Back to Journals » Neuropsychiatric Disease and Treatment » Volume 11
Association of metabotropic glutamate receptor 3 gene polymorphisms with schizophrenia risk: evidence from a meta-analysis
Authors Yang X, Wang G, Wang Y, Yue X
Received 22 November 2014
Accepted for publication 22 January 2015
Published 25 March 2015 Volume 2015:11 Pages 823—833
DOI https://doi.org/10.2147/NDT.S77966
Checked for plagiarism Yes
Review by Single anonymous peer review
Peer reviewer comments 3
Editor who approved publication: Professor Wai Kwong Tang
Xiaoqin Yang,1 Guiping Wang,2 Yaodong Wang,3 Xia Yue4
1Department of Bioinformatics, School of Life Science and Technology, Tongji University, Shanghai, 2Department of Pharmacy, College of Health Sciences, Guangzhou Medical University, 3Department of Cell Biology, School of Basic Medical Sciences, 4Department of Forensic Medicine, School of Basic Medical Sciences, Southern Medical University, Guangzhou, People’s Republic of China
Abstract: To date, the role of metabotropic glutamate receptor 3 (GRM3) rs274622, rs1468412, rs917071, rs6465084, and rs2299225 polymorphisms in schizophrenia remains controversial.
To provide a clearer picture for the effect of the five most studied GRM3 polymorphisms on risk of schizophrenia, this meta-analysis with eligible data from published studies was performed. Relevant case–control studies were retrieved by literature search and selected according to established inclusion criteria. Odds ratios with 95% confidence intervals were used to assess the strength of association. A total of 33 individual studies were identified and included in our meta-analysis: nine for rs1468412, with 5,314 cases and 6,147 controls; six for rs917071, with 2,660 cases and 3,517 controls; seven for rs274622, with 3,820 cases and 4,015 controls; five for rs2299225, with 3,492 cases and 3,735 controls; and six for rs6465084, with 4,960 cases and 5,613 controls. However, no significant association was found between these GRM3 polymorphisms and schizophrenia in the overall population. With respect to rs1468412 polymorphism, a finding of very borderline statistical significance emerged in dominant comparison model for non-Asian populations, calling for large-scale verification to assess the marginally elevated risk of schizophrenia. In conclusion, these GRM3 polymorphisms have limited effect on the risks of schizophrenia. Further large and well-designed studies are needed to confirm this conclusion.
Keywords: schizophrenia, GRM3, SNP, meta-analysis
Introduction
Schizophrenia is one of the most detrimental psychiatric disorders, with high heritability1 and a median incidence of 15.2/100,000 persons worldwide.2 Plenty of efforts have been made to understand its pathological mechanisms, and now it is suggested that genetic vulnerability with nongenetic factors can act in combination to lead to the development of schizophrenia.3 In recent years, with increased knowledge of human gene functions and architecture, it has become clear that differences in genetic backgrounds, such as single nucleotide polymorphisms (SNPs), could substantially influence schizophrenia risk.3–5
Glutamate has been implicated as the most ubiquitous excitatory neurotransmitter in the central nervous system. The glutamatergic neurotransmission plays important roles in various brain functions, including motor control, learning and memory, cognition, and neural development.6 Moreover, evidence has indicated that hypofunction of glutamatergic neurotransmission may be important in the etiology and pathophysiology of schizophrenia.7–9 As a member of the group II metabotropic glutamate receptors (mGluR), mGluR3 (encoded by the GRM3 gene) functions as the regulator of glutamate neurotransmission and synaptic plasticity.10,11 Given the prior evidence, GRM3, along with other glutamatergic genes, is a candidate gene for schizophrenia.
The human GRM3 gene rs274622, rs917071, rs1468412, rs6465084, and rs2299225 loci are five commonly studied polymorphisms for schizophrenia. The rs274622 locus, along with other polymorphisms, might be useful as a predictor of negative-symptom improvement in persons with schizophrenia treated with olanzapine.12 In combination with the rs757656 polymorphism, the rs917071 locus has been associated with alcohol dependence,13 which could have substantial influence on schizophrenic exacerbation.14 As for the rs1468412 locus, it has been suggested to be associated with worsened cognitive and symptom response to antipsychotic therapy for schizophrenia.15 For the rs2299225 locus, several genetic epidemiological studies have been performed to assess its association with schizophrenia risk.16–18 rs6465084 has been described as a functional polymorphism that may influence glutamate concentrations, and the minor allele of this SNP was associated with poor performance on several cognitive test.19,20 As well as helping verify whether these SNPs might contribute etiologically to the schizophrenia, rather than just pathophysiologically or therapeutically, several genetic association studies have been performed to identify common risk variants at the GRM3 gene locus. However, the evidence for the association between GRM3 polymorphisms and schizophrenia risk is still controversial. The most likely reasons for the inconsistencies among these studies could be tremendous differences in sample size, diverse ethnic background, incompatible statistical methods, and possible sampling bias. To provide a clearer picture for the effect of these five GRM3 polymorphisms on the risk of schizophrenia, we conducted this meta-analysis with eligible data from published studies.
Materials and methods
Search strategy
Systematic searches of the PubMed database (to November 2, 2014) were performed using the search terms: “GRM3”, “metabotropic glutamate receptor 3”, “polymorphism”, “SNP”, “variant”, and “schizophrenia”. The search was limited to human studies. All eligible studies were retrieved, and their bibliographies were checked for other relevant research. When the data set for the same patient population was used in several publications, only the most recent or complete study was included for further analysis. An additional manual search was also conducted to add any recently published studies.
Inclusion criteria
The following criteria were used for the study selection: 1) evaluate the association between five GRM3 polymorphisms (rs1468412, rs274622, rs917071, rs2299225, and rs6465084) and schizophrenia; 2) case–control studies; 3) sufficient data for estimating an odds ratio (OR) with 95% confidence interval (CI); and 4) studies with full-text articles in English.
Data extraction
Data were carefully evaluated and extracted from all eligible studies by two investigators independently according to the inclusion criteria listed above. When disagreement occurred, the third investigator was invited to discuss and check the data until a consensus was reached. The following elements were collected from eligible studies: first author’s name, year of publication, country, ethnicity, diagnostic criteria of schizophrenia cases, genotype frequency of cases and controls, and the results of Hardy–Weinberg equilibrium (HWE) test.
Statistical analysis
Departure from the HWE for the control group in each study was assessed with Pearson’s goodness-of-fit χ2 test with 1 df by an online program (http://ihg.gsf.de/cgi-bin/hw/hwa1.pl), and violation of the HWE was determined using a threshold of P<0.05. The strength of association between GRM3 polymorphisms and schizophrenia risk was measured by ORs with 95% CIs. Pooled ORs were calculated for homozygote comparison (aa versus AA, a being the minor allele and A the major allele), heterozygote comparison (Aa versus AA), dominant model (aa+Aa versus AA), and recessive model (aa versus Aa+AA), respectively. Studies that have no events in either arms (schizophrenia and control groups) were excluded from the corresponding comparison model. Heterogeneity among pooled studies was investigated by the χ2-based Cochran’s Q test21 and I2 statistics.22 To be more conservative, heterogeneity was considered to be present when the Cochran Q test P-value was less than 0.1, then a random-effect model (DerSimonian–Laird method)23 was utilized; otherwise, a fixed-effect model (Mantel–Haenszel method)24 was utilized. Inconsistency across studies was accessed by the I2 statistic, and I2 <25%, 25%–75%, and >75% were considered to represent low, moderate, and high degree of heterogeneity, respectively.22 To explore the source of heterogeneity among the studies of this meta-analysis, a Galbraith plot was generated to detect the potential outliers, which could largely contribute to the heterogeneity.25 Stratification analyses were performed to quantify the different effects of ethnicity. Sensitivity analysis was carried out through omitting individual study in turn to check the consistency of the results. Publication bias was evaluated by visual inspection of funnel plots and Egger’s test (P<0.05 was considered a significant publication bias).26
All statistical tests were performed with metafor (version: 1.9-4)27 and meta (version: 3.8-0; http://cran.r-project.org/web/packages/meta/) packages of R (version 3.1.1).
Results
Study characteristics
The initial literature search through the PubMed database yielded 42 published articles. When reviewed in full-text, eight had no usable reported data, two was not case–control studies, 12 were not concerned with schizophrenia risk, three were reviews, two were not GRM3 research, two were not concerned with these five GRM3 polymorphisms, and two were not in English. All these articles were excluded. Among the remaining eleven articles, studies presented separate ORs by different polymorphisms, ethnicity, or cohort, and each of them was considered separately for pooling analysis. Therefore, nine studies for rs1468412 polymorphism (5,314 cases and 6,147 controls),16–18,28–32 seven studies for rs274622 (3,820 cases and 4,015 controls),16–18,28,29,32 six studies for rs917071 (2,660 cases and 3,517 controls),16,28–32 six studies for rs6465084 (4,960 cases and 5,613 controls),20,30–34 and five studies for rs2299225 (3,492 cases and 3,735 controls)16–18,32 were included eventually. A flowchart of study selection is shown in Figure 1, and the main characteristics of eligible studies are summarized in Table 1.
  | Figure 1 Flowchart of study selection in this meta-analysis. |
Quantitative synthesis
The associations between these GRM3 polymorphisms and schizophrenia risks were analyzed and summarized ORs for schizophrenia risk of these five polymorphisms are shown in Table 2. The overall ORs by the fixed/random-effect model showed no significant association between GRM3 rs274622, rs917071, rs1468412, rs2299225 or rs6465084 polymorphism and schizophrenia risk (Figure 2). When stratified according to ethnicity, no significant association with increased schizophrenia risk was observed in all subgroups for rs274622, rs917071, and rs6465084 polymorphisms. As for rs1468412, only the dominant model for the non-Asian subgroup showed a slightly significant association (OR: 1.15; 95% CI: 1.01–1.30; P=0.04), while the other comparison models showed no relationship with schizophrenia risk for the Asian or the non-Asian populations (Table 2).
  | Figure 2 Forest plots for GRM3 polymorphisms and schizophrenia risk. |
Heterogeneity analysis
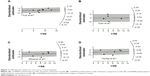
Galbraith plots were created to graphically assess the sources of heterogeneity in the comparison models for GRM3 polymorphisms. For the dominant model of rs1468412 (Figure 3A), rs2299225 (Figure 3B), and rs6465084 (Figure 3C), when the most obvious outliers were excluded, heterogeneity significantly decreased (dominant model for rs1468412: P=0.37, I2=7.8%; dominant model for rs2299225: P=0.23, I2=30.9%; dominant model for rs6465084: P=0.20, I2=33.3%), and the results remained stable for both overall and subgroup analyses (data not shown). As for the heterozygote model of the rs6465084 polymorphism (Figure 3D), after removing the outlier, the heterogeneity among studies was also alleviated, although still existed (heterozygote model of rs6465084: P=0.05, I2=58.1%).
Publication bias and sensitivity analysis
Begg’s funnel plot and Egger’s test were done to assess the publication bias of studies. The shape of the funnel plots (Figure 4) did not show any obvious asymmetry. The statistical results of Egger’s test still did not reveal publication bias for rs274622 polymorphism (homozygote model, P=0.794; heterozygote model, P=0.680; dominant model, P=0.656; recessive model, P=0.830), rs917071 polymorphism (homozygote model, P=0.185; heterozygote model, P=0.682; dominant model, P=0.470; recessive model, P=0.215), rs6465084 (homozygote model, P=0.678; heterozygote model, P=0.882; dominant model, P=0.624; recessive model, P=0.701) and rs2299225 (homozygote model, P=0.088; heterozygote model, P=0.822; dominant model, P=0.893; recessive model, P=0.094). For the rs1468412 polymorphism, the heterozygote model (P=0.026) and dominant model (P=0.013) showed publication bias, but not the homozygote model (P=0.120) or recessive model (P=0.285). However, when the outlier detected by the Galbraith plot (Figure 3A) was omitted, Egger’s test P-value turned to no significance (heterozygote model, P=0.300; dominant model, P=0.267).
  | Figure 4 Funnel plot for publication bias in studies on GRM3 polymorphisms and schizophrenia risk. |
The influences of each individual study on the overall ORs for the five polymorphisms were evaluated. The results showed the pooled ORs of these polymorphisms were not materially altered by the omission of any individual study (Figure 5).
  | Figure 5 Sensitivity analyses of the summary odds ratios of the association between GRM3 polymorphisms and schizophrenia risk. |
Discussion
Common genetic polymorphisms or mutation in the glutamatergic neurotransmission may lead to profound influence to most aspects of normal brain function and involvement in many neuropathologic conditions. In recent years, interest in the genetic susceptibility to neuropathogenesis has led to a growing attention to the study of polymorphisms of genes involved in schizophrenia. To date, many molecular epidemiological studies have been performed to explore the role of GRM3 polymorphisms on schizophrenia susceptibility, but the results are still far from conclusive. Moreover, several studies were plagued by their limited sample size, which subsequently led to statistical power too low to unveil the effects that may exist. Therefore, we performed this meta-analysis with eligible studies in order to improve the statistical power and accurately determine the effect size of the association between GRM3 polymorphisms and schizophrenia risk.
Our results showed that GRM3 rs1468412, rs274622, rs917071, rs2299225, and rs6465084 polymorphisms were not associated with schizophrenia risk when all studies were pooled together. Ethnicity is usually regarded as a potential factor that may influence the risk of common disease by different genetic backgrounds and environmental exposures, while the stratified analyses in the present study allowed us to infer the ethnic specificity of the association between these SNPs and schizophrenia. In further subgroup analysis stratified by ethnicity, none of the GRM3 rs274622, rs917071, or rs6465084 polymorphisms showed any significant association with schizophrenia risk. With respect to rs1468412, although an increased risk was observed in the dominant model for non-Asian populations, given the limited studies and sample sizes for this subgroup, the borderline statistical significance (P=0.04) in only one single comparison model is not enough to indicate any underlying association. Further large and well-designed studies are needed to confirm this.
Heterogeneity between studies should be noted, because this may potentially affect the strengths of the meta-analysis. In the present study, heterogeneity was found in the heterozygote comparison model for the rs6465084 polymorphism, and in the dominant comparison model for the rs1468412, rs6465084, and rs2299225 polymorphisms. A Galbraith plot was utilized to identify which of the included studies may be sources of heterogeneity. After removal of those identified outliers, the heterogeneity among studies was obviously alleviated, while the main conclusions remained stable. Furthermore, the publication bias for the heterozygote model and dominant model of rs1468412 significantly decreased after removing the outlier. The sensitivity analysis did not show significant alteration of our meta-analyses for any of the five polymorphisms, indicating the robustness of our results. Taken together, these results strengthened the reliability of the conclusions from this meta-analysis.
Although comprehensive analysis provided pooled data on a substantial number of cases and controls and objectively evaluated the influence of five GRM3 polymorphisms on the risk of schizophrenia, there are still some limitations inherited from the involved studies and our analysis strategies. First of all, the number of studies and subjects included in the current meta-analysis were still relatively small and most of the published studies were performed in Asian-descent and Caucasian-descent populations. It is critical that larger and well-designed multiethnic studies with detailed subtyping data should be performed to reevaluate the association and further explore the possible ethnic specificity. In addition, our analysis did not consider the possibility of gene–environment interactions or the linkage disequilibrium between gene polymorphisms, due to lack of original data. Finally, although all cases and controls of each study were recruited with similar inclusion criteria, some other potential factors not taken into account, such as lifestyle, may have biased our results. Owing to these limitations listed, further rigorous multicentric studies with large samples, multiple races and ethnicities, standardized unbiased genotyping methods, and well-matched controls are needed to validate the findings. In addition, other information, including but not limited to gene–gene interaction, gene–environment interaction, and lifestyle factors, should also be taken into account to evaluate the combined effect on schizophrenia.
Conclusion
In conclusion, this meta-analysis suggests that there is lack of evidence proving the association of GRM3 rs1468412, rs274622, rs917071, rs6465084, and rs2299225 polymorphisms with increased risk of developing schizophrenia.
Acknowledgments
This work was supported by a grant from the National Natural Science Foundation of China (grant 81102301) and a PhD startup fund for scientific research at Guangzhou Medical University (2011C06).
Author contributions
Xiaoqin Yang and Xia Yue contributed to the design of the project, and wrote the manuscript; Xiaoqin Yang and Guiping Wang conducted the research; Guiping Wang and Yaodong Wang contributed to the design of the project and data acquisition; all authors critically revised the manuscript and approved the final version to be published.
Disclosure
The authors report no conflicts of interest in this work.
References
Cannon TD, Kaprio J, Lönnqvist J, Huttunen M, Koskenvuo M. The genetic epidemiology of schizophrenia in a finnish twin cohort: A population-based modeling study. Archives of General Psychiatry. 1998;55(1):67–74. | ||
McGrath J, Saha S, Chant D, Welham J. Schizophrenia: a concise overview of incidence, prevalence, and mortality. Epidemiologic Reviews. 2008;30:67–76. | ||
Giusti-Rodriguez P, Sullivan PF. The genomics of schizophrenia: update and implications. The Journal of Clinical Investigation. 2013;123(11):4557–4563. | ||
Pan Y, Yao J, Wang B. Association of dopamine D1 receptor gene polymorphism with schizophrenia: a meta-analysis. Neuropsychiatric Disease and Treatment. 2014;10:1133–1139. | ||
Liu L, Fan D, Ding N, et al. The relationship between DRD2 gene polymorphisms (C957T and C939T) and schizophrenia: A meta-analysis. Neuroscience Letters. 2014;583C:43–48. | ||
Raju D, Smith Y. Differential Localization of Vesicular Glutamate Transporters 1 and 2 in the Rat Striatum. In: Bolam JP, Ingham C, Magill P, eds. The Basal Ganglia VIII. Vol 56: Springer US; 2005:601–610. | ||
Meador-Woodruff JH, Healy DJ. Glutamate receptor expression in schizophrenic brain. Brain Research Reviews. 2000;31(2–3):288–294. | ||
Olney JW, Farber NB. Glutamate receptor dysfunction and schizophrenia. Archives of General Psychiatry. 1995;52(12):998–1007. | ||
Tsai G, Coyle JT. Glutamatergic mechanisms in schizophrenia. Annual Review of Pharmacology and Toxicology. 2002;42(1):165–179. | ||
Harrison PJ, Weinberger DR. Schizophrenia genes, gene expression, and neuropathology: on the matter of their convergence. Mol Psychiatry. 2004;10(1):40–68. | ||
Harrison P, Lyon L, Sartorius L, Burnet P, Lane T. The group II metabotropic glutamate receptor 3 (mGluR3, mGlu3, GRM3): expression, function and involvement in schizophrenia. Journal of Psychopharmacology. 2008;22(3):308–322. | ||
Bishop JR, Ellingrod VL, Moline J, Miller D. Association between the polymorphic GRM3 gene and negative symptom improvement during olanzapine treatment. Schizophrenia Research. 2005;77(2–3):253–260. | ||
Xia Y, Wu Z, Ma D, et al. Association of Single-Nucleotide Polymorphisms in a Metabotropic Glutamate Receptor GRM3 Gene Subunit to Alcohol-Dependent Male Subjects. Alcohol and Alcoholism. 2014;49(3):256–260. | ||
Gerding LB, Labbate LA, Measom MO, Santos AB, Arana GW. Alcohol dependence and hospitalization in schizophrenia. Schizophrenia Research. 1999;38(1):71–75. | ||
Bishop JR, Reilly JL, Harris MS, et al. Pharmacogenetic associations of the type-3 metabotropic glutamate receptor (GRM3) gene with working memory and clinical symptom response to antipsychotics in first-episode schizophrenia. Psychopharmacology. 2014. | ||
Jia W, Zhang R, Wu B, et al. Metabotropic glutamate receptor 3 is associated with heroin dependence but not depression or schizophrenia in a Chinese population. PloS One. 2014;9(1):e87247. | ||
Albalushi T, Horiuchi Y, Ishiguro H, et al. Replication study and meta-analysis of the genetic association of GRM3 gene polymorphisms with schizophrenia in a large Japanese case-control population. American journal of medical genetics. Part B, Neuropsychiatric genetics: the official publication of the International Society of Psychiatric Genetics. 2008;147(3):392–396. | ||
Chen Q, He G, Chen Q, et al. A case-control study of the relationship between the metabotropic glutamate receptor 3 gene and schizophrenia in the Chinese population. Schizophrenia Research. 2005;73(1):21–26. | ||
Egan MF, Straub RE, Goldberg TE, et al. Variation in GRM3 affects cognition, prefrontal glutamate, and risk for schizophrenia. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(34):12604–12609. | ||
Mössner R, Schuhmacher A, Schulze-Rauschenbach S, et al. Further evidence for a functional role of the glutamate receptor gene GRM3 in schizophrenia. European Neuropsychopharmacology: the Journal of the European College of Neuropsychopharmacology. 2008;18(10):768–772. | ||
Cochran WG. The Combination of Estimates from Different Experiments. Biometrics. 1954;10(1):101–129. | ||
Higgins JP, Thompson SG, Deeks JJ, Altman DG. Measuring inconsistency in meta-analyses. BMJ (Clinical Research Ed.). 2003;327(7414):557–560. | ||
DerSimonian R, Laird N. Meta-analysis in clinical trials. Controlled Clinical Trials. 1986;7(3):177–188. | ||
Mantel N, Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. Journal of the National Cancer Institute. 1959;22(4):719–748. | ||
Galbraith RF. Graphical Display of Estimates Having Differing Standard Errors. Technometrics. 1988;30(3):271–281. | ||
Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ (Clinical Research Ed.). 1997;315(7109):629–634. | ||
Viechtbauer W. Conducting Meta-Analyses in R with the metafor Package. Journal of Statistical Software. 2010;36(i03). | ||
Bishop JR, Wang K, Moline J, Ellingrod VL. Association analysis of the metabotropic glutamate receptor type 3 gene (GRM3) with schizophrenia. Psychiatric Genetics. 2007;17(6):358. | ||
Fujii Y, Shibata H, Kikuta R, et al. Positive associations of polymorphisms in the metabotropic glutamate receptor type 3 gene (GRM3) with schizophrenia. Psychiatric Genetics. 2003;13(2):71–76. | ||
Jönsson EG, Saetre P, Vares M, et al. DTNBP1, NRG1, DAOA, DAO and GRM3 polymorphisms and schizophrenia: an association study. Neuropsychobiology. 2009;59(3):142–150. | ||
Norton N, Williams HJ, Dwyer S, et al. No evidence for association between polymorphisms in GRM3 and schizophrenia. BMC Psychiatry. 2005;5:23. | ||
Tochigi M, Suga M, Ohashi J, et al. No association between the metabotropic glutamate receptor type 3 gene (GRM3) and schizophrenia in a Japanese population. Schizophrenia Research. 2006;88(1–3):260–264. | ||
Mounce J, Luo L, Caprihan A, Liu J, Perrone-Bizzozero NI, Calhoun VD. Association of GRM3 polymorphism with white matter integrity in schizophrenia. Schizophrenia Research. 2014;155(1–3):8–14. | ||
Nunokawa A, Watanabe Y, Kitamura H, et al. Large-scale case-control study of a functional polymorphism in the glutamate receptor, metabotropic 3 gene in patients with schizophrenia. Psychiatry and Clinical Neurosciences. 2008;62(2):239–240. |
 © 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.
© 2015 The Author(s). This work is published and licensed by Dove Medical Press Limited. The full terms of this license are available at https://www.dovepress.com/terms.php and incorporate the Creative Commons Attribution - Non Commercial (unported, v3.0) License.
By accessing the work you hereby accept the Terms. Non-commercial uses of the work are permitted without any further permission from Dove Medical Press Limited, provided the work is properly attributed. For permission for commercial use of this work, please see paragraphs 4.2 and 5 of our Terms.