hardy Weinberg Equilibrium in Sudanese Tuberculosis Patients and Healthy Controls |
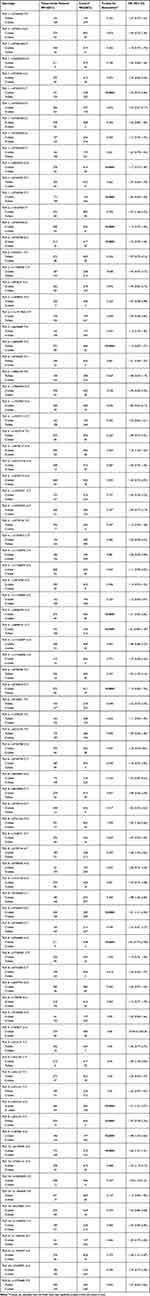 |
Table 4 Allele Frequencies of Tuberculosis Patients in Comparison to a Matching Healthy Control Population |
To determine if there was an association between any of the studied SNPs and tuberculosis, the allele frequencies between the control population and the patient population were compared with the Fisher Exact test. It appeared that in the tuberculosis population the TLR1 (rs5743557, rs4833095, rs5743596), TLR2 (rs5743704, rs5743708, rs3804099), TLR4 (rs4986790, rs4986791), TLR6 (rs5743810), TLR8 (rs3764879, rs3764880), TLR9 (rs352165, rs352167, rs187084) and TLR10 (rs4129009) were significantly more often encountered (p<0.0001) (Table 4) than in the control population and were associated with tuberculosis in the Sudanese population (Table 4). For the other polymorphisms tested, no association with tuberculosis was found in the population tested (Table 4).
Discussion
Association Between TLR1, TLR2, TLR4, TLR6, TLR8, TLR9 and TLR10 Genes Polymorphisms and Tuberculosis
Tuberculosis is a complex disease in which environmental, immunological and genetic factors are contributed. It has been estimated that 10% of the infected population with M. tuberculosis may develop TB disease sometime in their life, suggesting that the majority of those infected are endowed with a protective immune response. Previous association studies have indicated a potential involvement of genetic variation within innate immune response genes as risk factors for TB. In particular, TLR1,39 TLR2,40–45 TLR4,46 with TLR847 and TLR930 genetic variants have been associated with TB, suggesting that TLR-mediated responses may be important for protection to M. tuberculosis infection in humans.
TLRs are a family of PRRs consisting of 12 members in human and other mammals. TLRs play a crucial role in the recognition of M. tuberculosis, this immune activation occurs only in the presence of functional TLRs. Variants of TLRs may influence their expression, function and alters the recognition or signaling mechanism, which leads to the disease susceptibility.48,49 The polymorphisms of TLRs have been hypothesized to affect the tuberculosis susceptibility. However, the direct evidence remains controversial.
Therefore, in the present study, we genotyped 86 SNPs including TLR1, TLR2, TLR4, TLR6, TLR8, TLR9 and TLR10 in the Sudanese tuberculosis patients to determine whether they are associated with susceptibility to TB in Sudanese tuberculosis patients.
Our results describes novel mutations in the TLR1, TLR2, TLR4, TLR6, TLR8, TLR9 and TLR10 genes and describes their association with pulmonary tuberculosis infection. Studies of the genetic factors involved in complex diseases have not yet provided clear explanations for the onset of such diseases, though they may help to identify the risk factor to get the infection.
Our results revealed that TLR1 rs5743557, TLR1 rs4833095, and TLR1 rs5743596 allele were more frequently found in the patients population compared to the healthy controls population. The association of the TLR1 rs5743557, TLR1 rs4833095 and TLR1 rs5743596 allele with tuberculosis were also found in other populations originating from Caucasian, Indian population and South Asian for TLR1 rs483309532,50 and, East Asian Population for TLR1 rs5743557,32 but not in populations originating from in North China32 and African American subpopulation.39 Further, in vitro experiments showed that the TLR1 rs4833095 is in strong linkage disequilibrium (LD) with TLR1 rs5743618 and determining how these two SNPs contribute to TB susceptibility has proven difficult. TLR1 rs5743618 -GT genotype was related to reduction in surface expression of TLR1 in monocytes and granulocytes. In addition, after stimulated by inactivated H37Rv, samples from children with the rs5743618-GT genotype showed a decreased production of TNF-α and CXCL10, invariable production of IL-6 and IL-8 and increased production of IL-10.51 In Spanish population, rs5743618 G allele and GG genotype influenced the susceptibility to PTB;33 in Han Chinese population, Ma et al reported that rs5743618 was not associated with adult TB.39 Zhang et al32 conducted a meta-analysis on TLR1 rs5743618 and also found no association with TB susceptibility, which corresponds with our results.32 Salie et al performed an association study of 23 polymorphisms in five TLR genes (TLR1, TLR2, TLR4, TLR8, and TLR9) in TB cases and healthy controls in a South African population. The study found that TLR1 rs5743618, TLR8 rs3764879, and TLR8 rs3764880 polymorphisms were associated with TB susceptibility in both sexes.52
Our results also revealed that TLR2 rs5743708, TLR2 rs3804099 were more frequently found in the patient population compared to the healthy control population. The association of the TLR2 rs5743708, with tuberculosis were also found in other populations originating from Asian and Hispanic populations40,53,54 but not European subgroup.52 Zhang et al32 showed a significant association with TB susceptibility for the rs5743708 A allele and AA genotype across different ethnic groups in Asians and Europeans, but decreased risk in the Hispanic population.32
In contrast, the TLR2 rs5743708 and TLR2 rs5743704 were not associated with TB meningitis in an Indian population.36,55 TLR2 rs3804099 was associated with susceptibility to TB meningitis rather than with susceptibility to pulmonary TB in a case-control study of a Chinese cohort.56 Another study investigated possible associations of 16 polymorphisms of six TLR genes and TIRAP with TB susceptibility in a Chinese population. It found that TLR2 rs5743708, polymorphism was associated with pulmonary TB.53
Several studies have demonstrated critical role of TLR4 in M. tuberculosis recognition and verified necessity of these TLR for development of a protective response against M. tuberculosis infection.41 Variants in TLR4, rs4986790 and rs4986791, were investigated for their association with susceptibility or resistance to pulmonary tuberculosis.27,28,30,34,46
The results of our study are consistent with the reports of Najmi et al in India28 Ferwerda et al in Tanzania46 and Pulido et al in Spain57 but not similar to that reported previously in other Sudanese31 and South India58 Asian populations34 European Caucasians,34 North and South American34,39, African populations34,39, Gambian TB population59, a south-eastern Chinese population,60 Mexico,35 and USA.30
It is known that for TLR6 rs5743810 SNP have a protective effect against TB development. The T allele was found by Shey et al61 to reduce NF-kB signalling which led to an altered level of IL–6 production, while Randhawa et al62 showed that it leads to increased IFN-γ production and thus protection against M. tuberculosis. These functional studies correlate with the results found in this meta-analysis as well as that of Zhang et al32 where the T allele and TT genotype was also associated with resistance to TB disease.
The TLR8 polymorphism plays a significant part in the immune response in regulating the induction of interferon (IFN) and inflammatory cytokines.47,52 Our results revealed that TLR8 rs3764879 and rs3764880 allele were more frequently found in the patients population compared to the healthy controls population. The association of the TLR8 rs3764879 and rs3764880 alleles with tuberculosis were also found in other populations originating from Russian and Indonesian populations.47 Furthermore, associations have also been found in Turkish male children,63 Pakistan population,64 and South African population.52 However, neither Kobayashi et al27 nor Chimusa et al65 showed any association between rs3764880 and TB susceptibility.
Ethnic differences in TLR polymorphisms may in part reflect the ethnic diversity of host TB susceptibility. Davila et al found that around 30% of the Indonesian male subjects carried the A allele (rs3764880) associated with risk for TB, whereas this same allele was present in 78% of Russian patients.47 34.3% of the Turkish male children with pulmonary TB had the A allele associated with risk for TB, which is similar to Indonesian population.63 The rs3764880 polymorphism of TLR8 was observed more than 16% among the healthy Chinese adult population by Cheng et al.66
The gene of TLR9 is located on chromosome 3p21.3. The total length of TLR9 gene is approximate 5 kb. Its coding gene has two exons, and the major coding region is in the second exon.53,67 TLR9 is an intracellular pathogen recognition receptor (PRR) that recognizes non-methylated cytosine-phosphate-guanine (CpG) motifs in bacterial DNA.53,67 Based on NCBI SNP database, twelve SNPs have been identified for TLR9 gene. Studies have indicated certain race population with special genotype of TLR9 polymorphism might have higher risk for TB. Our results revealed that TLR9 rs352165, rs352167 and rs187084 allele were more frequently found in the patients population compared to the healthy controls population. Sanchez et al found that TLR9 rs352165, rs352167 were not associated with TB risk in a Colombian population.37
In previous study, a meta-analysis was performed to assess the association between seven extensively studied TLR9 polymorphisms (rs187084, rs352165, rs5743836, rs5743842, rs352139, rs352140 and rs352167) and TB risk. The analysis revealed an association between certain TLR9 polymorphism and TB risk. The studies included Indians, Iranian and West African, Indonesians, Vietnamese, Chinese and Mexicans. The results showed that rs187084 and rs5743836 polymorphisms were not associated with TB risk, while the association between rs352139 polymorphism and TB risk may vary by race.68,69
Bharti et al found that rs187084 locus may be associated with susceptibility to TB in Indian population.69 The SNP rs187084 and rs5743836 SNPs located in the promoter are the most important and have been associated with various inflammatory diseases located in the promoter of TLR9 gene.70–72 Previous functional analyses have shown that both rs187084 and rs5743836 SNPs influence the transcription of TLR9 by regulation of promoter activity.70,72,73 Some studies found that the rs187084 in TLR9 showed no association with TB in Vietnam and Iran.74,75
Our result revealed that rs5743836, rs164637, rs352139, rs352140, rs352143, and rs352162 allele may not be risk factors for susceptibility to pulmonary tuberculosis in Sudanese populations. On the other hands rs352139 has been strongly associated with susceptibility to TB in Indonesian72 and Vietnamese populations,27,73 African-Americans and in Mexican Amerindians but not with Caucasians and African patients from Guinea-Bissau.30 Sanders et al found that TLR9 rs5743836 and TLR9 rs352140 alleles have protective effect against meningococcal meningitis in Dutch children.76
The rs5743836 in TLR9 showed a strong association with tuberculosis in African-Americans and Caucasians,31 while the association was not found in Vietnam75 or Mexico population74 and Chinese population.53 Different variants of TLR9 rs352140 polymorphisms are described in the genome database38 and in in the Asian population, in Japan.77
TLR9 rs352142 polymorphism was positively associated with meningeal TB, while variant TWF2 rs352143 was associated with pulmonary TB in a Vietnamese cohort.75
Recently, a number of studies indicated that TLR10 serves as a modulatory pattern-recognition receptor with mainly inhibitory properties on TLR2-derived immune responses, which are involved in the progression of TB.39,40 Our result revealed that TLR10 rs4129009 that was associated with TB susceptibility in Sudanese tuberculosis patients. Various studies found that single-nucleotide polymorphisms in TLR10 were associated with susceptibility to tuberculosis in different ethnic groups. Ma et al demonstrated that polymorphisms of TLR10 were significantly associated with TB in African and European Americans.39
Bulat-Kardum et al found that the rs11096957 AA genotype was associated with a predisposition to TB in the Caucasian population.78 However, in the present study, we did not observe association between rs11096957 and risk of TB, no association was also found in Han Chinese population.79 This inconsistent result is likely due to the ethnic difference.
There are some limitations in our study which need to be mentioned. The sample size was relatively limited and the methods were limited to PCR-RFLP. Moreover, some genotypes and alleles of SNPs were found to have low frequency in the studied population which may limit the statistical power. Moreover, we did not correlate the TLR SNPs with demographic characteristics or with others clinical parameters. Moreover, functional exploration inferring these candidate SNPs should be conducted further to confirm our findings. With these limitations, the study provides evidence for the association of SNPs present in TLR1, TLR2, TLR4, TLR6, TLR8, TLR9 and TLR10 genes with pulmonary tuberculosis. The presence of associated SNPs with pulmonary tuberculosis may give us the clue that it may provide susceptibility to pulmonary tuberculosis.
In conclusion, since TLRs are the key modulator of inflammatory processes, the associated specific TLRs with PTB may be a potential target for attenuation of specific TLR pathways to reduce the severity of the disease and testing for TLR SNPs may be helpful for early prediction of the course of the disease and early identification of patients who at risk. This the first published results that studies 86 SNPs and their association to pulmonary tuberculosis. Future studies are warranted in a larger sample to replicate our findings and understand the mechanism of association of TLR polymorphism in PTB.
Abbreviations
TLRs, Toll-like Receptors; PCR-RFLP, polymerase chain reaction and restriction fragment length polymorphism; SNPs, single nucleotide polymorphism; HWE, Hardy-Weinberg equilibrium.
Data Sharing Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethics Approval and Consent to Participate
The present study was approved by the Ethics Committee of University of Khartoum, Khartoum, Sudan (5/2018).This study adheres to the Declaration of Helsinki (1964). Written informed consents were obtained from all participants in the study or legally responsible guardians for participants less than 18 years old.
Acknowledgments
We would like to thank all health centers’ staff for their kind collaboration and assistance. And also great thanks to all participants contributed to this work.
Disclosure
The authors declare that they have no competing interests in this work.
References
1. Chun X, Peijun T, Cheng D, et al. Vitamin D receptor geneFOKI polymorphism contributes to increasing the risk of HIV-negative tuberculosis: evidence from a meta-analysis. PLoS One. 2015;10:e0140634. doi:10.1371/journal.pone.0140634
2. World Health Organization. Global Tuberculosis Report 2020. Geneva, Switzerland: World Health Organization; 2020.
3. World Health Organization. Coronavirus disease 2019 (COVID-19) situation report – 197; 2020. Available from: https://www.who.int/docs/default-source/coronaviruse/situation-reports/20200804-covid-19-sitrep-197.pdf?sfvrsn=94f7a01d_2.
4. Faqihi F, Alharthy A, Noor AlFateh BA, Balahmar A, Karakitsos D. COVID-19 in a patient with active tuberculosis: a rare case-report. Respir Med Case Rep. 2020;31:101146.
5. Liu Y, Bi L, Chen Y, Wang Y, Fleming J, Yu Y. Active or latent tuberculosis increases susceptibility to COVID-19 and disease severity. MedRxiv BioRxiv. 2020. doi:10.1101/2020.03.10.20033795
6. Swaminathan S, Nagendran G. HIV and tuberculosis in India. J Biosci. 2008;33(4):527–537.
7. World Health Organization. TB/HIV FACTS 2009; 2009. Available from: https://www.who.int/tb/challenges/hiv/factsheet_hivtb_2009update.pdf.
8. Dirlikov E, Raviglione M, Scano F. Global tuberculosis control: toward the 2015 targets and beyond. Ann Intern Med. 2015;163(1):52–58.
9. Diedrich CR, Flynn JL. HIV-1/mycobacterium tuberculosis co-infection immunology: how does HIV-1 exacerbate tuberculosis? Infect Immun. 2011;79(4):1407–1417.
10. Jiang H, Zhou Y, Tang W. Maintaining HIV care during the COVID-19 pandemic. The lancet. 2020;3018(20):30105.
11. Fernando SL, Britton WJ. Genetic susceptibility to mycobacterial disease in humans. Immunol Cell Biol. 2006;84(2):125–137. doi:10.1111/j.1440-1711.2006.01420.x
12. Karlsson EK, Kwiatkowski DP, Sabeti PC. Natural selection and infectious disease in human populations. Nat Rev Genet. 2014;15(6):379–393.
13. Casanova JL, Abel L.Genetic dissection of immunity to mycobacteria: the human model. Annu Rev Immunol. 2002;20:581–620.
14. Guide SV, Holland SM. Host susceptibility factors in mycobacterial infection: genetics and body morphotype. Infect Dis Clin North Am. 2002;16:163–186.
15. Bellamy R, NuldaBeyers KP, McAdam WJ, et al. Genetic susceptibility to tuberculosis in Africans: a genome-wide scan. Proc Natl Acad Sci U S A. 2000;97:8005–8009.
16. Graham S, Cooke Sarah J, Keith P, et al. Mapping of a novel susceptibility locus suggests a role for MC3R and CTSZ in human tuberculosis. Am J Respir Crit Care Med. 2008;178:203–207.
17. Moller M, Hoal EG. Current findings, challenges and novel approaches in human genetic susceptibility to tuberculosis. Tuberculosis. 2010;90:71–83.
18. Mahasirimongkol S, Yanai H, Nishida N, et al. Genome-wide SNP-based linkage analysis of tuberculosis in Thais. Genes Immun. 2009;10:77–83.
19. Miller EN, Jamieson SE, Joberty C, et al. Genome-wide scans for leprosy and tuberculosis susceptibility genes in Brazilians. Genes Immun. 2004;5:63–67.
20. Mahasirimongkol S, Yanai H, Mushiroda T, et al. Genome-wide association studies of tuberculosis in Asians identify distinct at-risk locus for young tuberculosis. J Hum Genet. 2012;57:363–367.
21. Thye T, Vannberg FO, Wong SH, et al. Genome-wide association analyses identifies a susceptibility locus for tuberculosis on chromosome 18q11.2. Nat Genet. 2010;42:739–741.
22. Bafica A, Scanga CA, Feng CG, Leifer C, Cheever A, Sher A. TLR9 Regulates Th1 responses and cooperates with TLR2 in mediating optimal resistance to Mycobacterium tuberculosis. J Exp Med. 2005;202:1715–1724.
23. Carvalho NB, Oliveira FS, Duraes FV, et al. Toll-like receptor 9 is required for full host resistance to Mycobacterium avium infection but plays no role in induction of Th1 responses. Infect Immun. 2011;79:1638–1646.
24. Chen YC, Hsiao CC, Chen CJ, et al. Toll-like receptor 2 gene polymorphisms, pulmonary tuberculosis, and natural killer cell counts. BMC Med Genet. 2010;11:17.
25. Chow JC, Young DW, Golenbock DT, Christ WJ, Gusovsky F. Toll-like receptor-4 mediates lipopolysaccharide-induced signal transduction. J Biol Chem. 1999;274:10689–10692.
26. Azad AK, Sadee W, Schlesinger LS. Innate immune gene polymorphisms in tuberculosis. Infect Immun. 2012;80:3343–3359.
27. Kobayashi K, Yuliwulandari R, Yanai H, et al. Association of TLR polymorphisms with development of tuberculosis in Indonesian females. Tissue Antigens. 2012;79:190–197.
28. Najmi N, Kaur G, Sharma SK, Mehra NK. Human toll-like receptor 4 polymorphisms TLR4 Asp299Gly and Thr399Ile influence susceptibility and severity of pulmonary tuberculosis in the Asian Indian population. Tissue Antigens. 2010;76:102–109.
29. Thada S, Valluri V, Gaddam SL. Influence of toll like receptor gene polymorphisms to tuberculosis susceptibility in humans. Scand J Immunol. 2013;78:221–229.
30. Velez DR, Wejse C, Stryjewski ME, et al. Variants in toll-like receptors 2 and 9 influence susceptibility to pulmonary tuberculosis in Caucasians, African-Americans, and West Africans. Hum Genet. 2010;127:65–73.
31. Zaki HY, Leung KH, Yiu WC, Gasmelseed N, Elwali NE, Yip SP. Common polymorphisms in TLR4 gene associated with susceptibility to pulmonary tuberculosis in the Sudanese. Int J Tuberc Lung Dis. 2012;16:934–940.
32. Zhang Y, Jiang T, Yang X, et al. Toll-like receptor −1, −2, and −6 polymorphisms and pulmonary tuberculosis susceptibility: a systematic review and meta-analysis. PLoS One. 2013;8:e63357.
33. Ocejo-Vinyals JG. Human toll-like receptor 1 T1805G polymorphism and susceptibility to pulmonary tuberculosis in northern Spain. Int J Tuberc Lung Dis. 2013;17:652–654.
34. Tian T, Jin S, Dong J, Li G. Lack of association between toll-like receptor 4 gene Asp299Gly and Thr399Ile polymorphisms and tuberculosis susceptibility: a meta-analysis. Infect Genet Evol. 2013;14:156–160.
35. Rosas-Taraco AG, Revol A, Salinas-Carmona MC, Rendon A, Caballero-Olin G, Arce-Mendoza AY. CD14 C(−159)T polymorphism is a risk factor for development of pulmonary tuberculosis. J Infect Dis. 2007;196:1698–1706.
36. Biswas D, Gupta SK, Sindhwani G, Patras A. TLR2 Polymorphisms, Arg753Gln and Arg677Trp, are not associated with increased burden of tuberculosis in Indian patients. BMC Res Notes. 2009;2:162.
37. Sanchez D, Lefebvre C, Rioux J, Garcia LF, Barrera LF. Evaluation of toll-like receptor and adaptor molecule polymorphisms for susceptibility to tuberculosis in a Colombian population. Int J Immunogenet. 2012;39:216–223.
38. Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801.
39. Ma X, Liu Y, Gowen BB, Graviss EA, Clark AG, Musser JM. Full-exon resequencing reveals toll-like receptor variants contribute to human susceptibility to tuberculosis disease. PLoS One. 2007;2:e1318.
40. Ogus AC, Yoldas B, Ozdemir T, et al. The Arg753GLn polymorphism of the human toll-like receptor 2 gene in tuberculosis disease. Eur Clin Respir J. 2004;23:219.
41. Abel B, Thieblemont N, Quesniaux VJF, et al. Toll-like receptor 4 expression is required to control chronic Mycobacterium tuberculosis infection in mice. J Immun. 2002;169(6))::3155–3162.
42. Dalgic N. Arg753Gln polymorphism of the human Toll-like receptor 2 gene from infection to disease in pediatric tuberculosis. Hum Immunol. 2011;72:440.
43. Ben-Ali M, Barbouche MR, Bousnina S, Chabbou A, Dellagi K. Toll-like receptor 2 Arg677Trp polymorphism is associated with susceptibility to tuberculosis in Tunisian patients. Clin Diagn Lab Immunol. 2004;11:625.
44. Thuong NT, Hawn TR, Thwaites GE, et al. A polymorphism in human TLR2 is associated with increased susceptibility to tuberculous meningitis. Genes Immun. 2007;8:422.
45. Etokebe GE, Skjeldal F, Nilsen N, et al. Toll-like receptor 2 (P631H) mutant impairs membrane internalization and is a dominant negative allele. Scand J Immunol. 2010;71:369.
46. Ferwerda B. The toll-like receptor 4 Asp299Gly variant and tuberculosis susceptibility in HIV-infected patients in Tanzania. AIDS. 2007;21:1375.
47. Davila S, Hibberd ML, Hari Dass R, et al. Genetic association and expression studies indicate a role of toll-like receptor 8 in pulmonary tuberculosis. PLoS Genet. 2008;4:e1000218.
48. Quesniaux V, Fremond C, Jacobs M, et al. Toll-like receptor pathways in the immune responses to mycobacteria. Microbes Infect. 2004;6:946.
49. Jo EK, Yang CS, Choi CH, Harding CV. Intracellular signalling cascades regulating innate immune responses to Mycobacteria: branching out from Toll-like receptors. Cell Microbiol. 2007;9:1087.
50. Sinha E, Biswas SK, Mittal M, et al. Toll-like Receptor 1 743 A>G, 1805 T>G & Toll-like Receptor 6 745 C>T gene polymorphism and tuberculosis: a case control study of north Indian population from Agra (India). Hum Immunol. 2014;75:880–886. PMID: 24984237.
51. Dittrich N, Berrocal-Almanza LC, Thada S, et al. Toll-like receptor 1 variations influence susceptibility and immune response to Mycobacterium tuberculosis. Tuberculosis. 2015;95:328–335.
52. Salie M, Daya M, Lucas LA, et al. Association of Toll-like receptors with susceptibility to tuberculosis suggests sex-specific effects of TLR8 polymorphisms. Infect Genet Evol. 2015;34:221–229.
53. Wu L, Hu Y, Li D, Jiang W, Xu B. Screening toll-like receptor markers to predict latent tuberculosis infection and subsequent tuberculosis disease in a Chinese population. BMC Med Genet. 2015;16:19. PMID: 25928077. doi:10.1186/s12881-015-0166-1
54. Folwaczny M, Glas J, Torok HP, Limbersky O, Folwaczny C. Toll-like receptor (TLR) 2 and 4 mutations in periodontal disease. Clin Exp Immunol. 2004;135(2):330–335.
55. Rizvi I, Garg RK, Jain A, et al. Vitamin D status, Vitamin D receptor and Toll like receptor-2 polymorphisms in tuberculous meningitis: a case-control study. Infection. 2016;44:633–640.
56. Zhao Y, Bu H, Hong K, et al. Genetic polymorphisms of CCL1 rs2072069 G/A and TLR2 rs3804099 T/C in pulmonary or meningeal tuberculosis patients. Int J Clin Exp Pathol. 2015;8:12608–12620.
57. Pulido I, Leal M, Genebat M, Pacheco YM. The TLR4 ASP299GLY polymorphism is a risk factor for active tuberculosis in caucasian HIV-infected patients. Curr HIV Res. 2010;8(3):253–258.
58. Selvaraj P. Toll-like receptor and TIRAP gene polymorphisms in pulmonary tuberculosis patients of South India. Tuberculosis. 2010;90:306.
59. Newport MJ, Allen A, Awomoyi AA, et al. The toll-like receptor 4 Asp299Gly variant: no influence on LPS responsiveness or susceptibility to pulmonary tuberculosis in The Gambia. Tuberculosis. 2004;84:347.
60. Xue Y. Toll-like receptors 2 and 4 gene polymorphisms in a southeastern Chinese population with tuberculosis. International. J Immunogenet. 2010;37:135.
61. Shey MS, Randhawa AK, Bowmaker M, et al. Single nucleotide polymorphisms in toll-like receptor 6 are associated with altered lipopeptide- and mycobacteria-induced interleukin–6 secretion. Genes Immun. 2010;11:561–572. PMID: 20445564. doi:10.1038/gene.2010.14
62. Randhawa AK, Shey MS, Keyser A, et al. Association of human TLR1 and TLR6 deficiency with altered immune responses to BCG vaccination in South African Infants. PLoS Pathog. 2011;7. doi:10.1371/journal.ppat.1002174
63. Dalgic N, Tekin D, Kayaalti Z, Cakir E, Soylemezoglu T, Sancar M. Relationship between toll-like receptor 8 gene polymorphisms and pediatric pulmonary tuberculosis. Dis Markers. 2011;31(1):33–38.
64. Bukhari M, Aslam MA, Khan A, et al. TLR8 gene polymorphism and association in bacterial load in southern Punjab of Pakistan: an association study with pulmonary tuberculosis. Int J Immunogenet. 2015;42(1):46–51.
65. Chimusa ER, Zaitlen N, Daya M, et al. Genome-wide association study of ancestry-specific TB risk in the South African Coloured population. Hum Mol Genet. 2014;23(3):796–809.
66. Cheng PL, Eng HL, Chou MH, You HL, Lin TM. Genetic polymorphisms of viral infection-associated Toll-like receptors in Chinese population. Transl Res. 2007;150::311–318.
67. Nilsson D, Andiappan AK, Hallden C, et al. Toll-like receptor gene polymorphisms are associated with allergic rhinitis: a case control study. BMC Med Genet. 2012;13:66.
68. Chen Z, Wang W, Liang J, Wang J, Feng S, Zhang G. Association between toll-like receptors 9 (TLR9) gene polymorphism and risk of pulmonary tuberculosis: meta-analysis. BMC Pulm Med. 2015;15(1):1.
69. Bharti D, Kumar A, Mahla RS, et al. The role of TLR9 polymorphism in susceptibility to pulmonary tuberculosis. Immunogenetics. 2014;66(12):675–681.
70. Hamann L, Glaeser C, Hamprecht A, Gross M, Gomma A, Schumann RR. Toll like receptor (TLR)-9 promotor polymorphisms and at herosclerosis. Clin Chim Acta. 2006;364(1–2):303–307.
71. Roszak A, Lianeri M, Sowinska A, Jagodzinski PP. Involvement of Toll-like Receptor 9 polymorphism in cervical cancer development. Mol Biol Rep. 2012;39(8):8425–8430.
72. Ng MT, Van’t Hof R, Crockett JC, et al. Increase in NF-kappaB binding affinity of the variant C allele of the toll-like receptor 9–1237 T/C polymorphism is associated with Helicobacter pylori-induced gastric disease. Infect Immun. 2010;78(3):1345–1352.
73. Fischer J, Weber ANR, Bohm S, et al. Sex-specific effects of TLR9 promoter variants on spontaneous clearance of HCV infection. Gut. 2017;66(10):1829–1837.
74. Torres-García D, Cruz-Lagunas A, García-Sancho Figueroa MC, et al. Variants in toll-like receptor 9 gene influence susceptibility to tuberculosis in a Mexican population. J Transl Med. 2013;11:220.
75. Graustein AD, Horne DJ, Arentz M, et al. TLR9 gene region polymorphisms and susceptibility to tuberculosis in Vietnam. Tuberculosis. 2015;95:190–196.
76. Sanders MS, van Well GT, Ouburg S, Lundberg PS, van Furth AM, Morré SA. Single nucleotide polymorphisms in TLR9 are highly associated with susceptibility to bacterial meningitis in children. Clin Infect Dis. 2011b;52:475–480.
77. Wujcicka W, Gaj Z, Wilczy_nski J, Nowakowska D. Possible role of TLR4 and TLR9 SNPs in protection against congenital toxoplasmosis. Eur J Clin Microbiol Infect Dis. 2015;4:2121–2129.
78. Bulat-Kardum LJ, Etokebe GE, Lederer P, Balen S, Dembic Z. Genetic polymorphisms in the toll-like receptor 10, interleukin (IL)17A and IL17F genes differently affect the risk for tuberculosis in Croatian population. Scand J Immunol. 2015;82(1):63–69. doi:10.1111/sji.12300
79. Tang F, Li Z, Wang Y, et al. Toll-like receptor 1 and 10 polymorphisms, Helicobacter pylori susceptibility and risk of gastric lesions in a high-risk Chinese population. Infect Genet Evol. 2015;31:263–269. doi:10.1016/j.meegid.2015.02.005
 © 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2023 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.